Using CRISPR/ttLbCas12a for in planta Gene Targeting in A. thaliana
Patrick Schindele, Patrick Schindele, Holger Puchta, Holger Puchta, Laura Merker, Laura Merker
Abstract
CRISPR/Cas systems enable gene editing through the induction of site-specific DNA double-strand breaks (DSB). However, the nature of the induced modification highly depends on the mechanism used for DNA DSB repair. Non-homologous end joining (NHEJ)-mediated targeted mutagenesis induced by CRISPR/Cas is an already standardly applied tool, which can lead to various different kinds of mutations at a specific genomic site. Nevertheless, precise genome modification using homologous donor sequences is still challenging in plants. Applications depending on the less frequent homologous recombination (HR) require further improvements to create an attractive and efficient tool for general application in plants. Focusing on this issue, we developed the in planta gene targeting (ipGT) system, which is based on the simultaneous excision of a stably integrated, homologous donor sequence and the induction of a DSB within the target site. In recent years, several improvements were achieved enhancing gene targeting (GT) frequencies. After the successful application of Streptococcus pyogenes Cas9 (Sp Cas9) and Staphylococcus aureus Cas9 (Sa Cas9) for ipGT, we were able to further improve the system using Lachnospiraceae bacterium Cas12a (Lb Cas12a), which also enables cleavage in T-rich regions. Most recently, we tested an improved, temperature-tolerant version of Lb Cas12a (tt Lb Cas12a) for ipGT and were able to further increase GT efficiencies. Here, we describe the experimental procedure of the recently published ipGT system using tt Lb Cas12a in Arabidopsis thaliana in detail. © 2020 The Authors.
Basic Protocol 1 : Construction of CRISPR/tt Lb Cas12a expression vector to analyze ipGT efficiencies
Basic Protocol 2 : Achieving heritable GT plants
INTRODUCTION
Gene targeting (GT) is a precise method to modify genomic DNA by the use of a template DNA based on the DSB repair mechanism of homologous recombination (HR) (Puchta, Dujon, & Hohn, 1996). However, common CRISPR/Cas-mediated targeted mutagenesis relies on the error-prone non-homologous end joining (NHEJ) repair mechanism leading to site-specific insertions and deletions that cannot be predefined like GT modifications (Schindele, Dorn, & Puchta, 2020). In the case of GT, the integration of exactly defined modifications is feasible by using a donor sequence for HR containing the desired modification flanked by homologous regions to the target locus. For obtaining successful GT events, the DSB has to be repaired via HR using this donor sequence. To prevent the unintended cleavage of the repair template, additional silent mutations within the donor sequence are provided. In plants, HR frequencies are limited and thus GT efficiencies are low (Huang & Puchta, 2019). Therefore, we set up the ipGT system (see Fig. 1), in which the targeting nuclease not only induces the DSB within the target but also excises and thus activates the integrated donor sequence in order to elevate GT frequencies (Fauser et al., 2012).
