Purification of SRSF1 from E. coli for Biophysical and Biochemical Assays
Zihan Zhang, Zihan Zhang, Jun Zhang, Jun Zhang
intrinsically disordered proteins
NMR
phase separation
phosphorylation
RNA splicing
SR protein
SRSF1
Abstract
The Ser/Arg-rich splicing factors (SR proteins) constitute a crucial protein family in alternative splicing, comprising twelve members characterized by unique repetitive Arg-Ser dipeptide sequences (RS) and one to two RNA-recognition motifs (RRM). The RS regions of SR proteins undergo variable phosphorylation, resulting in unphosphorylated, partially phosphorylated, or hyper-phosphorylated states based on functional requirements. Despite the identification of the SR protein family over 30 years ago, the purification of native SR proteins in soluble form at large quantities has presented challenges due to their low solubility. This protocol delineates a method for acquiring soluble, full-length, unphosphorylated, hypo- and hyper-phosphorylated SRSF1, a prototypical SR family member. Notably, this protocol facilitates the purification of SRSF1 in ample quantities suitable for NMR, as well as various biophysical and biochemical studies. The methodologies and principles outlined herein are expected to extend beyond SRSF1 protein production and can be adapted for purifying other SR protein family members or SR-related proteins, such as snRNP70 and U2AF-35. Given the involvement of these proteins in numerous essential biological processes, this protocol will prove beneficial to researchers in related fields. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Purification of SRSF1 from E. coli
Support Protocol : Purification of ULP1
Basic Protocol 2 : Purification of hypo-phosphorylated SRSF1 from E. coli
Basic Protocol 3 : Purification of hyper-phosphorylated SRSF1 from E. coli
INTRODUCTION
Ser/Arg-rich splicing factors constitute a crucial protein family essential for alternative splicing. Comprising 12 members, this family shares one to two RNA-recognition motifs and repetitive Arg-Ser dipeptide regions (RS) ranging from 50 to 300 amino acids (Anko, 2014; Long & Caceres, 2009; Manley & Krainer, 2010; Pan et al., 2008; Zhou & Fu, 2013). Serine residues in RS regions of SR proteins undergo varying phosphorylation, leading to unphosphorylated, partially phosphorylated, or hyper-phosphorylated states, which is thought to account for functional diversity (Aubol et al., 2018; Gui et al., 1994). The RS region is also found in the much larger family of SR-related proteins, which contain the repetitive RS regions but not the other structural features (Blencowe et al., 1999; Cascarina & Ross, 2022). SR proteins rely on their RRM domains to specifically recognize cis regulatory RNA elements known as exonic splicing enhancers. Meanwhile, SR proteins also interact with U1-70K and U2AF35, recruiting the U1 and U2 spliceosome particles to the splicing sites, respectively (Chabot & Steitz, 1987; Moore et al., 1993; Zillmann et al., 1987). Therefore, binding of SR proteins typically promotes exon inclusion, although their regulatory functions vary depending on the location of cognate RNA for some RNA transcripts (Dembowski et al., 2012; Erkelenz et al., 2013).
SRSF1 (Serine/Arginine-Rich Splicing Factor 1, also known as ASF/SF2) is an archetype member of the SR protein family (Cho et al., 2011; Kohtz et al., 1994). Several in vivo studies have shown that SRSF1 is found in condensates (Azpurua et al., 2021; Fei et al., 2017; Hammarskjold & Rekosh, 2017; Haward et al., 2021; Ilik et al., 2020; Lamond & Spector, 2003; Li & Wang, 2021). Aberrant condensation behaviors have also been observed in disease states (Azpurua et al., 2021; Ilik et al., 2020; Li & Wang, 2021). Like many other splicing factors, SRSF1 modulates trafficking to the speckles (Tripathi et al., 2012). In addition to its essential roles in RNA splicing, SRSF1 is also found to be involved in nonsense-mediated mRNA decay (NMD) (Zhang & Krainer, 2004), nuclear export of mRNA (Huang et al., 2003), miRNA processing (Wu et al., 2010), genome stability (Li & Manley, 2005), and protein SUMOylation (Pelisch et al., 2010).
Despite the identification of the SR protein family over 30 years ago, the purification of native SR proteins in soluble form at large quantities has proven challenging due to their low solubility. This technical impasse has significantly impeded our understanding of functions of SR proteins, such as their RNA-binding specificity for pre-mRNA substrates, protein-interacting mechanism during spliceosome assembly, function modulation by RS phosphorylation, and phase separation behavior (Shepard & Hertel, 2009).
This protocol outlines procedures for obtaining soluble, full-length SRSF1 in the unphosphorylated (Basic Protocol 1), hypo- (Basic Protocol 2) and hyper-phosphorylated (Basic Protocol 3) states. These protocols use E. coli as a host to produce a high yield of proteins. The typical yield of per L of LB medium is 5 mg. Our previous publication has shown that adding the 4-mer ERER peptide solubilizes SRSF1 in different phosphorylation states and a concentration of 0.5 mM can be achieved. The purification procedure consists of three steps, the Ni-NTA affinity chromatography, the ion exchange chromatography, and the size exclusion chromatography. Other than the necessary plasmid encoding SRSF1 and its mutants, the plasmid encoding the CLK1 kinase is also required to produce hypo- and hyper-phosphorylated SRSF1. According to mass spectrometry, 18 to 22 phosphate groups are added to hyper-phosphorylated SRSF1 and 9 to 12 phosphate groups are added to hypo-phosphorylated SRSF1. Notably, this protocol enables the purification of SRSF1 in sufficient quantities for biophysical and structural biology characterization. The methodologies and principles outlined herein are not limited to SRSF1 and can be adapted for the purification of other SR proteins or SR-related proteins.
Basic Protocol 1: PURIFICATION OF SRSF1 FROM E. coli
