Inhibitor-free DNA extraction from soil and sediment samples
Dominik Buchner
Abstract
This protocol describes how to extract inhibitor-free DNA from soil and sediment samples. 5g of soil or up to 10g of sediment can be processed in one extraction, but there is also a miniaturized version for 250mg of input material, if less DNA is required. The protocol is based on the DNeasy PowerMax Soil Kit but costs much less. A lot of the buffers can be found in the following patent https://patents.google.com/patent/US7459548B2/en
Before start
Make sure all buffers are prepared before starting.
Steps
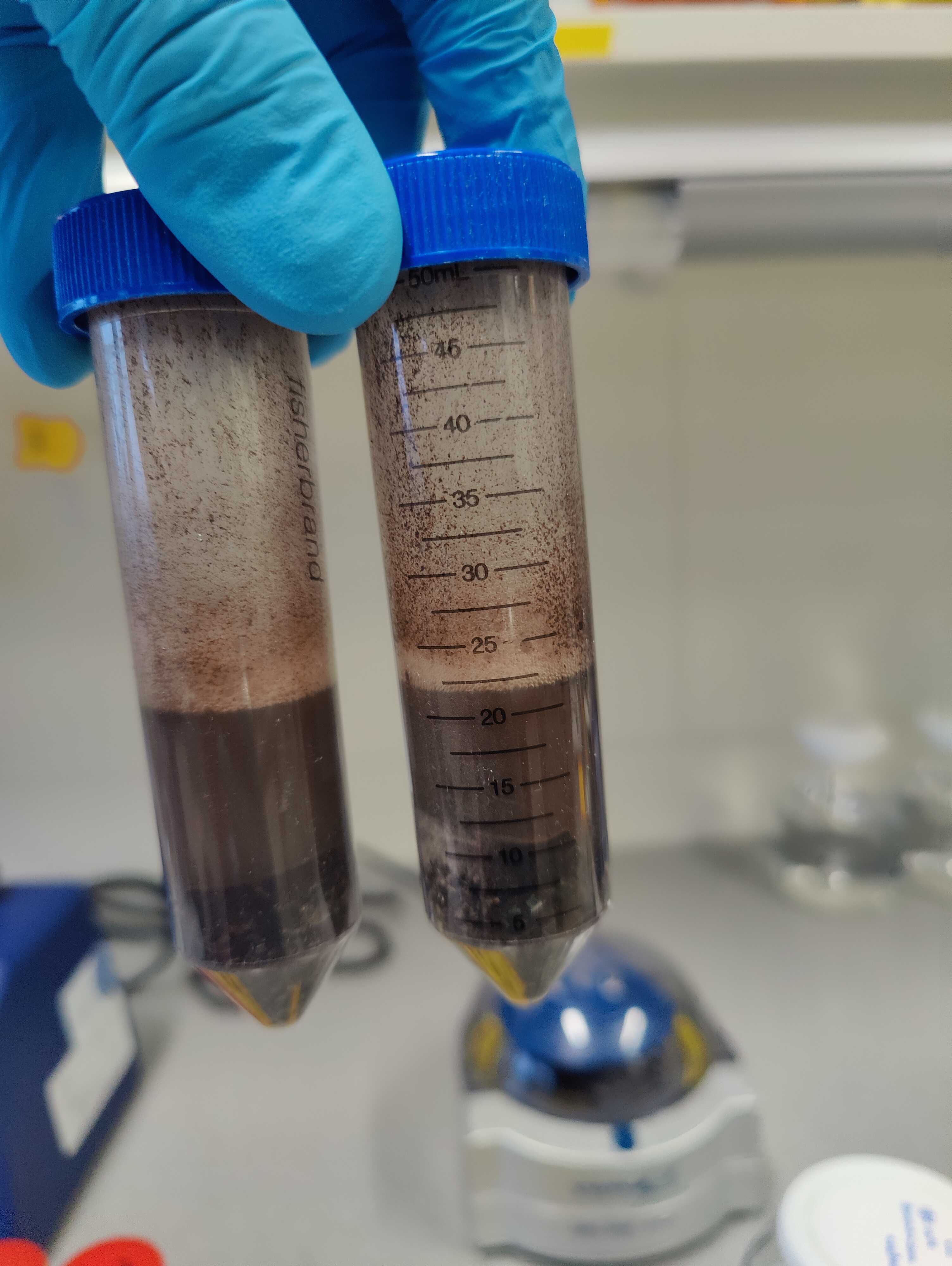
Protocol for up to 10 g of input material
Add 15mL and 1.2mL. Vortex shortly.
Add 5mL , vortex shortly, and incubate at 4°C for 0h 10m 0s .
Add 30mL . Vortex or invert to mix.
2500x g,20°C . Discard the flow-through. Repeat once to bind the complete sample volume.