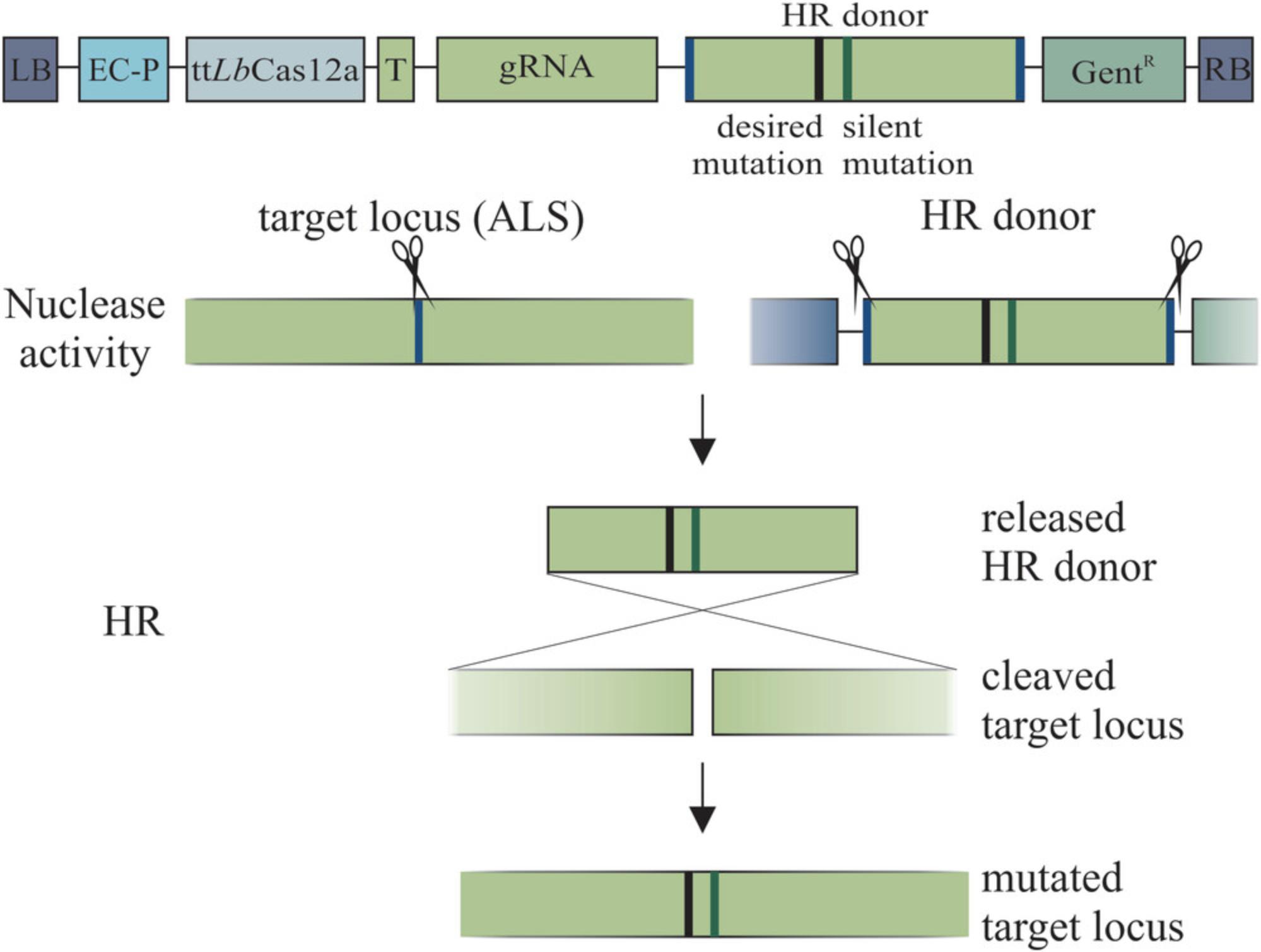
As CRISPR/Cas systems became a well-established and easy to handle programmable tool, they also became an auspicious tool for ipGT. The field of CRISPR/Cas is rapidly evolving and since the first adaptation of Sp Cas9, several new CRISPR/Cas systems and orthologues were adopted for their use in eukaryotes. We originally applied Sp Cas9 for ipGT in Arabidopsis with moderate success (Schiml, Fauser, & Puchta, 2014), but over the last years we started to test other nucleases. After showing that Sa Cas9 is more efficient than Sp Cas9 for NHEJ-mediated targeted mutagenesis in plants, we demonstrated that Sa Cas9 also improves ipGT efficiencies (Steinert, Schiml, Fauser, & Puchta, 2015; Wolter, Klemm, & Puchta, 2018). In particular, egg-cell specific expression of Cas proteins and the screening for the most efficient transgenic line turned out to be decisive for successful GT (Miki, Zhang, Zeng, Feng, & Zhu, 2018; Wolter et al., 2018). Recently, Cas12a was shown to be an efficient nuclease that also allows the targeting of T-rich sites (Zetsche et al., 2015). With Lb Cas12a, as most efficient ortholog in plants, another increase of GT frequency was accomplished (Wolter & Puchta, 2019). However, a problem for using Lb Cas12a is its temperature sensitivity. The enzyme has highly efficient cutting activities at 37°C but not at 22°C, which is required for plant cultivation. Therefore, our group developed a temperature-tolerant version of Lb Cas12a (tt Lb Cas12a) to enhance cutting efficiencies at lower temperatures (Malzahn et al., 2019; Schindele & Puchta, 2020). Just recently, we were able to demonstrate that tt Lb Cas12a outperformed the native enzyme in ipGT at 22°C by 2.4-fold and at 28°C by 1.7-fold (Merker, Schindele, Huang, Wolter, & Puchta, 2020).
