Software Tool for Automatic Quantification of Sarcomere Length and Organization in Fixed and Live 2D and 3D Muscle Cell Cultures In Vitro
Milena Bellin, Milena Bellin, Valeria V. Orlova, Valeria V. Orlova, Christine L. Mummery, Christine L. Mummery, Jeroen M. Stein, Jeroen M. Stein, Ulgu Arslan, Ulgu Arslan, Marnix Franken, Marnix Franken, Jessica C. de Greef, Jessica C. de Greef, Sian E. Harding, Sian E. Harding, Neda Mohammadi, Neda Mohammadi, Berend J. van Meer, Berend J. van Meer
Abstract
Sarcomeres are the structural units of the contractile apparatus in cardiac and skeletal muscle cells. Changes in sarcomere characteristics are indicative of changes in the sarcomeric proteins and function during development and disease. Assessment of sarcomere length, alignment, and organization provides insight into disease and drug responses in striated muscle cells and models, ranging from cardiomyocytes and skeletal muscle cells derived from human pluripotent stem cells to adult muscle cells isolated from animals or humans. However, quantification of sarcomere length is typically time consuming and prone to user-specific selection bias. Automated analysis pipelines exist but these often require either specialized software or programming experience. In addition, these pipelines are often designed for only one type of cell model in vitro. Here, we present an easy-to-implement protocol and software tool for automated sarcomere length and organization quantification in a variety of striated muscle in vitro models: Two dimensional (2D) cardiomyocytes, three dimensional (3D) cardiac microtissues, isolated adult cardiomyocytes, and 3D tissue engineered skeletal muscles. Based on an existing mathematical algorithm, this image analysis software (SotaTool) automatically detects the direction in which the sarcomere organization is highest over the entire image and outputs the length and organization of sarcomeres. We also analyzed videos of live cells during contraction, thereby allowing measurement of contraction parameters like fractional shortening, contraction time, relaxation time, and beating frequency. In this protocol, we give a step-by-step guide on how to prepare, image, and automatically quantify sarcomere and contraction characteristics in different types of in vitro models and we provide basic validation and discussion of the limitations of the software tool. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Staining and analyzing static hiPSC-CMs with SotaTool
Alternate Protocol : Sample preparation, acquisition, and quantification of fractional shortening in live reporter hiPSC lines
Support Protocol 1 : Finding the image resolution
Support Protocol 2 : Advanced analysis settings
Support Protocol 3 : Finding sarcomere length in non-aligned cells
INTRODUCTION
Healthy adult muscle tissue is characteristic of striated sarcomeres (the contractile machinery) which are aligned and well organized along the length of each cell and the tissue axis. However, in conditions such as toxicity, disease, and during development, sarcomeres are less organized and achieve a decreased contraction force (Ribeiro et al., 2015). Cardiomyocytes and skeletal muscle cells can now be derived routinely and with high efficiency from human induced pluripotent stem cells (hiPSCs) using a variety of protocols based on defined culture media and differentiation factors (Birket et al., 2015; Mills et al., 2017). However, hiPSC-derived cardiomyocytes (hiPSC-CMs) and hiPSC-derived skeletal muscle (hiPSC-SM) are often structurally and functionally immature which is why the induction of maturation via new methods is widely studied (Jalal, Dastidar, & Tedesco, 2021; Yang, Pabon, & Murry, 2014). This immaturity and contractile deficiency are associated with a reduced sarcomere length, which is ∼2.2 µm in adult human cardiomyocytes but usually around ∼1.65 µm in immature hiPSC-CMs (Cohn et al., 2019; Gerbin et al., 2021; Giacomelli et al., 2020; van der Velden et al., 1998). Similarly, many cardiac genetic diseases, such as hypertrophic cardiomyopathy, which is caused by mutations in myosin heavy chain or myosin binding protein C3, affect muscle sarcomere structure and organization such that there is loss of contractile force (Cohn et al., 2019; Wijnker et al., 2016). To use hiPSC-derived muscle in disease or toxicity models, it is therefore highly relevant to assess and quantify sarcomere length and organization, and preferably in combination with a measurement of the contractile properties.
Here, we describe an unbiased and rapid method for quantifying sarcomere length and organization in multiple muscle cell types and models, based on the SarcOmere Texture Analysis (SOTA) algorithm. SOTA was originally developed by Sutcliffe et al. (2018) to quantify sarcomere organization and length for static images in a MATLAB script. We made a user-friendly application in Python called SotaTool, which aims to enhance the performance of SOTA by implementing various background subtraction methods, adding an alignment index and including automated segmentation. In contrast to other approaches, the method does not require regions of interest (ROIs) to be pre-defined or selected, or images of fluorescent sarcomeres to be aligned manually (Cao, Manders, & Helmes, 2021; Pasqualin et al., 2016; Peterson, Kalda, & Vendelin, 2013). While current algorithms typically rely on manual user input per image to indicate the directionality of the sarcomeres, SotaTool detects the optimal angle automatically, reducing analysis time and selection bias. We show here how this software tool can be applied to many different datasets that contain sarcomeric striations: Immunostained cardiomyocytes, 3D tissue engineered skeletal muscles (3D TESMs), and phase-contrast images of primary neonatal rabbit CMs. The analysis gives information on the sarcomere organization and length. Aside from static images, this tool can also be used on videos or fluorescent reporter cells, broadening the output to sarcomere shortening, contraction duration, and beat rate (Fig. 1). The approach presented here addresses several problems associated with other methods: (1) underrepresentation of the entire image available for each muscle cell, (2) tendency for selection bias, and (3) time required for analyzing large datasets.
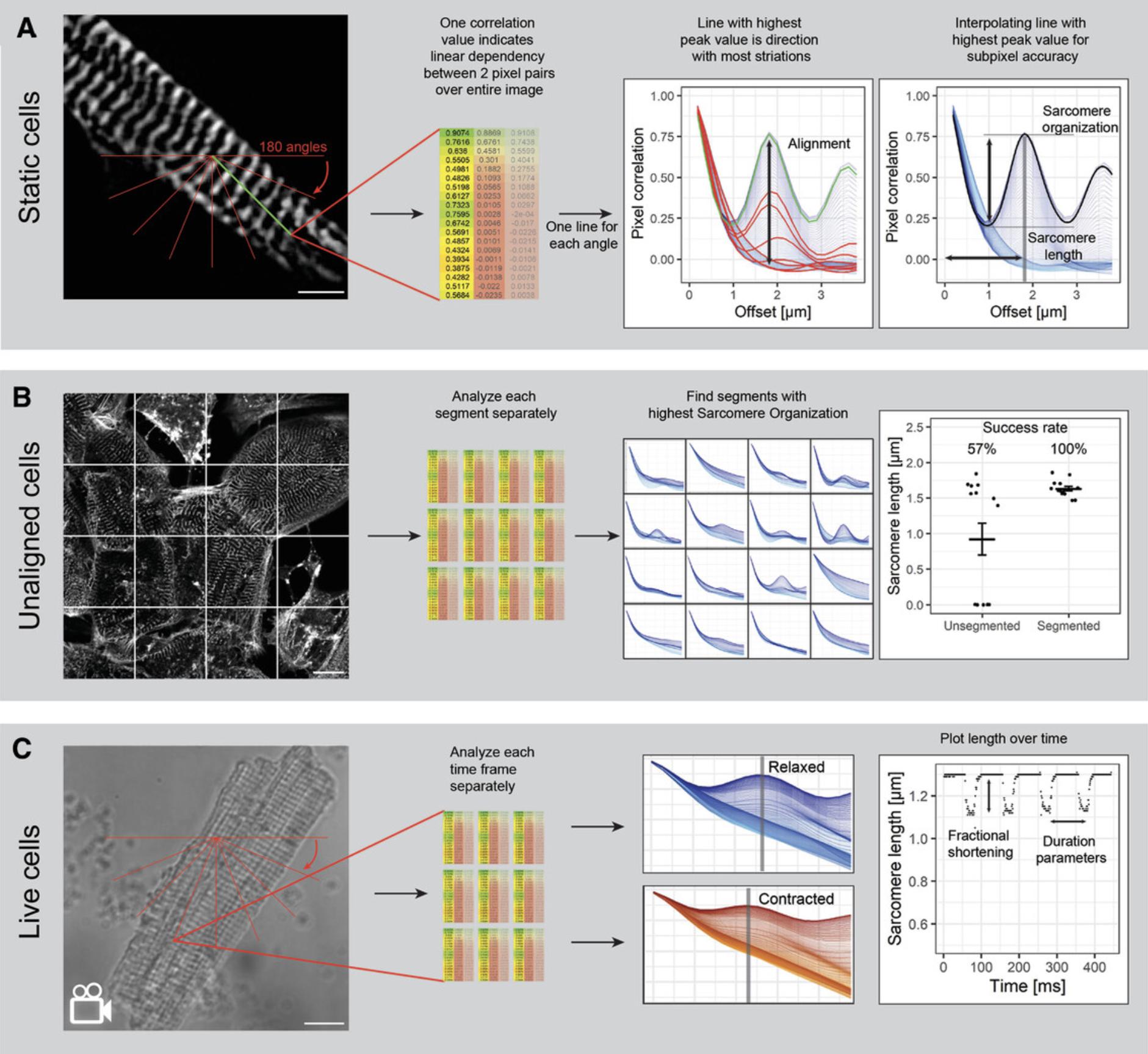
We describe step-by-step application and validation of this protocol as a sarcomere length quantification method across multiple in vitro platforms, from static 2D and 3D structures in fixed tissues and cells to fractional shortening in videos of contracting muscle. We also provide step-by-step protocols for preparation and imaging of hiPSC-CM and skeletal muscle cultures that support robust analysis (Fig. 1). We expect this new tool to reduce manual labor and time necessary for multiple muscle research applications, thus facilitating discoveries on how sarcomere structure is controlled and affected by drugs and disease.