This protocol describes the purification of unphosphorylated SRSF1 from E. coli. The plasmid encoding human SRSF1 is introduced into BL21-CodonPlus (DE3) competent cells, and the cells are grown in LB or M9 media to produce SRSF1.Following the protocol, pure and soluble unphosphorylated SRSF1 can be obtained with a yield of ∼5 mg per L.
Materials
-
LB medium (see recipe)
-
Buffers and stock solutions (see recipes):
- 5 M NaCl
- Kanamycin stock solution
- Chloramphenicol stock solution
- IPTG stock solution
- PMSF
- 1 M Tris, pH 7.5
- 1 M Arg/Glu with varying pH
- 5 M imidazole, pH 8.0
- 1 M TCEP
- 0.5 M EDTA
- 8 M Urea
- 20% NaN3
- Lysis buffer
- High-salt washing buffer
- Washing buffer
- Ni-NTA elution buffer
- Heparin column buffer A
- Heparin column buffer B
-
70% ethanol
-
pSMT3 plasmid encoding the human SRSF1 (Addgene #201056; see Fig. 1 for the construct boundaries)
-
BL21-CodonPlus (DE3) competent cells (Agilent, cat. no. 230245)
-
Lysozyme (Sigma-Aldrich, cat. no. L-6876)
-
Pierce protease inhibitor mini tablets (Thermo Scientific, cat. no. A32953)
-
Pierce phosphatase inhibitor (Thermo Scientific, cat. no. A32957)
-
H2O, deionized or Milli-Q
-
Protein ladder (Fisher, cat. no. BP3603-1)
-
ULP1 enzyme (see Support Protocol)
-
1 M acetic acid (Fisher, cat. no. A38C-212)
-
2.5-L glass baffled flasks
-
Water bath (Fisher, CPD02 or equivalent)
-
2-, 10-, 20-, 200-, and 1000-μl pipettes and tips
-
Ice
-
Incubator shakers (New Brunswick Excella E25 and New Brunswick I 24, or equivalents)
-
Plate spreader (Fisher, cat. no. 14-665-230)
-
Incubator (VWR)
-
Inoculating loops/needles (Fisher, cat. no. 22-363-595)
-
Nanodrop spectrophotometer (One C) or equivalent
-
2-, 10-, and 25-ml serological pipettes
-
Centrifuges (Avanti JXN-26, Allegra X-30R, and Sorvall Legend X1R, or equivalents)
-
Centrifuge bottles
-
50-ml conical centrifuge tube (Falcon, cat. no. 14-959-49A)
-
Scale
-
1.5-ml low protein binding Eppendorf tubes (ThermoFisher Scientific, cat. no. 90410)
-
Spatula
-
Sonicator (QSONICA Q500)
-
HisPur Ni-NTA resin (Thermo Scientific, cat. no. 88222)
-
5-ml HiTrap heparin HP column (Cytiva)
-
pH meter (Accumet AB150 or equivalent)
-
ÄKTA Start protein purification system (Cytiva) or equivalent
-
HiLoad 16/600 Superdex 75 pg SEC column (Cytiva)
-
Centrifugal filter units (Amicon Ultra-15 or 4)
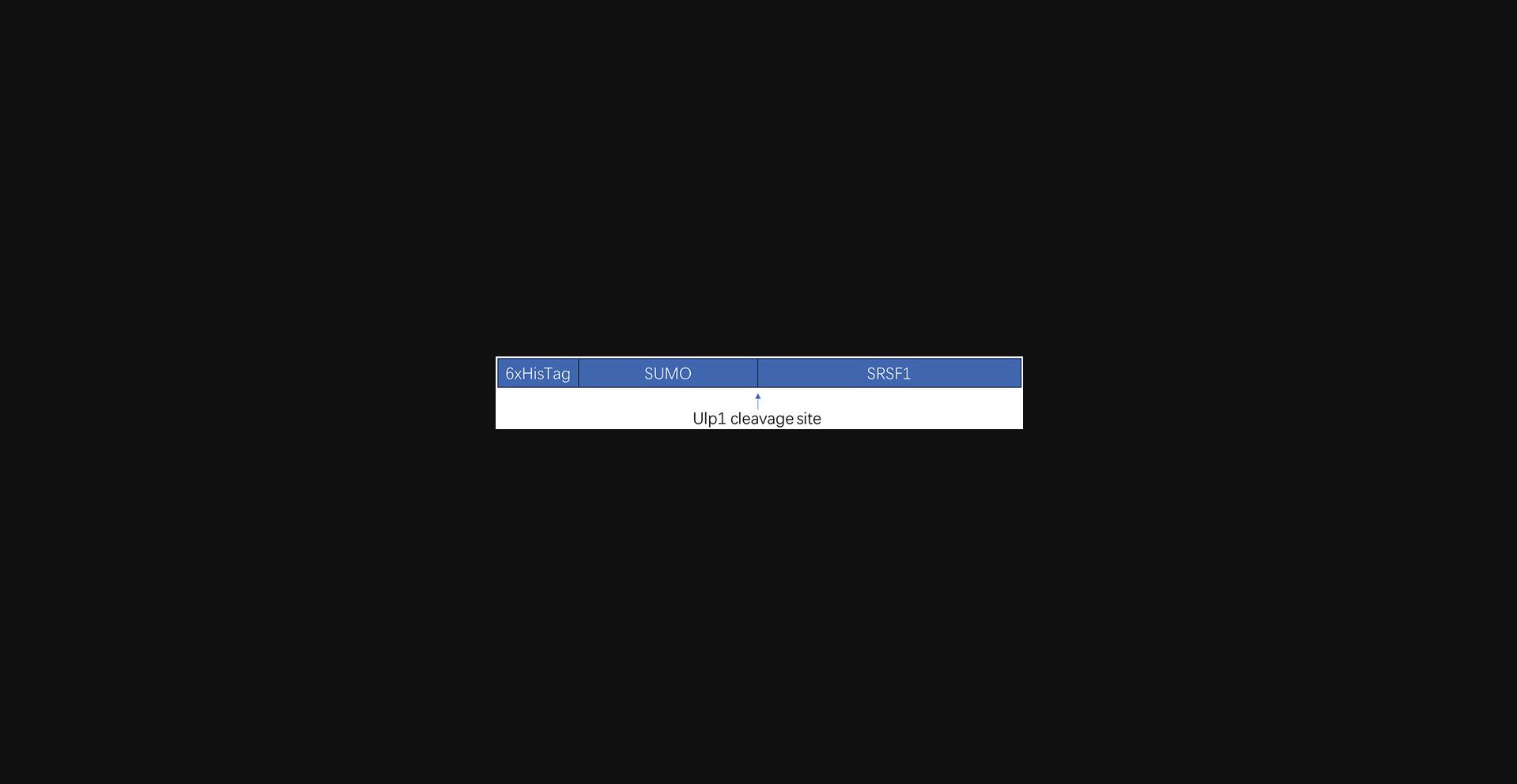
Day 1
Prepare media and buffers
1.Prepare 4 L autoclaved LB medium and a flask of 50 ml LB medium for the starter culture.
2.Prepare filtered buffers. Filter the buffers using vacuum filter units.
Transform the competent cells and spread the plate
3.Set a water bath to 42°C and the New Brunswick I 24 incubator shaker to 37°C. Wipe the working bench and 2- and 200-μl pipettes with 70% ethanol. Thaw the pSMT3 plasmid encoding human SRSF1 on ice.
4.Place a tube of BL21-CodonPlus (DE3) competent cells (20 to 50 μl) and thaw them on ice.
5.Add 1 μl pSMT3 plasmid into the competent cells and incubate the tube on ice for 15 min.
6.Heat shock the competent cells at 42°C for 45 s.
7.Put the cells on ice for 1 min.
8.Add 200 μl LB medium without antibiotics. Incubate the sample in the New Brunswick I 24 incubator shaker at 37°C for 1 hr.
9.Warm an LB agar plate with kanamycin (50 μg/ml) and chloramphenicol (50 μg/ml) in an incubator at 37°C.
10.Spread 100 μl competent cells onto the agar plate.
11.Incubate the agar plate upside down in the incubator at 37°C overnight.
Day 2
Grow and induce the cells
12.Pick colonies from the agar plate with inoculating loops and inoculate 50 ml LB medium for starter culture.
13.Shake the starter culture at 37°C for 2 hr at 220 rpm in the New Brunswick I 24 incubator.
14.Warm 4 L of LB medium supplemented with kanamycin (50 μg/ml) and chloramphenicol (50 μg/ml) in an incubator shaker at 37°C.
15.Add 10 ml starter culture to each flask of LB.
16.Continue to grow the cells at 37°C in the New Brunswick Excella E 25 incubator shaker until the OD600 reaches 0.6 to 0.8.
17.Induce the cells with 0.5 mM IPTG and adjust the shaker temperature to 22°C. Continue the cell culture with shaking at 220 rpm overnight.
Day 3
Harvest the cells
18.Using centrifuge bottles and the Avanti JXN-26 centrifuge, centrifuge the cells 15 min at 3000 × g , 4°C.
19.Remove the supernatant and collect cell pellets into a 50-ml centrifuge tube.
20.Weigh the cell pellet. Re-suspend the cells in 4 ml lysis buffer per gram of cells.
21.Add protease inhibitor tablets to the re-suspended cells.
22.Optional: Freeze/thaw the cells for three cycles.
Day 4
Purify the protein
23.Thaw the cells in a water bath at room temperature and add 1× PMSF immediately before sonication.
24.Sonicate the cells in a room temperature water bath.
25.Centrifuge the cell lysate in the Sorvall Legend X1R centrifuge 30 min at 23,710 × g , 25°C.
26.Prepare a Ni-NTA column during this time. Equilibrate the resin with 50 ml ddH2O, then with 25 ml elution buffer, and finally 25 ml washing buffer.
27.Make an electrophoresis sample for supernatant by mixing 5 μl cell lysate with 20 μl of 4× protein loading buffer and 55 μl water.
28.Load the supernatant to the equilibrated the Ni-NTA column at a flow rate of 2 to 3 ml/min. Make an electrophoresis sample for the flowthrough fraction by mixing 5 μl of the flowthrough sample with 20 μl of 4× protein loading buffer and 55 μl water.
29.Wash the resin with 200 ml high-salt washing buffer.
30.Wash the resin with 100 ml washing buffer.
31.Elute the sample with 30 ml elution buffer. Make an electrophoresis sample for the eluted fraction by mixing 20 μl eluted sample with 20 μl of 4× protein loading buffer and 40 μl water.
32.Add 1 mM PMSF to the eluted sample. Add 0.01 mg/ml of ULP1 to the eluted sample to cleave off the His-Sumo tag at 37°C for >1 hr. Make an electrophoresis sample by mixing 20 μl sample with 20 μl 4× protein loading buffer and 40 μl water.
33.Dilute the sample 3-fold with heparin column buffer A.
34.Elute the sample with a 100-ml gradient from 0% buffer B to 100% buffer B and collect fractions of the eluted sample.
35.Prepare electrophoresis samples for all peaks and perform SDS-PAGE (Fig. 2).