In this protocol, we focus on basic steps of the experimental procedure for using tt Lb Cas12a for ipGT in Arabidopsis thaliana , including the identification of the target site, design of the guide and HR donor sequence, the cloning of the T-DNA, the evaluation of the guide efficiency and the procedure of obtaining heritable GT plants.
Basic Protocol 1: CONSTRUCTION OF CRISPR/ttLbCas12a EXPRESSION VECTOR TO ANALYZE ipGT EFFICIENCIES
In this protocol, we describe the procedure to create tt Lb Cas12a constructs for ipGT in detail (see Fig. 2).
This protocol includes two steps: (1) Preparation and design; (2) Generation of the T-DNA.
Materials
- Aat II for the integration of the HR donor sequence
- Pac I for the integration of the HR donor sequence
- Plasmids (see recipe)
- ddH2O
- Bbs I for the integration of the annealed oligonucleotides
- Ligase for any conventional cloning step
- Escherichia coli , any standard cloning strain (e.g., NEB5α) for all cloning steps except for propagation of the destination vector before gateway cloning, where a ccdB -resistant strain has to be used
- Ampicillin-containing LB plates for colony selection
- Primers for PCR screening (see Table 1)
- LB medium for E. coli
- Plasmid preparation kit
- Gateway LR Clonase II for the transfer of the programmed guide to the binary vector
- TE buffer for Gateway reactions
- Proteinase K to stop Gateway reactions
- Spectinomycin-, rifampicin- and gentamicin-containing YEB plates for colony selection
- Agrobacterium tumefaciens , any standard transformation strain (e.g., GV3101)
- DNA polymerase for E. coli and A. tumefaciens colony PCRs
- dNTPs for E. coli and A. tumefaciens colony PCRs
- YEB medium for A. tumefaciens
- Arabidopsis thaliana plants, wild-type (ecotype Columbia-0) or mutant
| Oligonucleotides | Sequence (5′–3′) |
|---|---|
| M13-fw | TGTAAAACGACGGCCAGT |
| SS42 | TCCCAGGATTAGAATGATTAGG |
| SS102 | CACCATGTTATCACATCAATCC |
Preparation and design
1.Identification of the target site: Search for a potential target site with a protospacer adjacent motif (PAM; 5'-TTTV-guide-3') in the selected target locus in close proximity to the desired modification. Avoid sequences containing a set of five or more Ts as this is the termination sequence for RNA polymerase III.
2.Design of the HR donor: use 600 to 800 bp homologies on both flanks of the cleavage site. For excision of the HR donor, add the gRNA target sequence including the PAM to both ends of the donor sequence. To avoid Cas12a activity on the donor site, include silent mutations either in the PAM (preferred) or in the spacer sequence (nucleotides 4-7 proximal to the PAM are most vulnerable to mismatches). Additionally, add restriction sites (Aat II and Pac I) on the end of the HR donor, as they are required for inserting the HR donor sequence into your T-DNA (see Critical Parameters).
3.Generate your HR donor sequence: methods to generate your HR donor are overlap extension PCRs, Gibson assembly, site-directed mutagenesis PCRs, or gene synthesis.
Generation of the T-DNA: creating the entry vector
4.Order oligonucleotides for your target sequence. For tt Lb Cas12a the sequence should be 23-25 nt downstream of the PAM. Add “AGAT” at the 5′ end on the forward oligonucleotide and “GGCC” at the 5′ end on the reverse-complement oligonucleotide to ensure spacer integration into the gRNA expression cassette.