Basic Protocol: STAINING AND ANALYZING STATIC hiPSC-CMs WITH SotaTool
SotaTool is broadly applicable across multiple in vitro platforms in which sarcomeres are visible in contractile cells by phase contrast or fluorescent microscopy. This section describes the steps of plating, fixing, immunofluorescence staining, imaging, and analyzing 2D hiPSC-CM cultures (Fig. 2). For live cultures, the required steps are described in the next section. Here, we immunostained hiPSC-CMs for α-actinin but other sarcomeric proteins can also be stained. Better results were obtained with proteins that are part of the Z-discs or M-lines (e.g., titin, myomesin) than with A-band proteins (actin, troponin). Differentiation and immunostaining of skeletal muscle differs from the protocol described below and can be found in Iuliano et al. (2020). The acquisition and analysis (steps 29-49) are identical for all models.
Materials
-
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs), either from commercial sources (e.g., Ncardia, cat. no. K0104; Fujifilm, cat. no. 11713; Axol Bio, cat. no. ax2508) or differentiated in-house using a preferred protocol
-
Dulbecco's phosphate buffered saline, calcium, magnesium (DPBS; Thermo Fisher Scientific, cat. no. 14040091)
-
Dulbecco's phosphate buffered saline, no calcium, no magnesium (DPBS−; Thermo Fisher Scientific, cat. no. 14190094)
-
Multi Tissue Dissociation Kits (Miltenyi Biotec, cat. no. 130-110-204)
-
BSA-based polyvinyl alcohol essential lipids (BPEL; see Ng, Davis, Stanley, & Elefanty, 2008)
-
FBS (Biowest, cat. no. S00F910002)
-
Matrigel Basement Membrane Matrix, LDEV-Free (Corning, cat. no. 354234)
-
Paraformaldehyde (PFA; MilliporeSigma, cat. no. 158127-500G)
-
Triton X-100 (MilliporeSigma, cat. no. T8787)
-
Monoclonal anti-α-actinin (sarcomeric) antibody (MilliporeSigma, cat. no. A7811)
-
Donkey anti-Mouse IgG (H+L) (Thermo Fisher Scientific, cat. no. a-21203)
-
Tween 20 (MilliporeSigma, cat. no. P1379-100 ml)
-
4′,6-Diamidino-2-phenylindole, dihydrochloride (DAPI; Thermo Fisher Scientific, cat. no. D1306)
-
Immersion oil, Type-F (Leica, cat. no. 11944399)
-
Personal computer (Windows or macOS) with at least 6 GB of RAM
-
SP8 white light laser fluorescent confocal microscope (Leica) or equivalent, including well plate holder and 40× and/or higher objectives
-
Centrifuge (Eppendorf, cat. no. 5810R)
-
Aspirator
-
37°C humidified CO2 cell culture incubator (Sanyo)
-
Microscope for cell counting or an automated cytometer
-
Falcon tube (Thermo Fisher Scientific, cat. no. 14-959-53A)
-
5-, 10-, and 25-ml sterile plastic pipets (Greiner Bio-One, cat. no. 606180, 607180, and 760180, respectively)
-
15- and 50-ml polystyrene conical tubes (Corning Falcon, cat. no. 352097 and 352098, respectively)
-
PIPETMAN Classic P200 and P1000 (Gilson, cat. no. F123601 and F123602, respectively)
-
10-, 200-, and 1,000-μl filter tips (Corning, cat. no. 4807, 4810, and 4809, respectively)
-
6-well plate (Falcon, cat. no. 353046)
-
Falcon 96-well black/clear flat bottom microplate (Corning, cat. no. 353219)
Sample preparation
NOTE : The hiPSC-CM culture, dissociation, and replating steps are performed under aseptic conditions.
1.Thaw and culture commercially available hiPSC-CMs according to the manufacturer's instructions. Alternatively, use hiPSC-CMs differentiated using a preferred protocol (Breckwoldt et al., 2017; Correia et al., 2018; Current Protocols article: Giacomelli, Bellin, Orlova, & Mummery, 2017; Mummery et al., 2012). High (>80%) differentiation efficiency is recommended but not essential.
2.Coat a thin-bottom 96-well plate with a 50-µl drop of Matrigel (1:100 in DPBS for 1 hr at room temperature.
3.Prepare the dissociation reagent by adding 100 μl Enzyme T to 900 μl Buffer X from the multi tissue dissociation kit.
4.Wash cells with DPBS− once (1 ml per well for 6-well plates) and aspirate.
5.Add Enzyme T/Buffer X mix to the well and incubate 10 min in a humidified incubator (37°C, 5% CO2).
6.Add 1 ml BPEL plus 10% FBS to the well. Collect the entire volume with a P1000 pipet and mechanically disrupt the cells in suspension by gently pipetting up and down.
7.Collect all cells in a 15-ml Falcon tube.
8.Rinse the well with 2 ml BPEL plus 10% FBS and add to the Falcon tube. Repeat once.
9.Centrifuge cells at 300 × g for 3 min and remove supernatant.
10.Add 1 ml BPEL and resuspend.
11.Count cells manually or with an automatic cytometer.
12.Add 25,000 cells to the pre-coated 96-well plate in 200 µl BPEL.
13.Return to the humidified incubator and refresh BPEL after 24 hr.
Immunostaining hiPSC-CM monolayers in 96-well plates
14.After at least 3 days in culture, remove plates from the incubator and wash cells with cold DPBS−.
15.Fix cells with 4% PFA in 0.2 M DPBS, pH 7.4 for 10 min.
16.Wash three times with DPBS−.
17.Permeabilize with DPBS− with 0.1% Triton X-100 for 10 min.
18.Wash three times with DPBS−.
19.Block non-specific antibody binding for 1-2 hr with DPBS− plus 10% FBS.
20.Dilute primary antibody in DPBS− plus 10% FBS according to the manufacturer's instructions.