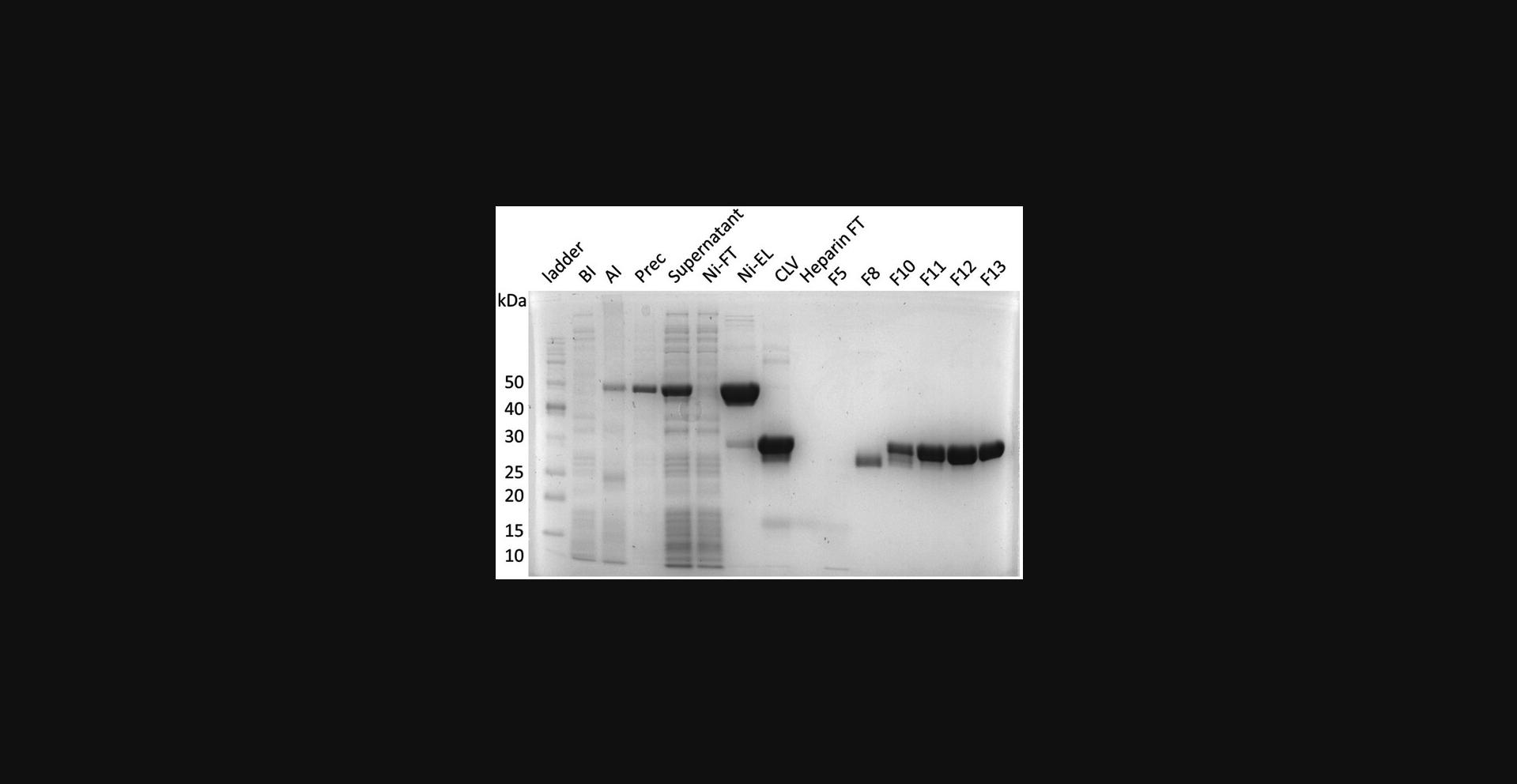
36.Concentrate protein to 4 to 5 mg/ml and load to a SEC column.
37.Concentrate the protein to a desired concentration using a centrifugal filter unit and aliquot to a suitable volume depending on your need. Freeze the samples at −80°C.
Support Protocol: PURIFICATION OF ULP1
This optional protocol introduces how to purify ULP1 that is used in the Basic Protocols to cleave the His-Sumo tag. Only Ni-NTA affinity chromatography is needed to obtain the ULP1 protease.
Additional Materials (also see Basic Protocol 1)
- pET28b plasmid DNA encoding the yeast ULP1
Day 1
Prepare media and buffers
1.Prepare 4 L of autoclaved LB medium and a flask of 50 ml LB medium for starter culture.
2.Prepare the buffers for ULP1 purification.
Transform the competent cells and spread the plate
3.Set a water to 42°C, and the New Brunswick I 24 incubator shaker to 37°C. Wipe the working bench, a 2-μl and a 200-μl pipettes with 70% ethanol.
4.Thaw a tube of competent cells (20 to 40 μl) and a tube of pET28b plasmid encoding ULP1 (pET28b-ULP1).
5.Add 1 μl pET28b-ULP1 to the BL21-CodonPlus (DE3) competent cells and incubate the tube on ice for 15 min.
6.Heat shock the competent cells in 42°C for 45 s.
7.Put the cells on ice for 1 min.
8.Add 200 μl fresh LB medium without antibiotics and incubate the cells in the New Brunswick I 24 incubator shaker at 37°C for 1 hr.
9.Warm an LB agar plate supplemented with kanamycin (50 μg/ml) and chloramphenicol (50 μg/ml) at 37°C.
10.Spread 100 μl of competent cells onto the agar plate.
11.Incubate the agar plate upside down in the incubator at 37°C overnight.
Day 2
Begin early since this day is lengthy.
Grow and induce the cells
12.Inoculate 50 ml LB medium as a starter culture using the colonies from the agar plate.
13.Incubate the starter culture with shaking at 37°C, 220 rpm for 2 hr in the New Brunswick I 24 incubator.
14.Warm 4 L LB medium in an incubator shaker at 37°C.
15.Add 10 ml starter culture to each L of LB supplemented with 50 μg/ml kanamycin and chloramphenicol.
16.Grow 4 L of cells at 37°C in the New Brunswick Excella E25 incubator shaker until OD600 reaches 0.6 to 0.8.
17.Induce the cells with 0.5 mM IPTG and adjust the temperature to 30°C. Continue the growth for 4 hr.
Harvest the cells
18.Centrifuge the cells using the Avanti JXN-26 centrifuge 15 min at 3000 × g , 4°C.
19.Remove the supernatant and collect cell pellets into a 50-ml centrifuge tube.
20.Weigh out the cell pellet. Use 4 ml lysis buffer per gram of cells. Re-suspend the cells in the lysis buffer.
21.Store the cells at −80°C.
Day 3
Purify the protein
22.Optional: Freeze/thaw the cells three times.
23.Add PMSF to 1 mM immediately after the freeze/thaw cycles.
24.Sonicate the cells in an ice-water bath.
25.Centrifuge the cell lysate in the Sorvall Legend X1R centrifuge 30 min at 23,710 × g , 4°C.
26.Prepare a Ni-NTA column during this time. Wash the resin sequentially with 50 ml ddH2O, 25 ml elution buffer, and finally 25 ml washing buffer.
27.Prepare an electrophoresis sample for the supernatant by mixing 5 μl lysate with 20 μl of 4× protein loading buffer and 55 μl water.
28.Load the supernatant to the Ni-NTA column at 4°C at a rate of 2 to 3 ml/min. Prepare an electrophoresis sample for the flowthrough fraction by mixing 5 μl of the flowthrough fraction, 20 μl of 4× protein loading buffer, and 55 μl water.
29.Wash the resin with 400 ml washing buffer.
30.Elute the sample with 40 ml elution buffer. Prepare an electrophoresis sample for the eluted sample by mixing 20 μl of the eluted fraction, 20 μl of 4× protein loading buffer, and 40 μl water.
31.Run the electrophoresis samples and check the purity of ULP1.
32.Measure the UV absorbance at 280 nm for the sample and record the A260/280 ratio.
33.Dilute or concentrate the protein to a concentration of 1 mg/ml at 280 nm.
34.Allocate ULP1 into 200-μl fractions and store at −80°C. ULP1 purified and stored according to this protocol has been tested to be active after 5 years of storage.
Basic Protocol 2: PURIFICATION OF HYPO-PHOSPHORYLATED SRSF1 FROM E. coli
This protocol describes how to purify human hypo-phosphorylated SRSF1 protein. To prevent phosphorylation of the C-terminal part of RS, serine residues after 225 are mutated to alanine to mimic a hypo-phosphorylated state. The purification process and buffer are the same as that for unphosphorylated SRSF1 with minor differences.
Additional Materials (also see Basic Protocol 1)
- pSMT3 plasmid encoding human SRSF1 mutant.
- Serine residues after 225 in SRSF1 are mutated to alanine to mimic a hypo-phosphorylated state. See Figure 3 for the construct.
- pETDuet-1 plasmid DNA encoding human CLK1 kinase (available upon request)
- Ampicillin stock solution (see recipe)
- 100 mM Na3VO4 (see recipe)

Day 1
Prepare media and buffers
1.Prepare 4 L autoclaved LB medium and a flask of 50 ml LB medium for starter culture.
2.Prepare filtered buffers. Filter the buffers using vacuum filter units.
Transform the competent cells and spread the plate
3.Set a water bath to 42°C and the New Brunswick I 24 incubator shaker to 37°C. Wipe the working bench and 2- and 200-μl pipettes with 70% ethanol. Thaw the pSMT3 plasmid encoding human SRSF1 mutant and pETDuet-1 plasmid DNA encoding the human CLK1 kinase on ice.
4.Place a tube of BL21-CodonPlus (DE3) competent cells (20 to 50 μl) and thaw them on ice.
5.Add 1 μl of each plasmid into the competent cells and incubate the tube on ice for 15 min.
6.Heat shock the competent cells at 42°C for 45 s.
7.Put the cells on ice for 1 min.