5.Dilute the oligonucleotides in ddH2O to a final concentration of 2 pmol/µl for each oligonucleotide in a total volume of 50 µl. Mix and incubate for 5 min at 95°C using a thermal cycler. Incubate another 20 min at room temperature for annealing of the oligonucleotides.
6.Digest the entry vector with Bbs I to prepare it for the integration of the annealed oligonucleotides. Purify the digested vector (e.g., by using a common DNA purification Kit) and dilute to a final concentration of 5 ng/µl.
7.Set up a ligation reaction (see a-e) and incubate as recommended by the manufacturer information.
-
2 µl digested entry vector
-
3 µl annealed oligonucleotides
-
1 µl T4 ligase buffer
-
1 µl T4 ligase
-
3 µl ddH2O
8.Transform the ligation product into E. coli for amplification.
9.Select for colonies on ampicillin-containing LB plates. Screen colonies by colony PCR using M13-fw and the reverse-complement oligonucleotide as primers (see Table 1).
10.Cultivate transformed colonies in liquid LB-amp medium.
11.Purify the plasmids (e.g., by using a common plasmid preparation kit) and validate them by sequencing with primer SS42 (see Table 1).
Generation of the T-DNA: creating the expression vector
12.Set up a reaction (see a-d) to transfer the validated gRNA expression cassette to pDe-EC-tt Lb Cas12 and pDe-ttLbCas12a by Gateway cloning in a total volume of 10 µl and incubate for 2 to 3 hr at room temperature. pDe-EC-tt Lb Cas12a is for the GT construct. pDe-tt Lb Cas12a is for evaluating the cleavage activity within the chosen target.
-
200 ng entry vector
-
150 ng destination vector
-
1 µl LR Clonase II
-
Fill up to 10 µl with TE buffer
13.Stop the Gateway reaction after incubation by adding 1 µl proteinase K and incubate 10 min at 37°C using a heating block.
14.Transform the Gateway reaction into E. coli for amplification.
15.Select for colonies on spectinomycin-containing LB plates. Screen for correct colonies by colony PCR using SS42/SS102 as primers (see Table 1).
16.Cultivate transformed colonies in liquid LB-spec medium.
17.Purify the plasmids and validate them by a restriction digestion.
18.Insert your HR donor sequence into the pDe-EC-tt Lb Cas12a vector via a restriction digestion with Aat II and Pac I.
19.Screen for correct clones by a suitable colony PCR and restriction digestion.
20.Validate the correctness of your final construct by full sequencing.
21.Transform your correct construct into A. tumefaciens (Wirth, Friesenegger, & Fiedler, 1989).
22.Select for colonies on spectinomycin-, rifampicin- and gentamicin-containing YEB plates. Validate the positive colonies by colony PCR using SS42/SS102.
23.Cultivate one correct colony in liquid YEB-spec-rif-gent medium.
24.Transform the final construct in A. thaliana via the floral dip method (Clough & Bent, 1998).
25.Harvest the T1 seeds after maturation.