21.Remove blocking solutions and add the solution with the antibody to the cells. Incubate cells overnight at 4°C or 3 hr at room temperature.
22.Remove the antibody and wash three times with DPBS− with 0.05% Tween 20 for 10 min.
23.Dilute the secondary antibody in DPBS− plus 10% FBS according to the manufacturer's instructions.
24.Add the secondary antibody to the cells and incubate 2 hr.
25.Wash cells five times with DPBS− with 0.05% Tween 20 for 5 min.
26.Counterstain with 1:500 DAPI for 10 min.
27.Wash with DPBS− with 0.05% Tween 20 twice.
28.Add a volume of DPBS− large enough to prevent drying.
Imaging the samples
This next step can be performed on various confocal microscope systems. The Leica SP8 is given as an example.
29.Turn on the Leica SP8 confocal microscopic system, the computer, and the Leica imaging suite.
30.Place the 96-well plate into the appropriate holder.
31.Select the appropriate objective.
32.Activate the appropriate laser light using the Leica Dye Assistant.
33.Make sure all other settings are optimal for the best resolution and minimal background.
34.Adjust objective so that the sarcomeres are in focus.
35.Acquire at least ten to fifteen images per condition.
36.Export each image as a separate TIFF.
Analysis of images with the SotaTool
NOTE : The next steps can be carried out on any computer (PC) and/or laptop with sufficient hardware (see Materials list), as the SotaTool is a stand-alone application requiring no installation. Computing time will depend on the resolution, size, and bit-depth of the raw images and the PC specifications. For one 1024 × 1024 TIFF with a bit depth of 16 and the default offset of 4 μm, the computing time is ∼50 s on a standard PC.
37.Transfer all TIFF images to a single folder.
38.Download the entire software folder of the SotaTool (www.github.com/steinjm/SotaTool) and place anywhere on the computer.
39.Double click on the executable file within the folder named SotaTool.exe.
40.Click ‘Browse’ to select the folder with the images to analyze (input directory, Fig. 3).
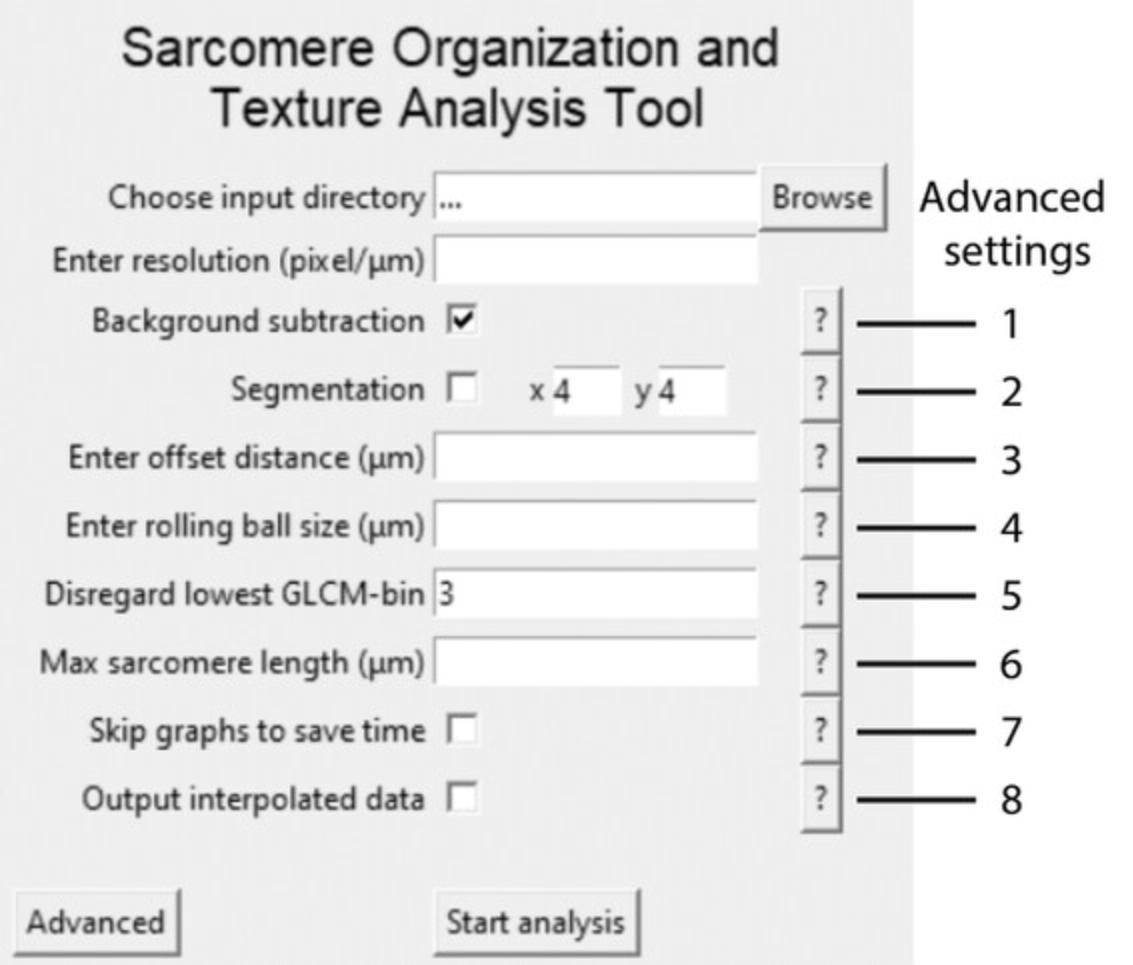
41.Input the resolution (pixels per micron).
42.Press ‘Start analysis’.
43.Check whether the output graphs are as expected.
Alternate Protocol: SAMPLE PREPARATION, ACQUISITION, AND QUANTIFICATION OF FRACTIONAL SHORTENING IN LIVE REPORTER hiPSC LINES
This protocol describes the analysis of videos of hiPSC-CMs expressing a mEGFP-tagged ACTN2 protein from an ACTN2-GFP WTC hiPSC line (Roberts et al., 2019). In choosing which tagged sarcomere proteins are best to use, the same recommendations apply as for static images (see introduction of Basic Protocol). ACTN2-reporter cells were differentiated using a modified version of Giacomelli et al. (2017). Analyses of phase-contrast images or live acquisitions of neonatal rabbit cardiomyocytes using SotaTool are identical; a protocol for the isolation of these cells can be found in MacQuaide, Ramay, Sobie, and Smith (2010).
Additional Materials (see also Basic Protocol)
-
hiPSC-CMs with a mEGFP-tagged ACTN2-reporter: Allen Institute for Cell Science ACTN2-GFP WTC iPSC line (AICS-0075-085, https://hpscreg.eu/cell-line/UCSFi001-A-4 with mEGFP inserted at ACTN2 C-terminus, obtained from Coriell Institute for Medical Research)
-
Andor Dragonfly 500 spinning disc inverted confocal microscope
-
Personal computer equipped with Andor Fusion software for microscopy and analysis (Oxford Instruments Andor, version 2.3)
-
On-stage incubator
-
Electric field stimulator (homemade or equivalent commercially available device, e.g., IonOptix myopacer cell stimulator)
Preparing the samples
The following steps are performed under aseptic conditions.
1.Perform steps 1-13 of Basic Protocol.
2.Thaw cells according manufacturer's instructions and plate on a thin-bottom 96-well plate.
3.Allow cells to recover for 24 hr and refresh medium.
Imaging the samples
NOTE : In order to extract sufficient data on the time dynamics and spatial resolution of the sarcomeric contraction, a frame rate of >50 frames per second (fps) is recommended for acquisition of live images on video. Note, this frame rate depends on the beating (and/or stimulation) frequency. For this purpose, we use an Andor Dragonfly spinning disc 500 with an on-stage incubator but an equivalent (widefield) system can be used as well.