8.Add 200 μl LB medium without antibiotics. Incubate the sample in the New Brunswick I 24 incubator shaker at 37°C for 1 hr.
9.Warm up an LB agar plate supplemented with kanamycin (50 μg/ml), chloramphenicol (50 μg/ml), and ampicillin (100 μg/ml) in an incubator at 37°C.
10.Spread 100 μl of competent cells onto the agar plate.
11.Incubate the agar plate upside down in the incubator at 37°C overnight.
Day 2
Grow and induce the cells
12.Pick colonies from the agar plate with inoculating loops and inoculate 50 ml LB medium supplemented with 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 100 μg/ml ampicillin for a starter culture.
13.Shake the starter culture at 37°C for 2 hr at 220 rpm in the New Brunswick I 24 incubator.
14.Warm 4 L LB medium supplemented with kanamycin (50 μg/ml), chloramphenicol (50 μg/ml), and ampicillin (100μg/ml) in an incubator shaker at 37°C.
15.Add 10 ml starter culture to each flask of LB.
16.Continue to grow the cells at 37°C in the New Brunswick Excella E25 incubator shaker until OD600 reaches 0.6 to 0.8.
17.Induce the cells with 0.5 mM IPTG and adjust the shaker temperature to 22°C. Continue the cell culture with shaking at 220 rpm overnight.
Day 3
Harvest the cells
18.Centrifuge cells in the Avanti JXN-26 centrifuge 15 min at 3000 × g , 4°C.
19.Remove the supernatant and collect cell pellets into a 50-ml centrifuge tube.
20.Weigh the cell pellet. Re-suspend the cells in lysis buffer with Na3VO4. Use 4 ml lysis buffer per gram of cells.
21.Add protease inhibitor tablets to the re-suspended cells.
22.Optional: Freeze/thaw the cell for three cycles.
Day 4
Purify the protein
23.Thaw the cells in a water bath at room temperature and add PMSF to 1 mM right before sonication.
24.Sonicate the cells in a room temperature water bath.
25.Centrifuge the cell lysate in the Sorvall Legend X1R centrifuge 30 min at 23,710 × g , 25°C.
26.Prepare a Ni-NTA column during this time. Equilibrate the resin with 50 ml ddH2O, then with 25 ml elution buffer, and finally 25 ml washing buffer.
27.Make an electrophoresis sample for the supernatant by mixing 5 μl cell lysate with 20 μl of 4× protein loading buffer and 55 μl water.
28.Load the supernatant to the equilibrated the Ni-NTA column at a flow rate of 2 to 3 ml/min. Make an electrophoresis sample for the flowthrough fraction by mixing 5 μl flowthrough sample with 20 μl of 4× protein loading buffer and 55 μl water.
29.Wash the resin with 200 ml high-salt washing buffer.
30.Wash the resin with 100 ml washing buffer.
31.Elute the sample with 30 ml elution buffer. Make an electrophoresis sample for the eluted fraction by mixing 20 μl eluted sample with 20 μl of 4× protein loading buffer and 40 μl water.
32.Add 1 mM PMSF to the eluted sample. Add 0.01 mg/ml ULP1 to the eluted sample to cleave off the His-Sumo tag at 37°C for >1 hr. Make an electrophoresis sample by mixing 20 μl sample with 20 μl 4× protein loading buffer and 40 μl water.
33.Dilute the sample 3-fold with heparin column buffer A.
34.Elute the sample with a 100-ml gradient from 0% buffer B to 100% buffer B and collect fractions of the eluted sample.
35.Prepare electrophoresis samples for all peaks and perform SDS-PAGE (Fig. 4).
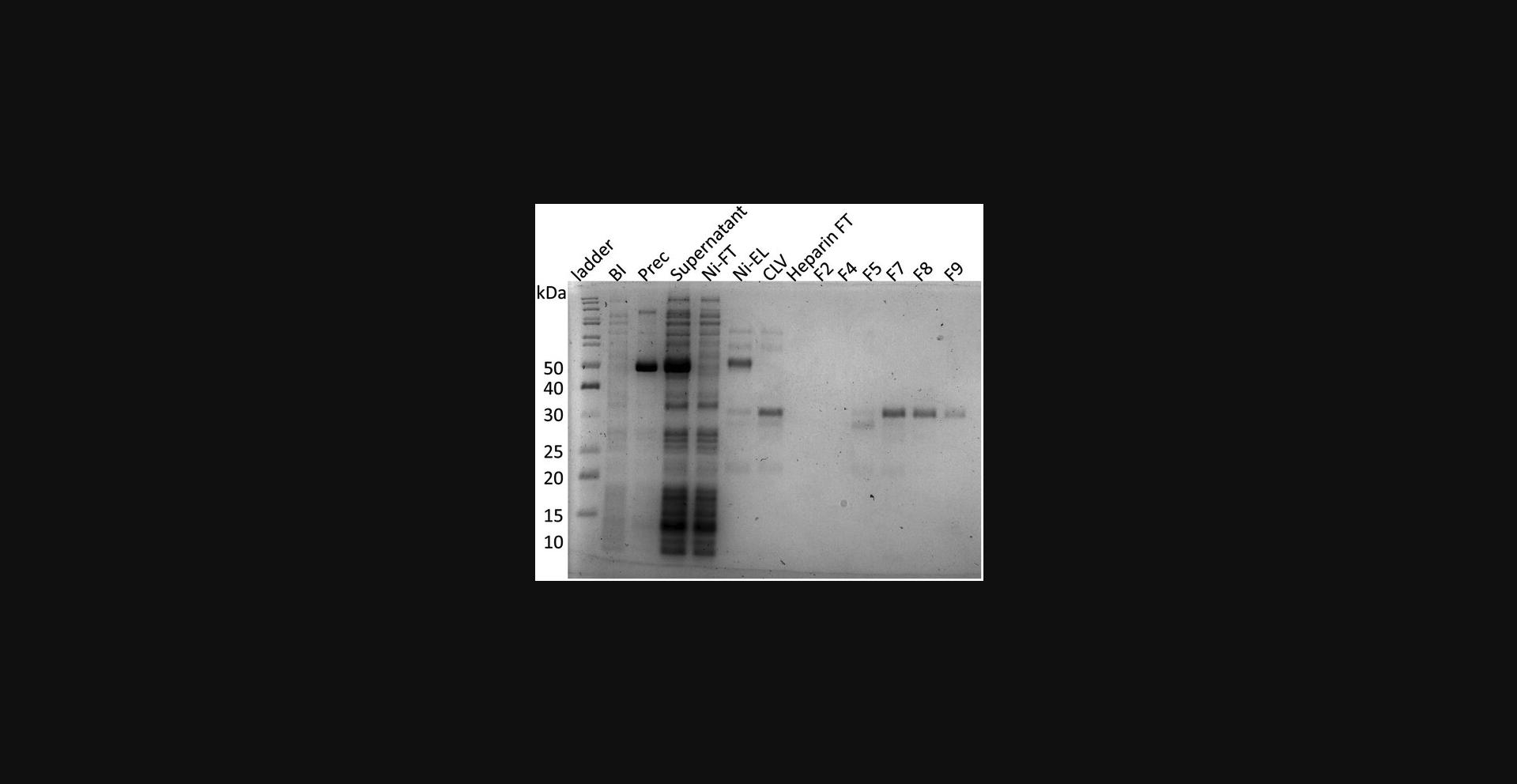
36.Concentrate protein to 4 to 5 mg/ml and load to a SEC column.
37.Concentrate the protein to a desired concentration using a centrifugal filter unit with molecular weight cutoff of 10 kDa and aliquot to a suitable volume depending on your need. Freeze the samples at −80°C.
Basic Protocol 3: PURIFICATION OF HYPER-PHOSPHORYLATED SRSF1 FROM E. coli
This protocol describes how to purify the human hyper-phosphorylated SRSF1 protein. The phosphorylation of SRSF1 is achieved by the CLK1 kinase. To minimize dephosphorylation by endogenous phosphatase after cell lysis, it is not recommended to stop the purification process until the ion exchange column step is completed.
Additional Materials (also see Basic Protocols 1 and 2)
-
pETDuet-1 plasmid encoding the human CLK1 kinase
-
Ampicillin stock solution (see recipe)
-
6 M guanidine HCl (see recipe)
-
Q column buffer A (see recipe)
-
Q column buffer B (see recipe)
-
5-ml HiTrap Q HP column (Cytiva)
NOTE : Steps from Day 1 to Day 3 are similar as that for purification of unphosphorylated SRSF1.
Day 1
Prepare media and buffers
1.Autoclave 4 L LB medium and a flask of 50 ml LB medium for a starter culture.
2.Prepare the buffers for protein purification and filter them using 0.22-μm membranes.
Transform the competent cells and spread the plate
3.Set a water bath to 42°C, and the New Brunswick I 24 incubator shaker to 37°C. Wipe the working bench and 2- and 200-μl pipettes with 70% ethanol.