Basic Protocol 2: ACHIEVING HERITABLE GT PLANTS
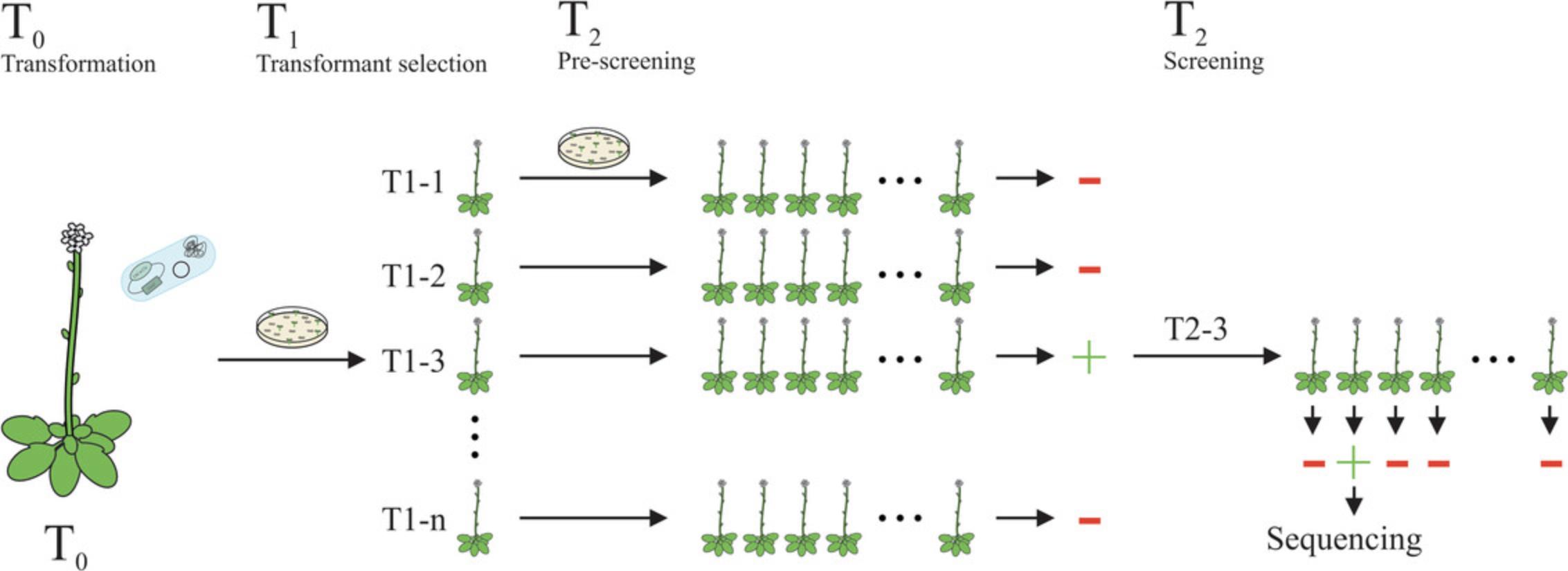
In this protocol, we describe the procedure to obtain heritable GT plants following A. thaliana transformation (see Fig. 2).
This protocol includes two steps: (1) Generating primary transformants; (2) Obtaining heritable GT plants.
Materials
-
T1 seeds (see Basic Protocol 1)
-
DNA polymerase for screening of GT plants
-
dNTPs for screening of GT plants
-
Gentamicin-containing GM plates for selecting transgenic plants
-
Additional reagents and equipment for extracting DNA via the rapid DNA extraction method (see Edwards, Johnstone, & Thompson, 1991)
Generating primary transformants
1.Sow seeds of the transformation on GM medium containing gentamicin to select for transgenic plants harboring the T-DNA.
2.Let the seeds maturate for 2 weeks at 22°C in a growth chamber.
3.For GT: Pick at least 40 T1 plants containing pDe-EC-tt Lb Cas12a and transfer them to soil until maturity.
4.For cleavage activity: Extract DNA from 20 primary transformants containing the pDe-tt Lb Cas12a construct via the rapid DNA extraction method (see Edwards et al., 1991). Analyze the mutagenesis efficiency by e.g., TIDE (see Critical Parameters).
5.Extract DNA from one leaf of each plant and set up a suitable PCR to verify the presence of your construct.
Obtaining heritable GT plants
6.Harvest the seeds of each T1 plant separately.
7.Sow about 100 seeds per line on GM medium.
8.Let the seeds maturate for 2 weeks at 22°C.
9.Extract DNA from one leaf of 100 plants per line as a pool.
10.Screen for positive GT events using a suitable PCR (see Critical Parameters).
11.Extract DNA from each plant of the positive identified T1 pools separately after another week of growth.
12.Analyze the T2 plants separately for heritable GT via PCR and confirm it by sequencing (see Critical Parameters).
13.Transfer positive T2 plants to soil and cultivate them to maturity.
14.Harvest seeds from each positive T2 line separately.