4.Start the Andor Dragonfly spinning disc confocal system, detectors, lasers, and on-stage incubator.
5.Wait for the on-stage incubator to achieve appropriate conditions (37°C, 5% CO2).
6.Mount the multi-well plate onto the microscope.
7.Start the Fusion software.
8.Perform steps 30-37 of Basic Protocol.
9.Place the electrodes of the field stimulator in the 96-well plate and pace the cells.
10.Check whether the cells are “following” the imposed pacing rate.
11.Acquire images.
12.Export each image as a separate TIFF.
Extracting the fractional shortening using the SotaTool
13.Perform steps 39-47 from Basic Protocol.
14.Open the sheet with the sarcomere lengths in any preferred data visualization software (e.g., Excel, R-Studio, Graphpad).
15.Plot the sarcomere length on the y -axis against the timestamp on the x -axis for each separately acquired movie.
16.Note the maximum sarcomere and minimum sarcomere length and calculate the difference.
17.Divide the distance of shortening by the maximum length × 100% for the fractional shortening of that cell.
Support Protocol 1: FINDING THE IMAGE RESOLUTION
This protocol describes the steps to determine resolution and offset distance from the image metadata.
Hardware
Personal computer (Windows or macOS) with at least 6 GB of RAM and access to the Internet
1.Download FIJI (or ImageJ) for the operating system of your computer from the following link: https://imagej.net/software/fiji/?Downloads).
2.Start the software.
3.Navigate to ‘File’ → ‘Open…’ and open the TIFF (or press CTR + O).
4.Navigate to ‘Image’ → ‘Show Info’ (or press CTR + I).
5.Note the resolution (pixels per micron).
Support Protocol 2: ADVANCED ANALYSIS SETTINGS
The following section describes the settings as shown in Figure 3.
Hardware
Personal computer (Windows or macOS) with at least 6 GB of RAM and access to the Internet
1.Background subtraction greatly enhances the image quality and therefore increases accuracy of the algorithm. This is turned on by default and always recommended when no prior background subtractions have been performed on the raw data.
2.Segmentation automatically segments the original images and saves them in a subfolder for subsequent analysis.
3.The offset distance is automatically defined as 4 μm but in this box the user can manually define the maximum offset distance. This greatly reduces computing time. It is crucial that the offset distance is not smaller than the expected sarcomere length.
4.Rolling ball size is automatically defined as 2 μm, which is optimized for hiPSC-CMs. Here, the user can manually override this ball size.
5.Gray-level co-occurrence matrix (GLCM)-filtering filters out the lowest gray values of the original image by disregarding the first columns and rows of the GLCM before calculating the correlation value. We have found the best default number to be three but the user can input a manual override here.
6.In this box, the user can insert the maximum expected sarcomere length. This feature is useful when there is noise in the image.
7.Skip graphs to save time. We do not recommend skipping the output graphs as this is the only way to verify the analysis has been performed correctly.
8.When this box is checked, an additional .csv file is exported in the output directory with the interpolated data.
Support Protocol 3: FINDING SARCOMERE LENGTH IN NON-ALIGNED CELLS
The following section describes the steps needed to take multiple measurements for non-aligned cells.
Additional Materials (also see Basic Protocol)
Personal computer (Windows or macOS) with at least 6 GB of RAM and access to the Internet
1.Perform Basic Protocol.
2.Check whether the program outputs values for sarcomere length or 0 values only.
3.Check the output graphs.
4.If there are no peaks visible, segmentation might be necessary to find proper sarcomere length values.
5.Repeat the analysis on the raw images but check ‘Segmentation’ before starting analysis.
6.Check the output sheet. For each image, the segment with the highest sarcomere organization score has the most reliable sarcomere length but the mean of multiple (properly analyzed) segments can also be used.
COMMENTARY
Background Information
Analysis of sarcomere length of static 2D muscle cells and fractional shortening of live cells in 3D is usually time consuming, requires considerable manual labor, and is highly prone to user-selection bias. For this reason, we created a versatile, robust, and rapid method for sarcomere length quantification of many different muscle models in an open-source stand-alone Python-based application as an extension of the algorithm developed by Sutcliffe et al. (2018).
This method is based on GLCM (Haralick, Shanmugam, & Dinstein, 1973), which translates images into arrays of i columns and j rows, one for each gray level of the original image (Fig. 4A). From one GLCM, a correlation value is calculated over the entire image, which is a measure of the linear dependency for gray-value combinations over a distance g (Fig. 4B). The algorithm calculates the correlation values for 180 angles (α) over 1-g pixel-pixel distances (Fig. 1A).
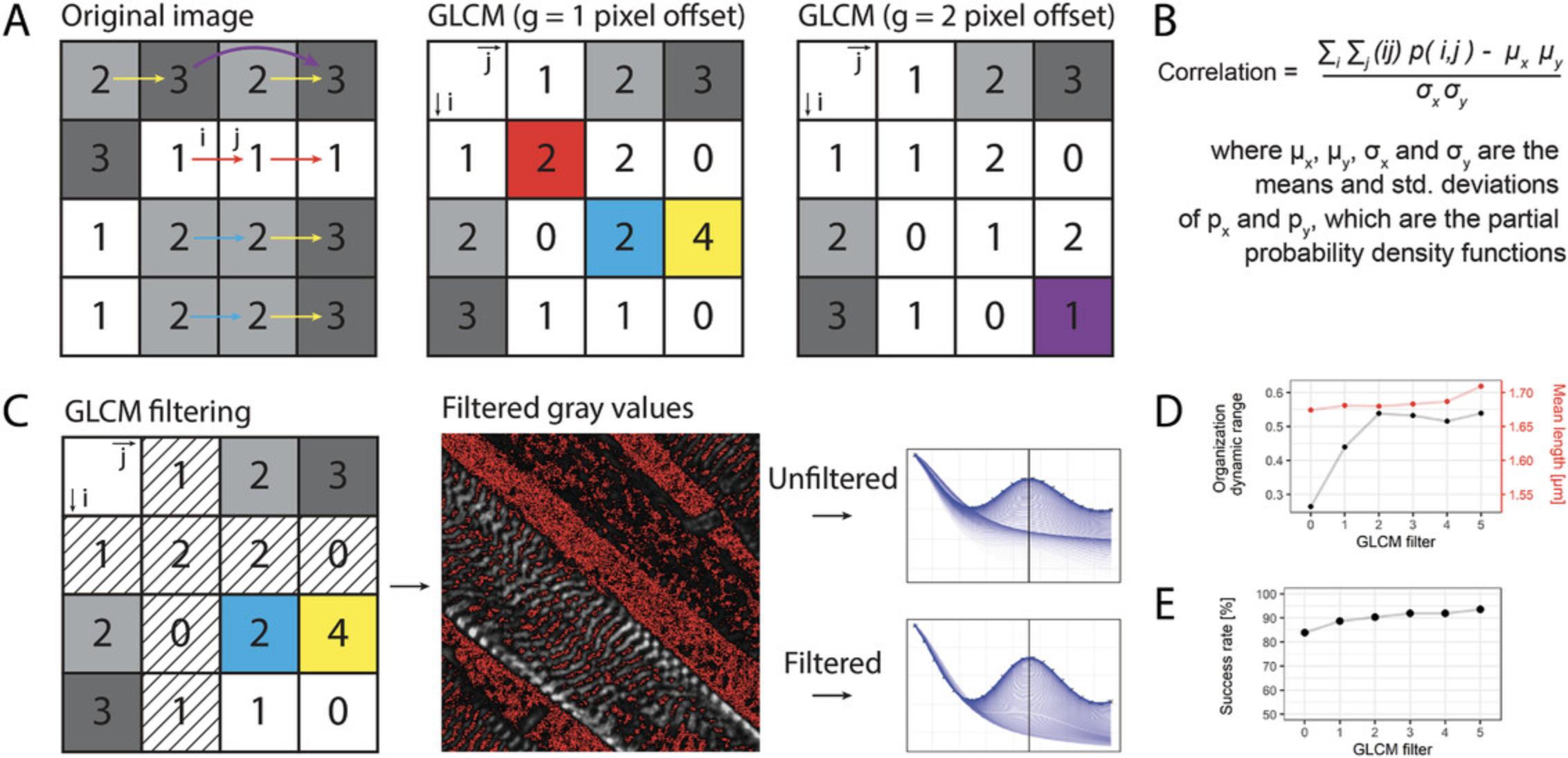
Repeated striations in the image (such as sarcomeres) result in damped oscillating correlation graphs. To extract the direction in which the striations are most clearly present, the algorithm finds and selects the angle α at which the correlation peak is maximum. At this point, the striation-striation distance (i.e., the x -axis offset of the highest peak) can be determined (Fig. 1). This x -axis offset is defined as the distance g in the angle α where the highest linear dependency between gray values over the entire image is found.