4.Thaw a tube of BL21-CodonPlus (DE3) competent cells (50 μl), a tube of the pSMT3 plasmid encoding SRSF1, and a tube of the pETDuet-1 plasmid encoding the human CLK1 kinase.
5.Add 1 μl of each plasmid into the competent cells and incubate the tube on ice for 15 min.
6.Heat shock the competent cells in 42°C for 60 s.
7.Incubate the cells on ice for 2 min
8.Add 200 μl fresh LB medium without antibiotics and incubate the cells at 37°C with shaking at 220 rpm for 1 hr in the New Brunswick I 24 incubator.
9.Warm a LB agar plate supplemented with kanamycin (50 μg/ml), chloramphenicol (50 μg/ml), and ampicillin (100 μg/ml) at 37°C.
10.Spread 100 μl cell culture onto the agar plate.
11.Incubate the agar plate upside down at 37°C overnight.
Day 2
Grow and induce the cells
12.Inoculate the 50 ml starter culture supplemented with 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 100 μg/ml ampicillin using the colonies from the agar plate.
13.Incubate the start culture at 37°C with shaking at 220 rpm for 2 hr in the New Brunswick I 24 incubator.
14.Warm 4 L of LB medium at 37°C before transferring the starter culture.
15.Add 10 ml starter culture to each L LB supplemented with 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 100 μg/ml ampicillin.
16.Culture cells at 37°C with shaking at 220 rpm in the New Brunswick Excella E25 incubator until OD600 reaches 0.6 to 0.8.
17.Induce the cells with 0.5 mM IPTG and adjust the temperature to 22°C. Shake overnight.
Day 3
Harvest the cells
18.Centrifuge cells in the Avanti JXN-26 centrifuge 15 min at 3000 × g , 4°C.
19.Remove the supernatant and collect cell pellets into a 50-ml centrifuge tube.
20.Weigh the cell pellet. Empirically, use 4 ml lysis buffer per gram of cells. Resuspend the cells in the lysis buffer.
21.Add protease inhibitor tablets to the resuspended cells.
22.Store the cells at −80°C.
Day 4
Purify the protein
23.Thaw the cells in a water bath at room temperature and add 1 mM PMSF immediately before sonication.
24.Sonicate the cells in an ice-water bath.
25.Centrifuge the cell lysate in the Sorvall Legend X1R centrifuge 40 min at 23,710 × g , 4°C.
26.Prepare 5 ml of Ni-NTA resin by sequentially washing it with 50 ml ddH2O, 25 ml elution buffer, and 25 ml washing buffer.
27.Prepare an electrophoresis sample for the supernatant by mixing 5 μl lysate, 20 μl of 4× protein loading buffer, and 55 μl water.
28.Load the supernatant to the equilibrated Ni-NTA column at 4°C at a rate of 2 to 3 ml/min. Collect the flowthrough. Prepare an electrophoresis sample for the flowthrough by mixing 5 μl flowthrough faction, 20 μl of 4× protein loading buffer, and 55 μl water.
29.Wash the resin with 200 ml high-salt washing buffer.
30.Wash the resin with 100 ml washing buffer.
31.Wash the resin with 20 ml low-salt washing buffer.
32.Elute the sample with 30 ml elution buffer. Prepare an electrophoresis sample for the eluted sample by mixing 20 μl eluted fraction, 20 μl of 4× protein loading buffer, and 40 μl water.
33.Add the PMSF and EDTA stocks to 1 mM to the eluted fraction. These two components inhibit protein degradation. Add 0.01 mg/ml ULP1 to cleave the His-Sumo tag at 37°C for 1 to 2 hr. Prepare an electrophoresis sample for the eluted sample by mixing 20 μl eluted fraction, 20 μl of 4× protein loading buffer, and 40 μl water.
34.Dilute the eluted sample 3-fold with Q column buffer A.
35.Elute the sample with a 100-ml gradient from 0% to 100% Q column buffer B. Add Arg/Glu to 0.4 M to make sure SRSF1 is soluble.
36.Run all the electrophoresis samples and check purity of the sample.
37.Concentrate the protein to 4 to 5 mg/ml using a centrifugal filter unit.
38.Equilibrate a Superdex 75 column into the SEC buffer.
39.Load the concentrated protein to SEC column.
40.Prepare SDS-PAGE samples for the main peaks from the SEC step.
41.Run all SDS-PAGE samples to confirm identity and purity (Fig. 5).
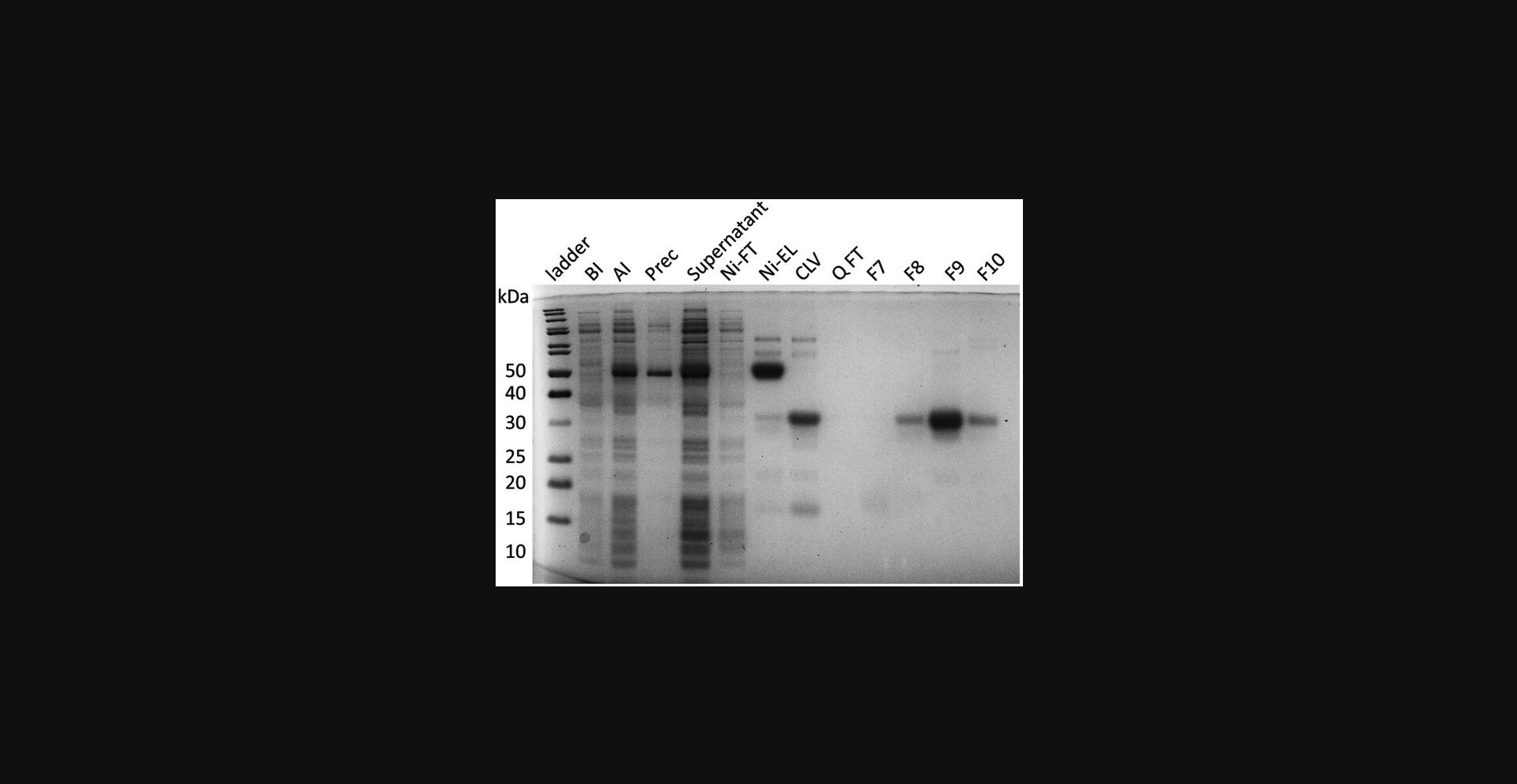
42.Concentrate the protein to a desired concentration. Aliquot and store the samples at −80°C.