15.Sow T3 seeds on GM medium.
16.Screen for loss of the T-DNA via PCR.
17.Confirm successful GT events to ensure stable inheritance.
REAGENTS AND SOLUTIONS
Plasmids
The plasmids are all freely available directly from the authors. The full sequence information is available at www.botanik.kit.edu/crispr.
pDe-EC-tt Lb Cas12a is the binary vector for a stable Agrobacterium tumefaciens -mediated A. thaliana transformation. The binary vector confers spectinomycin resistance to bacteria and gentamicin resistance to plants. It contains the A. thaliana codon-optimized tt Lb Cas12a ORF under the control of an egg-cell specific promoter (EC1.1/1.2 promoter). It is also a Gateway destination vector with a ccdB gene flanked by respective attachment sites for transferring the gRNA expression cassette to the binary vector. Restriction sites for inserting the HR donor sequence are located within the multiple cloning site.
pEn-RZ-Lb -Chimera is a Gateway entry vector containing an empty gRNA. Via TypIIS restriction enzymes, annealed oligonucleotides can be integrated to create a programmed gRNA. The gRNA is under the control of the Arabidopsis U6-26 promoter and flanked by the hammerhead (5′) and hepatitis delta virus (3′) ribozyme. The expression cassette is flanked by respective Gateway attachment sites to enable the transfer of the gRNA into the destination vector. The entry vector confers ampicillin resistance to bacteria.
pDe-tt Lb Cas12a is identical to pDe-EC-tt Lb Cas12a except for the expression of tt Lb Cas12a, which is controlled by the constitutive PcUBI 4-2 promoter.
COMMENTARY
Critical Parameters
For a successful application of ipGT, the design should be well considered. The most crucial point is the design of the donor sequence. The length of the homologies can vary, and larger homologies can increase HR frequencies. We recommend at least a size of 600 bp of homology for each end. Special attention is required for inserting silent mutations to avoid cleavage within the donor molecule. Here, the amino acid sequence must remain unchanged, but the silent mutation must be well-chosen to ensure tt Lb Cas12a is not able to induce a DSB. We recommend a modification of the PAM sequence as one mutation is usually enough to guarantee no cleavage. For choosing a suitable mutation in the PAM sequence, we recommend checking Gao et al. (2017).
To optimize the experimental success, we suggest testing cleavage activity first. This can be done by the determination of the efficiency of mutation induction by NHEJ at the respective site. For this, we recommend using TIDE analysis to find an optimal guide RNA. By using more efficient guides, HR frequencies can be increased.
For the screening of GT events via PCR, suitable primers should be selected. One primer should bind outside of the homology region and the other should only bind to the template mutation. This way, amplification of the T-DNA donor template is prevented and only the amplification of true positive GT events is feasible. Since this is a critical key point in applying ipGT, proper analysis of suitable primers and cycle numbers should be performed.
Troubleshooting
If problems occur in one of the steps mentioned above, the troubleshooting guide might help (see Table 2).
| Problem | Possible cause | Solution | |
|---|---|---|---|
| Generation of the T-DNA: creating the entry vector | No colonies | Restriction enzyme did not cleave | Increase incubation time of the digestion Use a larger digestion volume |
| Ligation did not work | Increase incubation time of the ligation Use a larger ligation volume Adjust the incubation temperature |
||
| Wrong selection media | Ensure using ampicillin-containing LB medium | ||
| No positive colonies | Mixture of digested and undigested plasmid | See “Restriction enzyme did not cleave” Purify digested plasmids via gel and repeat ligation |
|
| Generation of the T-DNA: creating the expression vector | No colonies | Gateway reaction did not work | Increase incubation time of the gateway reaction Ensure the correct quantity of used constructs Validate the sequence of the attachment sites of the entry and destination vector via sequencing |
| No positive colonies | Utilized wrong E. coli strain | Ensure not to use the ccdB-resistant strain | |
| Generating primary transformants | No primary transformants | A. thaliana transformation did not work | Ensure to follow Clough & Bent (1998) Check the OD before transformation |
| Wrong selection media | Ensure using gentamicin-containing GM medium for selection | ||
| No cleavage activity | Poor spacer design | Ensure that the target locus contains the PAM sequence Ensure that the spacer sequence does not contain the PAM sequence Ensure the spacer is not containing a set of Ts |
|
| Low cleavage activity | Weak spacer | Test other gRNA target sequences | |
| Obtaining heritable GT plants | No GT event | Weak spacer | Test other gRNA target sequences |
| Cleaved donor sequence | Ensure to integrate silent mutations in the donor sequence to completely avoid cleavage | ||
| No released donor sequence | Ensure that the PAM sequence is located at the ends of the donor sequence Ensure that the gRNA target sequence is located at the end of the donor sequence |
||
| Low number of positive lines | Scale up the number of analyzed plants |
Understanding Results
GT efficiencies should be presented as the average of positive GT lines in ratio to the total number of analyzed lines. The ipGT system was successfully applied with this protocol and the average GT efficiency that was accomplished using tt Lb Cas12a is 1.34%.