By using automatic and whole-image analysis, manual labor and selection bias are decreased, because no input is required from the user except for an optional maximum micrometer offset to minimize computing time. Most importantly, the selection of areas to measure and drawing of lines for fast Fourier transformation (FFT) analysis, as done in most current practices, is not required. Most current automated analysis software is reliant on segmentation or single point tracking of the sarcomere signal, which decreases throughput or robustness in 3D stainings of sarcomeres where there is often a higher signal/noise ratio (Toepfer et al., 2019). Aside from the sarcomere length and the direction of the striations, this algorithm also outputs a measure of sarcomere organization (the maximum peak value of the correlation graphs; Sutcliffe et al., 2018) and an alignment coefficient (the variation between all the curves on the previously calculated sarcomere length; Gerbin et al., 2021). Furthermore, two other Haralick features are incorporated in the output. The uniformity (also known as the angular second moment) is higher in uniform images, where 1 is a completely uniform image. The homogeneity (or the inverse difference moment) which represents image contrast, has a higher value for homogeneous images (Haralick et al., 1973; Sutcliffe et al., 2018).
One limitation of this algorithm is that images with morphologically adequate but misaligned sarcomeres do not produce correlation peaks. Therefore, we included an automated cropping step which segments the image in a user-defined manner. These segments are subsequently analyzed. In a dataset with non-aligned hiPSC-CMs cultured on standard tissue culture plastic, this increased the successful analysis of the images from 57% to 100% (see Fig. 1B). It results in a higher number of analyzed datapoints per image and requires post-processing user input to distinguish correctly from incorrectly analyzed segments. Another limitation of this method is that the metric for organization score has a narrow range, from 0 for disorganized to ∼0.3 for organized striations. We found that disregarding the first row and first column of the GLCM (where i and j are 0) greatly reduced the background and increased the baseline sarcomere organization score, and thereby the dynamic range of the output (Fig. 4D). This method also increased the success rate of peak detection and sarcomere length calculation in low-quality cells but did not affect the sarcomere length (Fig. 4E). It should be noted that altering the value for GLCM filtering (see Fig. 3, Advanced settings, step 5) and the value of the “rolling ball background subtraction” alters the sarcomere score and alignment but not the sarcomere length. For hiPSC-CMs, we found a filter of 3 is optimal but the user can override this in Advanced settings, step 5. To ensure broad applicability, the software does not take cell boundaries into account. Segmentation can be performed in tertiary software, such as CellProfiler (www.cellprofiler.org).
Multiple types of datasets containing striated images were analyzed and compared to other relevant methods. As shown in Figure 5, correlation of these datasets measured with SotaTool and the other methods resulted in a minimal R 2 of 0.862 among all tested datasets with p -values below .0001.
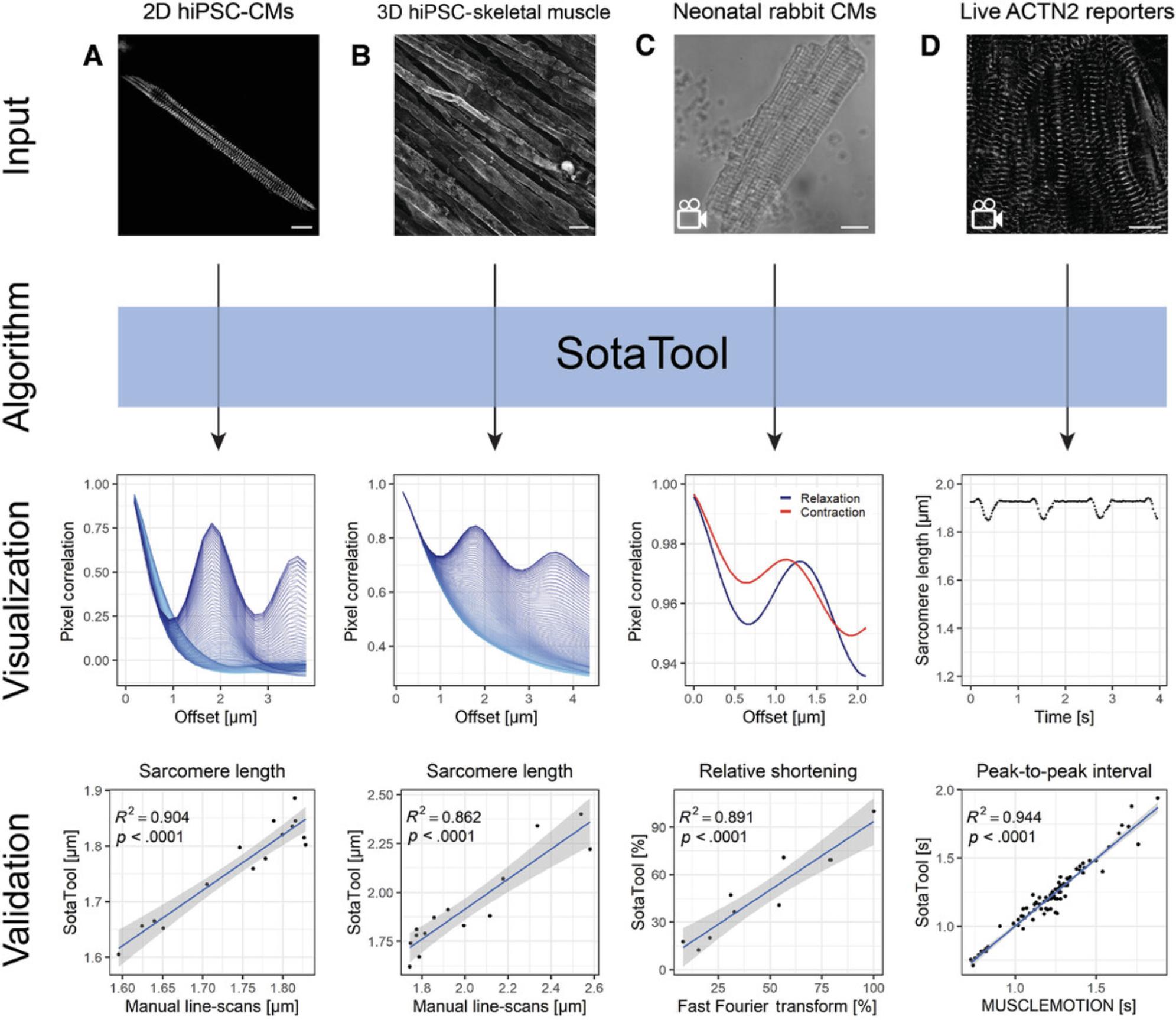
For the immunostained hiPSC-CMs cultured on micropatterned substrates (Birket et al., 2015), we performed manual length detection by creating manual line scans over the sarcomeres in ImageJ and measuring the average peak-to-peak distance. We found higher correlations between the methods and decreased absolute difference per datapoint upon increasing technical repeats of the manual detection (data not shown). This indicates that the sarcomere lengths resulting from the algorithm are closer to the ground truth as it takes all the striations in an entire image into account, whereas manual detection is limited by selection bias and sampling error. For the final analysis with ten manual line scans, the R 2 was 0.904 (Fig. 5A).
