SARS-CoV-2 Spike Gene N terminal Domain targeted Sequencing
Noor Saber Jawad, Nuha Joseph Kandala
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
Abstract
We use a simple and effective method for generating 757bp of the N terminal domain of thetheSARS-CoV-2 Spike gene for variant surveillance,
Steps
RNA Extraction
The Automated extraction was handled using the ExiPrep™ 96 Lite (A-5250, BIONEER) with the ExiPrep™ Viral DNA/RNA extraction kit (K-4614, BIONEER).
RT-PCR Amplification
TaqPath™ COVID‑19 CE‑IVD RT‑PCR Kit (Multiplex real-time RT-PCR test intended for qualitatively detecting nucleic acid from SARS‑CoV‑2) used for viral detection. follow the user manual recommendation as listed in the following link: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0019215_TaqPathCOVID-19_CE-IVD_RT-PCR%20Kit_IFU.pdf
Results can be distinguished to:
1- samples with three positive targets for (ORF1ab, N, and S genes)
2- sample with two positive targets for (ORF1ab and N genes), negative for theS gene. Failure of Spike gene amplification is referred to as S gene target failure (SGTF) or S gene signal dropdown.
3- SGTF resulted due to 69/70 codons deletion of Valine and Histidine, respectively.
C-CDNA Synthesis and Quality checking
Promega GoScript™ Reverse Transcription Mix with Random Primers system (A2800) is used to generate complementary DNA. following the same kit-recommended procedure.
Quality checking is considered for all steps using the Fluorometer Quantus using Quantifluor dye
Primers
the forward primer is: SubA_21587F: CCACTAGTCTCTAGTCAGTGTGTT
Reverse primer: SubA_22344R: CCAGCTGTCCAACCTGAAGA
these primers generate an amplicon of 757bp.
Primer's preparation:
These primers were supplied by Macrogen Company in a lyophilized form. Lyophilized primers were dissolved in nuclease-free water to give a final concentration of 100pmol/μl as a stock solution. A working solution of these primers was prepared by adding 10μl of primer stock solution (stored at freezer -20 C) to 90μl of nuclease-free water to obtain a working primer solution of 10pmol/μl.
Reagent preparation for Amplicon synthesis
Amplification reaction carried on using the flowing calculations:
1- 10 ul of GoTag Green Master Mix, Promega ( M7122 ).
2- 1 ul of Forward primer
3- 1ul of Reverse primer
4- 6 ul of nuclease-free water
5- 2ul of cDNA template.
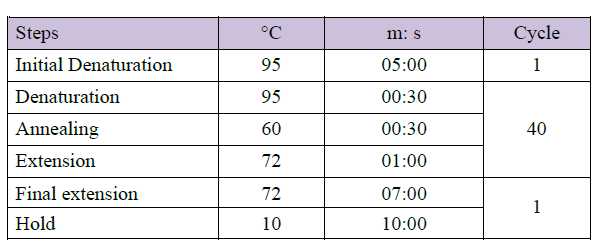
PCR adopted program and
Gel visualization
We use the classic gel visualization method through gel electrophoresis (100-1500 bp ladder gel marker) and gel documentation.
Sequencing
We referred our amplicons to a sequencing company (Macrogen, South Korea).