Add 10mL . 2500x g,20°C to wash and dry the column.
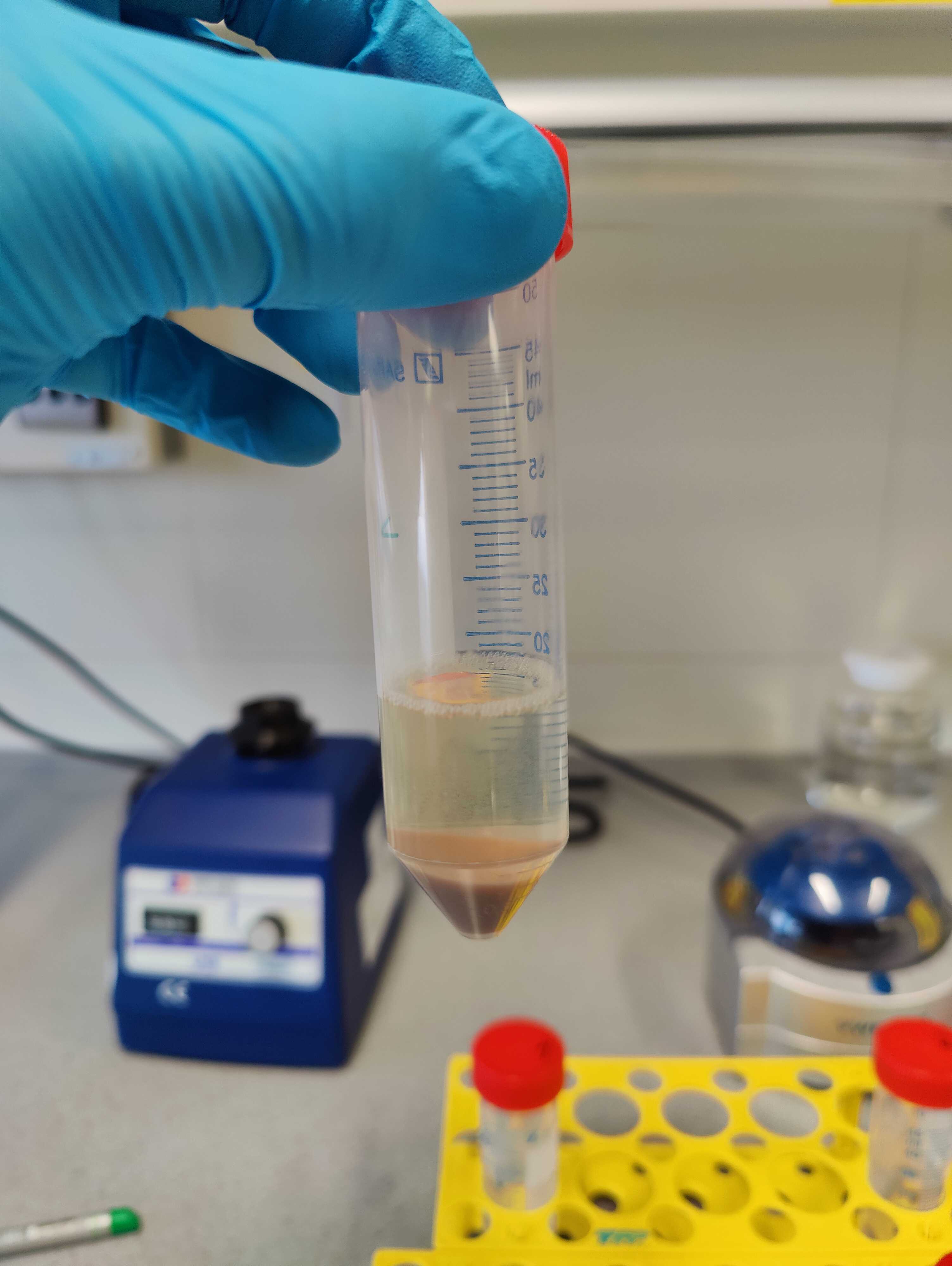
Transfer the column to a new tube. Add 1mL . Incubate for 0h 3m 0s at Room temperature .
2500x g,20°C to elute the DNA. DNA eluate should be completely colorless and ready to go for downstream analysis.
Protocol for up to 250 mg of input material
Prepare one 2 mL centrifuge tube per sample with 750 mg of garnet beads.
Add 250mg of soil or sediment sample.
Add 750µL and 60µL. Vortex shortly.
Place the samples on a Vortex adapter (e.g. Qiagen) and vortex at maximum speed for 0h 10m 0s .
10000x g,20°C . Transfer the supernatant to a new tube.
Add 250µL , vortex shortly, and incubate at 4°C for 0h 5m 0s .