We applied the same method of manual detection and SotaTool analysis of images of hiPSC-derived skeletal muscle fibers cultured in 3D stained for titin. These images were chosen because the sarcomeres differ morphologically from those in hiPSC-CMs. SotaTool was able to detect striations successfully and the sarcomere length scores corresponded to those using manual detection (R 2 = 0.862) taking 5 ROIs per image (Fig. 5B).
Videos of neonatal rabbit CMs with a frame rate of 100 fps were analyzed as single TIFF images (Fig. 5C). The resulting sarcomere lengths in contracted and relaxed state were subtracted, resulting in the fractional shortening. These were normalized and compared to the normalized shortening obtained using FFT analysis (Sala et al., 2018). The measurements from the two methods correlated with an R 2 of 0.891.
The live mEGFP-tagged ACTN2 reporter cells were recorded with a frame rate of 50 fps and analyzed in a similar fashion and the beat rate was quantified (Fig. 5D). After SotaTool analysis of all the separate frames, the peaks were analyzed by a custom script in R-studio, which correlated with MUSCLEMOTION (R 2 = 0.944; Sala et al., 2018). See Figure 8 for a representative acquisition of the reporter CMs along with the corresponding correlation graphs. Depending on the question, pacing of hiPSC-CMs might be considered. For example, the basal beat rate of hiPSC-CM can be variable between different differentiations or conditions which hampers comparison of absolute output parameters if they are not paced. On the other hand, to detect, for example, chronotropic changes, acquisition of images from spontaneously contracting cardiomyocytes are highly informative.
Analyzing live reporter cells with fluorescently tagged sarcomeres has several advantages over static cultures in which sarcomeres are immunostained. Firstly, live reporters can be monitored in real time over long periods, while immunostaining is a terminal end-point measurement. Secondly, in fixed cardiomyocytes, it is not possible to determine which cells are in systole, diastole, or an intermediate contraction/relaxation state; this may affect the sarcomere distance quantification. This is not an issue in live reporter cells where imaging is continuous. Moreover, PFA fixation can cause artefacts like cell shrinkage and disruption of cell organelles (Schnell, Dijk, Sjollema, & Giepmans, 2012). Of note though, live cell fluorescent microscopy at a high frame rate can cause phototoxicity in the cells or bleaching of the fluorescent signal and significantly more data-storage capacity and analysis time is needed for live cell image acquisition.
Critical Parameters
A single monolayer of cells is preferred because hiPSC-CMs are easier to image and analyze than hiPSC-CMs that have formed multiple layers. This is highly dependent on seeding density but selecting the correct hiPSC-CM seeding density occasionally requires some optimization. We recommend trying different seeding densities (e.g., 16k/cm2, 31k/cm2, 78k/cm2, 156k/cm2, 234k/cm2, 313k/cm2, where k = × 103) due to varying recovery efficiencies during passaging and following thawing after cryopreservation in different hiPSC-lines. Aside from the seeding density, the morphology can vary upon use of different coating proteins, e.g., Geltrex, fibronectin, or laminin instead of Matrigel.
For proper quantification of sarcomere length, the pixel size is the largest factor influencing the final result. Therefore, the images should not be “binned” during or after imaging. Moreover, an objective of over 40× is preferred for imaging. Due to the interpolation method of the algorithm, images derived using lower magnification objectives can also be analyzed to a certain extent but artefacts and incorrectly analyzed images may occur. For successful comparison of multiple datasets, the settings of the camera, laser, and SotaTool should always be identical. The same applies to the antibody dilutions and the incubation times during immunostaining.
Troubleshooting
After successful analysis, the output graph should look like Figure 6A. If there are insufficient recurring anisotropic striation patterns in the image, the output will look like Figure 6B. In this case, the user should opt for the automated segmentation step during analysis (see Support Protocol 3). If this still does not result in successful analysis of images, the user might use ImageJ or Photoshop to manually select an area of interest. Note that the organization parameters of manually cropped images cannot be compared to the organization of other unsegmented images. In certain cases, a recurrent noise signal can lead to output graphs like Figure 6C: Choosing maximum offset or maximum sarcomere distance will solve this. If an output graph looks like Figure 6D, most likely the pixel size is entered instead of the resolution, which should be in pixels/μm. If the value for the maximum sarcomere length is set too low, the output will look like 6E.
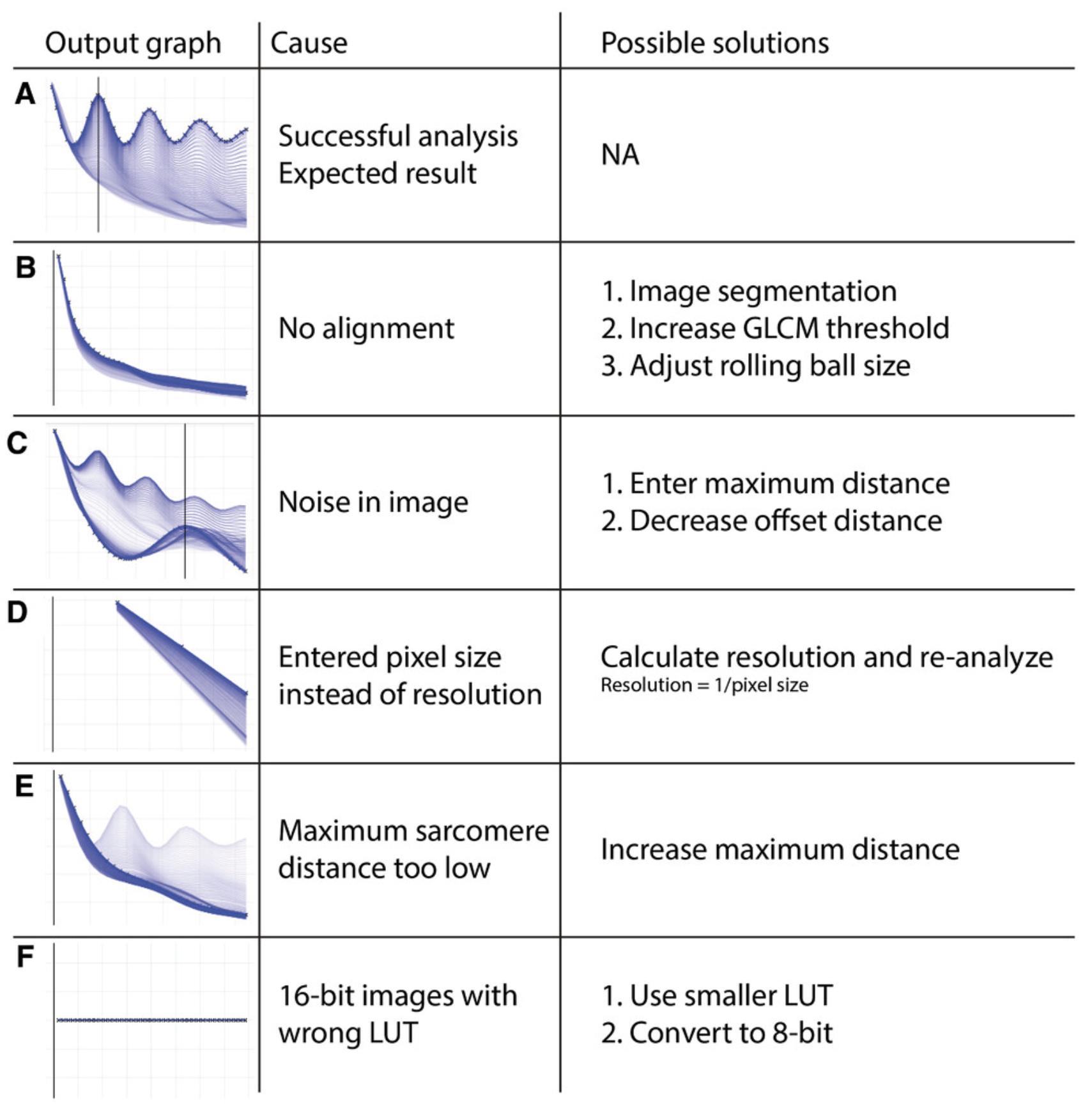
The GLCM is created by binning the original image in 256 gray values. When images with a bit depth higher than 8 are analyzed, this can result in all gray values being in the same bin (Figure 6F). These images should be pre-processed in ImageJ. Navigate to ‘Image’ → ‘Adjust’ → ‘Brightness/Contrast…’ and adjust the brightness and the contrast to the required level and press “Apply”. Save the image and re-run the analysis. Preferably, the same range is chosen across a larger dataset.