REAGENTS AND SOLUTIONS
LB medium, 1 L
- 10 g NaCl (Fisher, cat. no. S271-10)
- 10 g tryptone (Fisher, cat. no. BP1421-2)
- 5 g yeast extract (Fisher, cat. no. BP9727-2)
- Bring volume to 1 L with ddH2O
- Autoclave
- Store up to 2 months at room temperature
Stock solutions for buffer preparation
Ampicillin stock solution, 100 mg/ml, 1000×, 10 ml
- 1 g ampicillin (ACROS cat. no. 61177-0250)
- Dissolve in 10 ml Milli-Q water
- Sterilize using a syringe filter unit
- Store up to 6 months at −20°C
Arg/Glu with varying pH, 1 M, 1 L
- 174.2 g arginine (Thermo scientific cat. no. 104990025)
- Dissolve in 700 ml ddH2O or Milli-Q water
- Add glutamate (ACROS, cat. no. 156210010) to desired pH
- Bring volume to 1 L with ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter
- Store up to 2 months at room temperature or 4°C
Chloramphenicol stock solution, 50 mg/ml, 1000×, 10 ml
- 0.5 g chloramphenicol (ACROS cat. no. AC227920250)
- Dissolve in 10 ml Milli-Q water
- Sterilize using a syringe filter unit
- Store up to 6 months at 4°C
EDTA, 0.5 M, 10 ml
- 1.86 g EDTA disodium (Fisher, cat. no. BP120-1)
- Dissolve in 10 ml of Milli-Q water
- Adjust pH to 8.0 with NaOH
- Filter using a 0.22-μm membrane filter
- Autoclave
- Store up to 1 year at room temperature or 4°C
Guanidine HCl, 6 M, 100 ml
- 57.3 g guanidine hydrochloride (Thermo Fisher Scientific, cat. no. A13543.0I)
- Dissolve in 100 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter
- Store at up to 1 year at room temperature
Imidazole pH 8.0, 5 M, 100 ml
- 34.04 g imidazole (ACROS, cat. no. 122020020)
- Dissolve in 70 ml ddH2O or Milli-Q water
- Adjust pH to 8.0 with HCl
- Bring volume to 100 ml with ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter
- Store up to 1 year at room temperature or 4°C
IPTG stock solution, 1 M, 2000×, 10 ml
- 2.383 g IPTG (Fisher, cat. no. BP1755-100)
- Dissolve in 10 ml Milli-Q water
- Sterilize using a syringe filter unit
- Store up to 6 months at −20°C
Kanamycin stock solution, 50 mg/ml, 1000×, 10 ml
- 0.5 g kanamycin (Fisher, cat. no. 50-213-386)
- Dissolve in 10 ml Milli-Q water
- Sterilize using a syringe filter unit
- Store up to 1 year at 4°C
NaCl, 5 M, 1 L
- 292.2 g NaCl (Fisher, cat. no. S271-10)
- Dissolve in 1 L ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm membrane filter
- Store up to 1 year at room temperature or 4°C
NaN3, 20% (w/v), 50 ml
- 10 g NaN3 (Fisher, cat. no. BP922I-500)
- Dissolve in 50 ml ddH2O or Milli-Q water
- Store up to 1 year at room temperature
Na3VO4, 100 mM, 100 ml
- 3.68 g Na3VO4 (Alfa Aesar, cat. no. 81104)
- Dissolve in 100 ml ddH2O or Milli-Q water
- Adjust to pH 10.0, then boil until colorless
- Cool down to room temperature
- Repeat pH-adjusting, boiling, and cooling several cycles until the pH remains stable at 10
- Aliquot and store up to 1 year at −20°C
PMSF, 100 mM, 100×, 10 ml
- 1.742 g PMSF (Sigma-Aldrich, cat. no. P-7626)
- Add anhydrous ethanol to 10 ml
- Store up to 6 months at −20°C
TCEP, 1 M, 10 ml
- 2.5 g TCEP (Biosynth, cat. no. FT01756)
- Dissolve in 10 ml ddH2O or Milli-Q water
- Sterilize by filtration using a 0.22-μm syringe filter
- Aliquot into 1 ml fractions
- Store up to 2 months at −20°C
Tris·HCl pH 7.5, 1 M, 1 L
- 121.14 g Tris (Fisher, cat. no. BP152-1)
- Dissolve in 1 L ddH2O or Milli-Q water
- Adjust pH to 7.5 with HCl
- Sterilize by filtration using a 0.22-μm membrane filter
- Store up to 1 year at room temperature or 4°C
Urea, 8 M, 100 ml
- 48.48 g urea (Thermo Scientific, cat. no. 036429.A4)
- Dissolve in 100 ml ddH2O or Milli-Q water
- Use heat to aid in dissolving
- Sterilize by filtration using a 0.22-μm membrane filter
- Store at up to 1 year at room temperature
Purification of un- and hypo-phosphorylated SRSF1 (Basic Protocols 1 and 2, respectively)
Heparin column buffer A, 500 ml
- 50 ml of 1 M Arg/Glu pH 7.5 (100 mM)
- 0.5 ml of 20% NaN3 (0.02% w/v)
- 0.5 ml of 1 M TCEP (1 mM)
- Adjust to pH 4.6 using acetic acid
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Heparin column buffer B, 500 ml
- 34.84 g arginine (400 mM)
- 100 ml 5 M NaCl (1 M)
- 0.5 ml 20% NaN3 (0.02% w/v)
- 0.5 ml 1 M TCEP (1 mM)
- Adjust to pH 11.5 using NaOH
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
High-salt washing buffer, 200 ml
- 25 ml of 8 M Urea
- 1 ml of 5 M imidazole pH 8.0
- 0.2 ml of 1 M TCEP
- Bring volume to 200 ml with 5 M NaCl
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Lysis buffer, 200 ml
- 4 ml of 1 M Tris·HCl pH 7.5 (20 mM)
- 30 ml of 1 M Arg/Glu pH 7.5 (150 mM)
- 1 ml of 5 M imidazole pH 8.0 (25 mM)
- 80 ml of 5 M NaCl (2 M)
- 0.2 ml of 1 M TCEP (1 mM)
- 200 mg lysozyme (Sigma-Aldrich, cat. no. L-6876) (1 mg/ml)
- Bring volume up to 200 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
- For hypo-phosphorylated SRSF1, add 0.2 ml of 100 mM Na3VO4 (0.1 mM)
Ni-NTA elution buffer, 100 ml
- 50 ml of 1 M Arg/Glu pH 6.5 (500 mM)
- 5 ml of 5 M imidazole pH 8.0 (250 mM)
- 10 μl of 1 M TCEP (1 mM)
- Adjust to pH 6.5 using HCl or NaOH
- Bring volume to 100 ml with ddH2O or Milli-Q water
- 1 tablet of protease inhibitor cocktail
- Prepare fresh
pH-adjusting buffer
- For 11 ml assuming 20 ml pooled peaks fractions:
- 5 ml of 1 M Arg/Glu pH 6.5 (25% volume)
- 6 ml of 1 M acetic acid (30% volume)
- Store up to 2 months at 4°C
SEC buffer, 500 ml
- 200 ml of 1 M Arg/Glu pH 7.5 (0.4 M)
- 0.1 ml of 1 M TCEP (0.2 mM)
- Adjust to pH 7.5 using HCl or NaOH
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Washing buffer, 100 ml
- 2 ml of 1 M Tris·HCl pH 7.5 (20 mM)
- 0.5 ml of 5 M imidazole pH 8.0 (25 mM)
- 20 ml of 5 M NaCl (1 M)
- 0.1 ml of 1 M TCEP (1 mM)
- Bring volume to 100 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Purification of hyper-phosphorylated SRSF1 (Basic Protocol 3)
High-salt washing buffer, 200 ml
- 33 ml of 6 M guanidine HCl
- 1 ml of 5 M imidazole pH 8.0
- 40 μl of 1 M TCEP
- Bring volume to 200 ml with 5 M NaCl
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Low-salt washing buffer, 20 ml
- 0.4 ml of 1 M Tris·HCl pH 7.5 (20 mM)
- 0.1 ml of 5 M imidazole pH 8.0 (25 mM)
- 5 μl of 1 M TCEP (0.2 mM)
- Adjust to pH 8.0 using HCl or NaOH
- Bring volume to 20 ml with ddH2O or Milli-Q water
- Store up to 2 months at 4°C
Lysis buffer, 200 ml
- 4 ml of 1 M Tris·HCl pH 7.5 (20 mM)
- 20 ml of 1 M Arg/Glu pH 7.5 (100 mM)
- 1 ml of 5 M imidazole pH 8.0 (25 mM)
- 80 ml of 5 M NaCl (2 M)
- 0.2 ml of 1 M TCEP (1 mM)
- 100 mg lysozyme (Sigma-Aldrich, cat. no. L-6876) (0.5 mg/ml)
- 2 ml of 100 mM Na3VO4 (1 mM)
- Adjust to pH 8.5 using HCl or NaOH
- Bring volume to 200 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Ni-NTA elution buffer, 30 ml
- 7.5 ml of 1 M Arg/Glu pH 8.5 (250 mM)
- 3 ml of 5 M Imidazole pH 8.0 (500 mM)
- 0.3 ml of 100 mM Na3VO4 (1 mM)
- 30 μl of 1 M TCEP (1 mM)
- Adjust to pH 8.5
- Bring volume to 30 ml with ddH2O or Milli-Q water
- 1 protease inhibitor cocktail
- Prepare fresh
Q column buffer A, 500 ml
- 4.355 g arginine (50 mM)
- 1 ml of 0.5 M EDTA (1 mM)
- 0.5 ml of 20% NaN3 (0.02% w/v)
- 200 μl of 1 M TCEP (0.4 mM)
- Adjust to pH 9.5 with glutamic acid
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Q column buffer B, 500 ml
- 50 ml of 1 M Arg/Glu pH 8.5 (100 mM)
- 100 ml of 5 M NaCl (1 M)
- 1 ml of 0.5 M EDTA (1 mM)
- 0.5 ml of 20% NaN3 (0.2% w/v)
- 200 μl of 1 M TCEP (0.4 mM)
- Adjust to pH 8.5 using HCl or NaOH
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
SEC buffer, 500 ml
- 400 ml of 1 M Arg/Glu pH 7.5 (0.8 M)
- 0.1 ml of 1 M TCEP (0.2 mM)
- Adjust to pH 7.5 using HCl or NaOH
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Washing buffer, 100 ml
- 2 ml of 1 M Tris·HCl pH 7.5 (20 mM)
- 0.5 ml of 5 M imidazole pH 8.0 (25 mM)
- 20 ml of 5 M NaCl (1 M)
- 20 μl of 1 M TCEP (0.2 mM)
- Adjust to pH 8.0 using HCl or NaOH
- Bring volume to 100 ml with ddH2O or Milli-Q water
- Filter using a 0.22-μm filter membrane
- Store up to 2 months at 4°C
Purification of ULP1 (Support Protocol)
Lysis buffer, 500 ml