Time Considerations
Before starting the experiments, we recommend sufficient time for preparation and design. The general time required for performing the ipGT experiment itself to obtain a targeted homozygous Arabidopsis line is approximately 6 to 9 months.
Acknowledgments
Open access funding enabled and organized by Projekt DEAL. This work was funded by the EU Horizon 2020 Project 760331-2 “NEWCOTIANA.”
Author Contributions
Laura Merker : Visualization; writing-original draft; writing-review & editing. Patrick Schindele : Writing-review & editing. Holger Puchta : Writing-review & editing.
Literature Cited
- Clough, S. J., & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal , 16, 735–743. doi: 10.1046/j.1365-313×.1998.00343.x.
- Edwards, K., Johnstone, C., & Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research , 19, 1349. doi: 10.1093/nar/19.6.1349.
- Fauser, F., Roth, N., Pacher, M., Ilg, G., Sánchez-Fernández, R., Biesgen, C., & Puchta, H. (2012). In planta gene targeting. Proceedings of the National Academy of Sciences of the United States of America , 109, 7535–7540. doi: 10.1073/pnas.1202191109.
- Gao, L., Cox, D. B. T., Yan, W. X., Manteiga, J. C., Schneider, M. W., Yamano, T., … Zhang, F. (2017). Engineered Cpf1 variants with altered PAM specificities. Nature Biotechnology , 35, 789–792. doi: 10.1038/nbt.3900.
- Huang, T.-K., & Puchta, H. (2019). CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Reports , 38, 443–453. doi: 10.1007/s00299-019-02379-0.
- Malzahn, A. A., Tang, X., Lee, K., Ren, Q., Sretenovic, S., Zhang, Y., … Qi, Y. (2019). Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biology , 17, 9. doi: 10.1186/s12915-019-0629-5.
- Merker, L., Schindele, P., Huang, T.-K., Wolter, F., & Puchta, H. (2020). Enhancing in planta gene targeting efficiencies in Arabidopsis using temperature-tolerant CRISPR/LbCas12a. Plant Biotechnology Journal , doi: 10.1111/pbi.13426.
- Miki, D., Zhang, W., Zeng, W., Feng, Z., & Zhu, J.-K. (2018). CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nature Communications , 9, 1967. doi: 10.1038/s41467-018-04416-0.
- Puchta, H., Dujon, B., & Hohn, B. (1996). Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America , 93, 5055–5060. doi: 10.1073/pnas.93.10.5055.
- Schiml, S., Fauser, F., & Puchta, H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. The Plant Journal , 80, 1139–1150. doi: 10.1111/tpj.12704.
- Schindele, A., Dorn, A., & Puchta, H. (2020). CRISPR/Cas brings plant biology and breeding into the fast lane. Current Opinion in Biotechnology , 61, 7–14. doi: 10.1016/j.copbio.2019.08.006.
- Schindele, P., & Puchta, H. (2020). Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnology Journal , 18, 1118–1120. doi: 10.1111/pbi.13275.