If the program directly stops running without an output folder being made, this might indicate a faulty input folder, or one with restricted or password-protected access. If an output folder is made but has no content, make sure the file extension is .tif.
Understanding Results
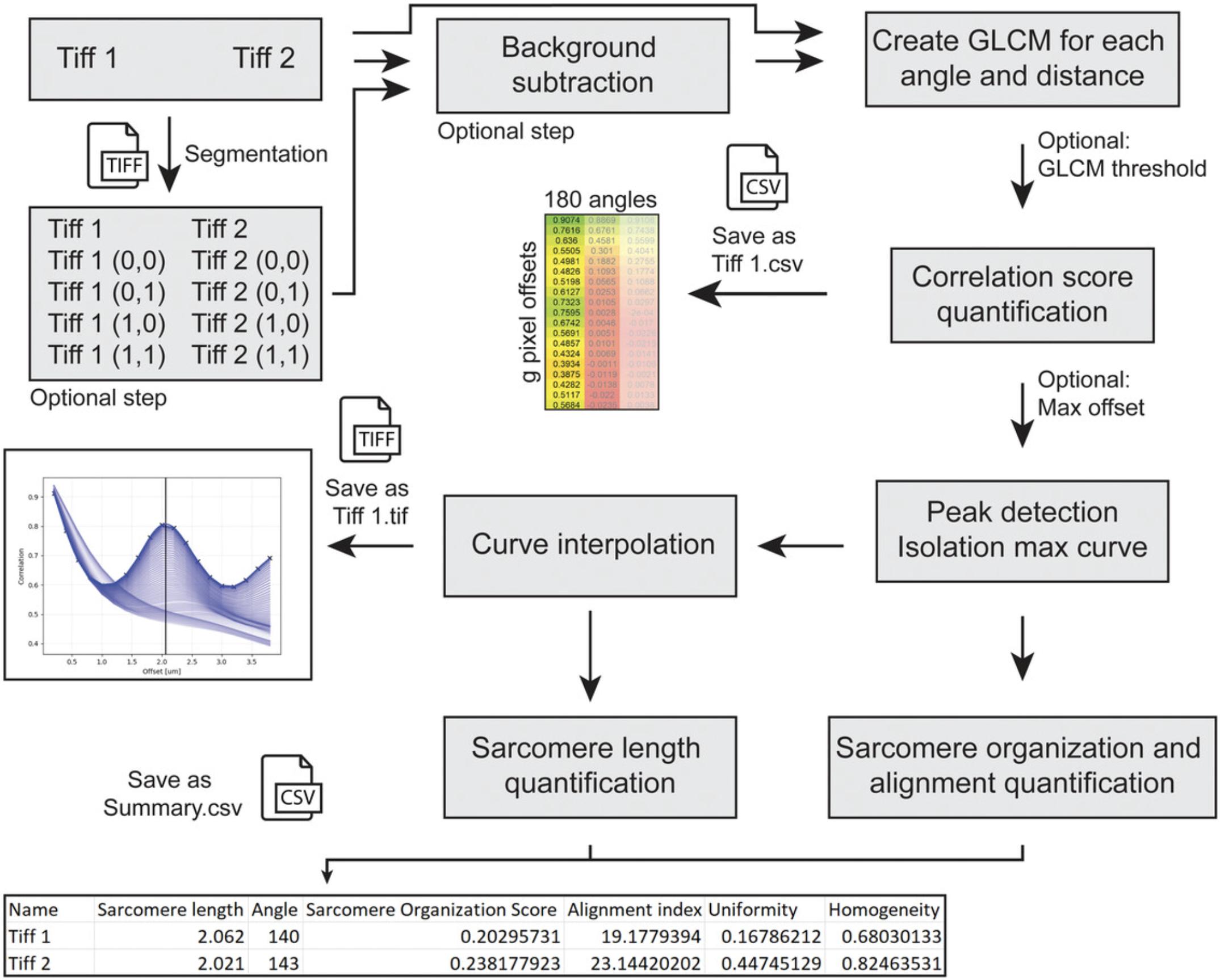
This protocol describes an automated analysis pipeline of multiple types of data. An overview of the input, output, and algorithm steps are given in Figure 7. Broadly, after two optional pre-processing steps (background subtraction and segmentation), the GLCM is calculated, and the raw data is saved per image as a comma delimited file (.csv). The curve with the highest peak is isolated from all the generated curves and the sarcomere organization score calculated as the difference between the height of the peak and its preceding minimum. At the offset distance of the maximum peak, the variance between all curves is taken as an alignment index. The sarcomere length is calculated from the maximum peak after interpolating with a bicubic spline, for sub-pixel accuracy. Per image, a graph is saved containing (1) all curves of all angles, (2) an overlay of the interpolated maximum graph, (3) its raw datapoints, and (4) a trendline visualizing the sarcomere length for that image. For the entire dataset, another .csv file is saved containing the file names, sarcomere lengths, angles, sarcomere organization score, and alignment index. Optionally, the interpolated data can be saved per image as .csv (see Fig. 3, Advanced settings, step 8).
Open Research
Data Availability Statement
The standalone application, source code, instructions, sample images, and supporting scripts can be found on www.github.com/steinjm/SotaTool.