- 12.5 ml of 1 M Tris·HCl pH 7.5 (25 mM)
- 2.5 ml of 5 M imidazole pH 8.0 (25 mM)
- 200 ml of 5 M NaCl (2 M)
- 50 μl of 1 M TCEP (0.1 mM)
- 100 mg lysozyme (Sigma-Aldrich, cat. no. L-6876) (0.2 mg/ml)
- Adjust to pH 7.5 using HCl or NaOH
- Bring volume to 500 ml with ddH2O or Milli-Q water
- Store up to 2 months at 4°C
Ni-NTA elution buffer, 100 ml
- 2.5 ml of 1 M Tris·HCl pH 7.5 (25 mM)
- 5 ml of 5 M imidazole pH 8.0 (250 mM)
- 20 ml of 5 M NaCl (1 M)
- 20 μl of 1 M TCEP (0.2 mM)
- 20% (v/v) glycerol
- Adjust to pH 7.5 using HCl or NaOH
- Bring volume to 100 ml with ddH2O or Milli-Q water
- Store up to 2 months at 4°C
Washing buffer, 400 ml
- 10 ml of 1 M Tris·HCl pH 7.5 (25 mM)
- 2 ml of 5 M imidazole pH 8.0 (25 mM)
- 160 ml of 5 M NaCl (2 M)
- 40 μl of 1 M TCEP (0.1 mM)
- Adjust to pH 7.5 using HCl or NaOH
- Bring volume to 400 ml with ddH2O or Milli-Q water
- Store up to 2 months at 4°C
COMMENTARY
In this protocol, we delineate the procedure for expressing, purifying, and dissolving SRSF1 in various phosphorylated states. Both unphosphorylated and hypo-phosphorylated SRSF1 can be purified in a similar way, while hyper-phosphorylated SRSF1 necessitates the use anion exchanging chromatography. The typical yield of purified SRSF1 is ∼2 to 3 mg of proteins per L of LB or M9 media. However, the protein yield may vary based on several factors, including the freshness of the transformed cells, the types of media employed, the expeditiousness of protein purification, and the condition of the cells. Therefore, it is advisable to use freshly transformed cells. Additionally, we recommend expediting the protein purification process to avoid protein degradation. In addition to these suggestions, the following parameters deserve special consideration in the purification of SRSF1.
Critical Parameters
Our recent study has revealed that SRSF1 undergoes phase separation in a physiological buffer (Fargason et l., 2023). We discovered that the interaction between the RS region and the conserved RRM sites, composed of acidic and aromatic residues, is responsible for the phase separation of SRSF1. Given that these interacting regions are conserved throughout the SR protein family, phase separation appears to be a common feature in this protein family (Fargason et al., 2023).
Phase separation poses significant challenges for the biophysical and biochemical characterization of proteins. Protein droplets, being denser than buffers, cause phase-separating proteins to precipitate during centrifugation or extended incubation, commonly employed in protein purification and concentration. Although phase separation droplets can be re-dissolved, those of some proteins age and harden over time, eventually transforming into irreversible aggregates.
The most critical parameter for the purification of SRSF1 is the concentration of Arg/Glu. SRSF1 exhibits solubility at high concentrations of Arg/Glu. However, Arg concentrations exceeding 100 mM are incompatible with the Ni-NTA resin, as Arg can elute His-tagged proteins like imidazole, albeit less effectively. Additionally, high concentrations of Arg interfere with protein binding to ion exchange columns. Therefore, a balance between SRSF1 solubility and ion exchange loading is necessary. The second crucial parameter for SRSF1 purification is temperature. While high temperatures favor the solubility of many phase-separating proteins, they also accelerate protein degradation. Consequently, our protocol incorporates room temperature in some steps for the purification of unphosphorylated SRSF1.
For the expression of hyper-phosphorylated SRSF1, we co-transform E. coli with the CLK1- and SRSF1-plasmids. It is imperative to express sufficient CLK1 to obtain satisfactory hyper-phosphorylated SRSF1. Based on our experience, the use of fresh commercial competent cells is critical for this purpose, as unhealthy competent cells typically yield large fractions of partially or unphosphorylated SRSF1.
The expression level of SRSF1 is high in the E. coli strain we utilize, and codon optimization is unnecessary. However, the expression of other SR proteins may require codon optimization due to codon bias among different organisms.
Troubleshooting
See Table 1 for a list of problems, possible causes and solutions for SRSF1 purification.
| Problem | Possible cause | Solution |
|---|---|---|
| Few or no colonies observed on agar plates; this usually happens for purification of phosphorylated SRSF1 when co-transformation is needed | Wrong antibiotics are used in the plate; the ratio of plasmids and competent cells are not optimized | Remake plate with correct antibiotics; use fresh commercial competent cells; the amount of plasmid for co-transformation is between 50-100 ng for each plasmid, and the competent cell volume should be 50-100 μl |
| Protein degradation | Freeze/thaw steps are too slow; protease inhibitors are not used; the purification between the Ni-NTA step to the ion exchange step is too slow | Accelerate the thaw step by inverting the sample in a water bath and freezing it immediately after completely thawed; add protease inhibitor cocktail tablet and PMSF; PMSF should be added right before lysis; avoid unnecessary waiting time in the purification procedure |
| Low protein yield after Ni-NTA column | Arg-rich proteins tend to bind to Ni-NTA tighter than other proteins; protein elution is incomplete; the plate with transformed cells has been stored too long | Double check a correct buffer (imidazole and Arg concentration) is used; elute another two times and check the eluted sample by SDS-PAGE gel; use cells transformed within 1 week; we noticed a significant decrease in the protein expression level if transformed cells were stored at 4°C for >2 weeks |
| Purification chromatography traces are extremely bumpy | Air bubbles are trapped in column | Prime the column with lots of water |
| The protein is eluted early than expected in the ion exchange step | Buffers are not placed in right inlets; residual high-salt buffers are in the AKTA machine; pH of the buffers is not accurate | Verify the buffers and the A/B inlets; flush tubing with buffer B first and then buffer A; ensure buffer inlet A and B are completely filled with the corresponding buffers; verify the pH and conductivities of buffers used |
| Contamination bands are observed | The Ni-NTA, ion exchange column, or SEC column is dirty; the washing step is too fast in the Ni-NTA step; the gradient of the ion exchange step is to steep; ULP1 cleavage is not complete | Clean all columns according to the protocol or the user manuals; the flow rate in the Ni-NTA washing step should be <10 ml/min; lower the gradient; give enough cleavage time described in the protocol; it takes ∼1 hr to finish the cleavage at 37°C; lower cleavage temperature needs longer cleavage time |
| Protein loss during concentration | Molecular weight cutoff (MWCO) of the concentrator is too high or membrane leakage; protein phase separation/aggregation | MWCO of the concentrator should be <½ of the protein MW; use a proper centrifuge speed recommended by the manual to avoid damaging the membrane; ensure correct concentrations of Arg/Glu are used to solubilize proteins; the sample should be inverted to mix for every 15 min centrifugation; avoid centrifugation longer than 15 min |
Time Considerations
Basic Protocols 1 to 3
The timeframes for purification of SRSF1 in different phosphorylation states are essentially the same. A rough time arrangement has been provided in the Basic Protocols. The following are details of the time consideration.
Preparation of media and buffers will take 2 to 3 hr if the commonly used stock solutions in a biochemistry lab are available. If not, making all buffers will take 1 or 2 days.
Transformation of competent cells takes 1.5 hr plus an overnight incubation for the cells to grow on the plate.
Growing cells takes 6 to 8 hr. This includes growing start culture, bulky culture, and monitoring cell growth.