- Steinert, J., Schiml, S., Fauser, F., & Puchta, H. (2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. The Plant Journal , 84, 1295–1305. doi: 10.1111/tpj.13078.
- Wirth, R., Friesenegger, A., & Fiedler, S. (1989). Transformation of various species of gram-negative bacteria belonging to 11 different genera by electroporation. Molecular & General Genetics MGG, 216, 175–177.
- Wolter, F., Klemm, J., & Puchta, H. (2018). Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. The Plant Journal , 94, 735–746. doi: 10.1111/tpj.13893.
- Wolter, F., & Puchta, H. (2019). In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. The Plant Journal , 100, 1083–1094. doi: 10.1111/tpj.14488.
- Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., … Zhang, F. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell , 163, 759–771. doi: 10.1016/j.cell.2015.09.038.
Citing Literature
Number of times cited according to CrossRef: 8
- Jing Li, Dali Kong, Yongping Ke, Wenjie Zeng, Daisuke Miki, Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis, BMC Biology, 10.1186/s12915-024-01810-7, 22 , 1, (2024).
- Toshisangba Longkumer, Louis Grillet, Hao‐Yi Chang, Tài Chiến Lường, Chih‐Yun Chen, Hadi Putra, Wolfgang Schmidt, Paul E. Verslues, Insertion of YFP at P5CS1 and AFL1 shows the potential, and potential complications, of gene tagging for functional analyses of stress‐related proteins, Plant, Cell & EnvironmentPlant, Cell & EnvironmentPlant, Cell & Environment, 10.1111/pce.14861, 47 , 6, (2011-2026), (2024).
- Long T. Nguyen, Nicolas C. Macaluso, Noah R. Rakestraw, Dylan R. Carman, Brianna L.M. Pizzano, Raymond C. Hautamaki, Santosh R. Rananaware, Isabel E. Roberts, Piyush K. Jain, Harnessing noncanonical crRNAs to improve functionality of Cas12a orthologs, Cell Reports, 10.1016/j.celrep.2024.113777, 43 , 2, (113777), (2024).
- Zhengjing Zhang, Wenjie Zeng, Wenxin Zhang, Jing Li, Dali Kong, Lei Zhang, Rui Wang, Fangnan Peng, Zhe Kong, Yongping Ke, Heng Zhang, Chanhong Kim, Huiming Zhang, Jose Ramón Botella, Jian-Kang Zhu, Daisuke Miki, Insights into the molecular mechanisms of CRISPR/Cas9-mediated gene targeting at multiple loci in Arabidopsis, Plant Physiology, 10.1093/plphys/kiac431, 190 , 4, (2203-2216), (2022).
- Michelle Rönspies, Patrick Schindele, Rebecca Wetzel, Holger Puchta, CRISPR–Cas9-mediated chromosome engineering in Arabidopsis thaliana, Nature Protocols, 10.1038/s41596-022-00686-7, 17 , 5, (1332-1358), (2022).
- Md Mahmudul Hassan, Guoliang Yuan, Yang Liu, Mobashwer Alam, Carrie A. Eckert, Gerald A. Tuskan, John F. Golz, Xiaohan Yang, Precision genome editing in plants using gene targeting and prime editing: existing and emerging strategies, Biotechnology Journal, 10.1002/biot.202100673, 17 , 10, (2022).
- Anis Meschichi, Mathieu Ingouff, Claire Picart, Marie Mirouze, Sophie Desset, Franck Gallardo, Kerstin Bystricky, Nathalie Picault, Stefanie Rosa, Frédéric Pontvianne, ANCHOR: A Technical Approach to Monitor Single-Copy Locus Localization in Planta, Frontiers in Plant Science, 10.3389/fpls.2021.677849, 12 , (2021).
- Daisuke Miki, Rui Wang, Jing Li, Dali Kong, Lei Zhang, Jian-Kang Zhu, Gene Targeting Facilitated by Engineered Sequence-Specific Nucleases: Potential Applications for Crop Improvement, Plant and Cell Physiology, 10.1093/pcp/pcab034, 62 , 5, (752-765), (2021).