Literature Cited
- Birket, M. J., Ribeiro, M. C., Kosmidis, G., Ward, D., Leitoguinho, A. R., van de Pol, V., … Mummery, C. L. (2015). Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Reports , 13(4), 733–745. doi: 10.1016/j.celrep.2015.09.025
- Breckwoldt, K., Letuffe-Brenière, D., Mannhardt, I., Schulze, T., Ulmer, B., Werner, T., … Hansen, A. (2017). Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nature Protocols , 12(6), 1177–1197. doi: 10.1038/nprot.2017.033
- Cao, L., Manders, E., & Helmes, M. (2021). Automatic detection of adult cardiomyocyte for high throughput measurements of calcium and contractility. PLoS One , 16(9), e0256713. doi: 10.1371/journal.pone.0256713
- Cohn, R., Thakar, K., Lowe, A., Ladha, F. A., Pettinato, A. M., Romano, R., … Hinson, J. T. (2019). A contraction stress model of hypertrophic cardiomyopathy due to sarcomere mutations. Stem Cell Reports , 12(1), 71–83. doi: 10.1016/j.stemcr.2018.11.015
- Correia, C., Koshkin, A., Duarte, P., Hu, D., Carido, M., Sebastião, M. J., … Serra, M. (2018). 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnology and Bioengineering , 115(3), 630–644. doi: 10.1002/bit.26504
- Gerbin, K. A., Grancharova, T., Donovan-Maiye, R. M., Hendershott, M. C., Anderson, H. G., Brown, J. M., … Gunawardane, R. N. (2021). Cell states beyond transcriptomics: Integrating structural organization and gene expression in hiPSC-derived cardiomyocytes. Cell Systems , 12(6), 670–687.e10. doi: 10.1016/j.cels.2021.05.001
- Giacomelli, E., Bellin, M., Orlova, V. V., & Mummery, C. L. (2017). Co-differentiation of human pluripotent stem cells-derived cardiomyocytes and endothelial cells from cardiac mesoderm provides a three-dimensional model of cardiac microtissue. Current Protocols in Human Genetics , 95(1), 21.9.1–21.9.22. doi: 10.1002/cphg.46
- Giacomelli, E., Meraviglia, V., Campostrini, G., Cochrane, A., Cao, X., van Helden, R. W. J., … Mummery, C. L. (2020). Human-iPSC-derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell , 26(6), 862–879.e11. doi: 10.1016/j.stem.2020.05.004
- Haralick, R. M., Shanmugam, K., & Dinstein, I. (1973). Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics , SMC-3(6), 610–621. doi: 10.1109/TSMC.1973.4309314
- Iuliano, A., Wal, E., Ruijmbeek, C. W. B., in ‘t Groen, S. L. M., Pijnappel, W. W. M. P., Greef, J. C., & Saggiomo, V. (2020). Coupling 3D printing and novel replica molding for in house fabrication of skeletal muscle tissue engineering devices. Advanced Materials Technologies , 5, 2000344. doi: 10.1002/admt.202000344
- Jalal, S., Dastidar, S., & Tedesco, F. S. (2021). Advanced models of human skeletal muscle differentiation, development and disease: Three-dimensional cultures, organoids and beyond. Current Opinion in Cell Biology , 73(August), 92–104. doi: 10.1016/j.ceb.2021.06.004
- Koc, A., Sahoglu Goktas, S., Akgul Caglar, T., & Cagavi, E. (2021). Defining optimal enzyme and matrix combination for replating of human induced pluripotent stem cell-derived cardiomyocytes at different levels of maturity. Experimental Cell Research , 403(2), 112599. doi: 10.1016/j.yexcr.2021.112599
- MacQuaide, N., Ramay, H. R., Sobie, E. A., & Smith, G. L. (2010). Differential sensitivity of Ca2+ wave and Ca2+ spark events to ruthenium red in isolated permeabilised rabbit cardiomyocytes. The Journal of Physiology , 588(23), 4731–4742. doi: 10.1113/jphysiol.2010.193375
- Mills, R. J., Titmarsh, D. M., Koenig, X., Parker, B. L., Ryall, J. G., Quaife-Ryan, G. A., … Hudson, J. E. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proceedings of the National Academy of Sciences , 114(40), E8372–E8381. doi: 10.1073/pnas.1707316114
- Mummery, C. L., Zhang, J., Ng, E. S., Elliott, D. A., Elefanty, A. G., & Kamp, T. J. (2012). Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes. Circulation Research , 111(3), 344–358. doi: 10.1161/CIRCRESAHA.110.227512
- Ng, E. S., Davis, R., Stanley, E. G., & Elefanty, A. G. (2008). A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nature Protocols , 3(5), 768–776. doi: 10.1038/nprot.2008.42
- Pasqualin, C., Gannier, F., Yu, A., Malécot, C. O., Bredeloux, P., & Maupoil, V. (2016). SarcOptiM for ImageJ: High-frequency online sarcomere length computing on stimulated cardiomyocytes. American Journal of Physiology-Cell Physiology , 311(2), C277–C283. doi: 10.1152/ajpcell.00094.2016
- Peterson, P., Kalda, M., & Vendelin, M. (2013). Real-time determination of sarcomere length of a single cardiomyocyte during contraction. American Journal of Physiology-Cell Physiology , 304(6), C519–C531. doi: 10.1152/ajpcell.00032.2012
- Ribeiro, M. C., Tertoolen, L. G., Guadix, J. A., Bellin, M., Kosmidis, G., D'Aniello, C., … Passier, R. (2015). Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro – Correlation between contraction force and electrophysiology. Biomaterials , 51, 138–150. doi: 10.1016/j.biomaterials.2015.01.067
- Roberts, B., Hendershott, M. C., Arakaki, J., Gerbin, K. A., Malik, H., Nelson, A., … Gunawardane, R. N. (2019). Fluorescent gene tagging of transcriptionally silent genes in hiPSCs. Stem Cell Reports , 12(5), 1145–1158. doi: 10.1016/j.stemcr.2019.03.001
- Sala, L., van Meer, B. J., Tertoolen, L. G. J., Bakkers, J., Bellin, M., Davis, R. P., … Mummery, C. L. (2018). Musclemotion. Circulation Research , 122(3), e5–e16. doi: 10.1161/CIRCRESAHA.117.312067
- Schnell, U., Dijk, F., Sjollema, K. A., & Giepmans, B. N. G. (2012). Immunolabeling artifacts and the need for live-cell imaging. Nature Methods , 9(2), 152–158. doi: 10.1038/nmeth.1855
- Sutcliffe, M. D., Tan, P. M., Fernandez-Perez, A., Nam, Y.-J., Munshi, N. V., & Saucerman, J. J. (2018). High content analysis identifies unique morphological features of reprogrammed cardiomyocytes. Scientific Reports , 8(1), 1258. doi: 10.1038/s41598-018-19539-z
- Toepfer, C. N., Sharma, A., Cicconet, M., Garfinkel, A. C., Mücke, M., Neyazi, M., … Seidman, C. E. (2019). SarcTrack: An adaptable software tool for efficient large-scale analysis of sarcomere function in hiPSC-Cardiomyocytes. Circulation Research , 124(8), 1172–1183. doi: 10.1161/CIRCRESAHA.118.314505
- van der Velden, J., Klein, L. J., van der Bijl, M. J. A. M., Huybregts, M. A., Stooker, W., Witkop, J., … Stienen, G. J. M. (1998). Force production in mechanically isolated cardiac myocytes from human ventricular muscle tissue. Cardiovascular Research , 38(2), 414–423. doi: 10.1016/S0008-6363(98)00019-4
- Wijnker, P. J. M., Friedrich, F. W., Dutsch, A., Reischmann, S., Eder, A., Mannhardt, I., … Carrier, L. (2016). Comparison of the effects of a truncating and a missense MYBPC3 mutation on contractile parameters of engineered heart tissue. Journal of Molecular and Cellular Cardiology , 97, 82–92. doi: 10.1016/j.yjmcc.2016.03.003
- Yang, X., Pabon, L., & Murry, C. E. (2014). Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circulation Research , 114(3), 511–523. doi: 10.1161/CIRCRESAHA.114.300558
Citing Literature
Number of times cited according to CrossRef: 3
- Phong D. Nguyen, Iris Gooijers, Giulia Campostrini, Arie O. Verkerk, Hessel Honkoop, Mara Bouwman, Dennis E. M. de Bakker, Tim Koopmans, Aryan Vink, Gerda E. M. Lamers, Avraham Shakked, Jonas Mars, Aat A. Mulder, Sonja Chocron, Kerstin Bartscherer, Eldad Tzahor, Christine L. Mummery, Teun P. de Boer, Milena Bellin, Jeroen Bakkers, Interplay between calcium and sarcomeres directs cardiomyocyte maturation during regeneration, Science, 10.1126/science.abo6718, 380 , 6646, (758-764), (2023).
- Ulgu Arslan, Marcella Brescia, Viviana Meraviglia, Dennis M. Nahon, Ruben W.J. van Helden, Jeroen M. Stein, Francijna E. van den Hil, Berend J. van Meer, Marc Vila Cuenca, Christine L. Mummery, Valeria V. Orlova, Vascularized hiPSC-derived 3D cardiac microtissue on chip, Stem Cell Reports, 10.1016/j.stemcr.2023.06.001, 18 , 7, (1394-1404), (2023).
- Dylan Mostert, Bart Groenen, Leda Klouda, Robert Passier, Marie-Jose Goumans, Nicholas A. Kurniawan, Carlijn V. C. Bouten, Human pluripotent stem cell-derived cardiomyocytes align under cyclic strain when guided by cardiac fibroblasts, APL Bioengineering, 10.1063/5.0108914, 6 , 4, (2022).