10000x g,20°C . Transfer the supernatant to a new tube.
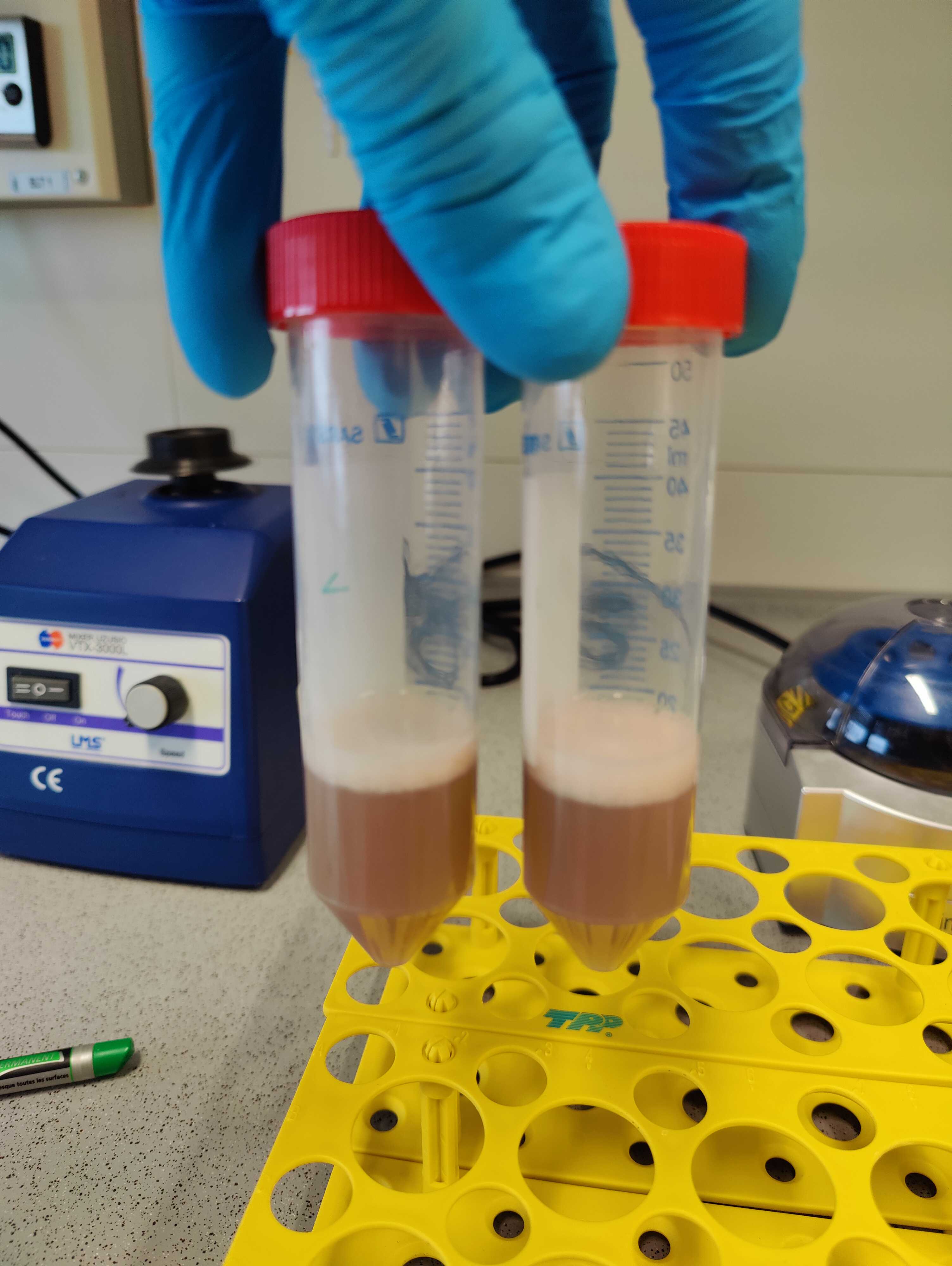
Add 200µL . A precipitate may form. Vortex shortly, incubate at 4°C for 0h 5m 0s .
10000x g,20°C . Transfer 600µL of the supernatant to a new tube.
Add 1200µL . Vortex to mix.
Load 650µL of the mixture to a mini spin column (e.g. Epoch Life Science).
10000x g,20°C . Discard the flow-through. Repeat two times to bind the complete sample volume.
Add 500µL . 10000x g,20°C to wash the column. Discard the flow-through.
10000x g,20°C to dry the column. Transfer the spin column to a clean 1.5 mL microcentrifuge tube.
Add 50µL . Incubate for 0h 3m 0s at Room temperature .
10000x g,20°C to elute the DNA. DNA eluate should be completely colorless and ready to go for downstream analysis.