Harvesting cells takes 4 to 5 hr. This includes collecting cells into centrifuge bottles (∼15 min), centrifugation (∼20 min), collecting cell pellets (∼10 min), and freeze/thaw cycles (3 to 4 hr).
Purification of protein takes 8 hr total. Specifically, thawing and lysing cells takes 1 hr; separating soluble fractions takes 1 hr; The Ni-NTA column step (including priming Ni-NTA column, loading, washing and eluting sample, and preparing SDS-PAGE samples) takes up to 2 to 3 hr. The ion exchange chromatography step (including priming column, loading sample to the ÄKTA machine, and collecting and concentrating samples) takes 3 to 4 hr. If a SEC column is used, another 4 to 5 hr are needed. This includes concentrating protein samples before SEC column, running the SEC column and concentrating protein samples. Equilibrating the SEC column is not included here because it can be performed after the ion exchange chromatography step.
Support Protocol
Time considerations for purification of ULP1 is similar as that for Basic Protocols 1 to 3. The difference is that purification of ULP1 does not need ion exchange chromatography or SEC. Purification of the protein takes 4 to 5 hr. Thawing and lysing cells takes 1 hr; separating soluble fractions takes 1 hr; the Ni-NTA column step (including priming the Ni-NTA column, loading, washing, and eluting sample, and preparing SDS-PAGE samples) takes up to 2 to 3 hr.
Acknowledgments
This work is supported by U.S. National Science Foundation, MCB (MCB2024964 to J.Z.) and U.S. National Institutes of Health (R35GM147091-01 to J.Z.).
Author Contributions
Zihan Zhang : Data curation; investigation; methodology; writing original draft. Jun Zhang : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; validation; writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
All data are included in this protocol or can be found in our recent eLife research article (Fargason et al., 2023).
Literature Cited
- Anko, M. L. (2014). Regulation of gene expression programmes by serine-arginine rich splicing factors. Seminars in Cell & Developmental Biology, 32, 11–21. https://doi.org/10.1016/j.semcdb.2014.03.011
- Aubol, B. E., Keshwani, M. M., Fattet, L., & Adams, J. A. (2018). Mobilization of a splicing factor through a nuclear kinase-kinase complex. Biochemical Journal , 475(3), 677–690. https://doi.org/10.1042/bcj20170672
- Azpurua, J., El-Karim, E. G., Tranquille, M., & Dubnau, J. (2021). A behavioral screen for mediators of age-dependent TDP-43 neurodegeneration identifies SF2/SRSF1 among a group of potent suppressors in both neurons and glia. PLoS Genetics , 17(11), e1009882. https://doi.org/10.1371/journal.pgen.1009882
- Blencowe, B. J., Bowman, J. A., McCracken, S., & Rosonina, E. (1999). SR-related proteins and the processing of messenger RNA precursors. Biochemistry and Cell Biology , 77(4), 277–291.
- Cascarina, S. M., & Ross, E. D. (2022). Expansion and functional analysis of the SR-related protein family across the domains of life. RNA , 28(10), 1298–1314. https://doi.org/10.1261/rna.079170.122
- Chabot, B., & Steitz, J. A. (1987). Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Molecular and Cellular Biology , 7(1), 281–293. https://doi.org/10.1128/mcb.7.1.281-293.1987
- Cho, S., Hoang, A., Sinha, R., Zhong, X. Y., Fu, X. D., Krainer, A. R., & Ghosh, G. (2011). Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proceedings of the National Academy of Sciences of the United States of America , 108(20), 8233–8238. https://doi.org/10.1073/pnas.1017700108
- Dembowski, J. A., An, P., Scoulos-Hanson, M., Yeo, G., Han, J., Fu, X. D., & Grabowski, P. J. (2012). Alternative splicing of a novel inducible exon diversifies the CASK guanylate kinase domain. Journal of Nucleic Acids , 2012, 816237. https://doi.org/10.1155/2012/816237
- Erkelenz, S., Mueller, W. F., Evans, M. S., Busch, A., Schoneweis, K., Hertel, K. J., & Schaal, H. (2013). Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA , 19(1), 96–102. https://doi.org/10.1261/rna.037044.112
- Fargason, T., De Silva, N. I. U., King, E., Zhang, Z., Paul, T., Shariq, J., Zaharias, S., & Zhang, J. (2023). Peptides that mimic RS repeats modulate phase separation of SRSF1, revealing a reliance on combined stacking and electrostatic interactions. eLife , 12, e84412. https://doi.org/10.7554/eLife.84412
- Fei, J., Jadaliha, M., Harmon, T. S., Li, I. T. S., Hua, B., Hao, Q., Holehouse, A. S., Reyer, M., Sun, Q., Freier, S. M., Pappu, R. V., Prasanth, K. V., & Ha, T. (2017). Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. Journal of Cell Science , 130(24), 4180–4192. https://doi.org/10.1242/jcs.206854
- Gui, J. F., Lane, W. S., & Fu, X. D. (1994). A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature , 369(6482), 678–682. https://doi.org/10.1038/369678a0
- Hammarskjold, M. L., & Rekosh, D. (2017). SR proteins: To shuttle or not to shuttle, that is the question. Journal of Cell Biology , 216(7), 1875–1877. https://doi.org/10.1083/jcb.201705009
- Haward, F., Maslon, M. M., Yeyati, P. L., Bellora, N., Hansen, J. N., Aitken, S., Lawson, J., von Kriegsheim, A., Wachten, D., Mill, P., Adams, I. R., & Caceres, J. F. (2021). Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. eLife , 10, e65104. https://doi.org/10.7554/eLife.65104
- Huang, Y., Gattoni, R., Stévenin, J., & Steitz, J. A. (2003). SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Molecular Cell , 11(3), 837–843. https://doi.org/10.1016/s1097-2765(03)00089-3
- Ilik, I. A., Malszycki, M., Lubke, A. K., Schade, C., Meierhofer, D., & Aktas, T. (2020). SON and SRRM2 are essential for nuclear speckle formation. Elife , 9, e60579. https://doi.org/10.7554/eLife.60579
- Kohtz, J. D., Jamison, S. F., Will, C. L., Zuo, P., Luhrmann, R., Garcia-Blanco, M. A., & Manley, J. L. (1994). Protein-protein interactions and 5'-splice-site recognition in mammalian mRNA precursors. Nature , 368(6467), 119–124. https://doi.org/10.1038/368119a0
- Lamond, A. I., & Spector, D. L. (2003). Nuclear speckles: A model for nuclear organelles. Nature Reviews Molecular Cell Biology , 4(8), 605–612. https://doi.org/10.1038/nrm1172
- Li, K., & Wang, Z. (2021). Speckles and paraspeckles coordinate to regulate HSV-1 genes transcription. Communications Biology , 4(1), 1207. https://doi.org/10.1038/s42003-021-02742-6
- Li, X., & Manley, J. L. (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell , 122(3), 365–378. https://doi.org/10.1016/j.cell.2005.06.008
- Long, J. C., & Caceres, J. F. (2009). The SR protein family of splicing factors: Master regulators of gene expression. Biochemical Journal , 417(1), 15–27. https://doi.org/10.1042/BJ20081501
- Manley, J. L., & Krainer, A. R. (2010). A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes & Development, 24(11), 1073–1074. https://doi.org/10.1101/gad.1934910
- Moore, M. J., Query, C. C., & Sharp, P. A. (1993). Splicing of precursors to mRNA by the spliceosome. In R. F. Gesteland., & J. F. Atkins (Ed.), The RNA World. Cold Spring Harbor Laboratory Press.
- Pan, Q., Shai, O., Lee, L. J., Frey, B. J., & Blencowe, B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics , 40(12), 1413–1415. https://doi.org/10.1038/ng.259
- Pelisch, F., Gerez, J., Druker, J., Schor, I. E., Muñoz, M. J., Risso, G., Petrillo, E., Westman, B. J., Lamond, A. I., Arzt, E., & Srebrow, A. (2010). The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proceedings of the National Academy of Sciences of the United States of America , 107(37), 16119–16124. https://doi.org/10.1073/pnas.1004653107
- Shepard, P. J., & Hertel, K. J. (2009). The SR protein family. Genome Biology , 10(10), 242. https://doi.org/10.1186/gb-2009-10-10-242
- Tripathi, V., Song, D. Y., Zong, X., Shevtsov, S. P., Hearn, S., Fu, X. D., Dundr, M., & Prasanth, K. V. (2012). SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Molecular Biology of the Cell , 23(18), 3694–3706. https://doi.org/10.1091/mbc.E12-03-0206
- Wu, H., Sun, S., Tu, K., Gao, Y., Xie, B., Krainer, A. R., & Zhu, J. (2010). A splicing-independent function of SF2/ASF in microRNA processing. Molecular Cell , 38(1), 67–77. https://doi.org/10.1016/j.molcel.2010.02.021
- Zhang, Z., & Krainer, A. R. (2004). Involvement of SR proteins in mRNA surveillance. Molecular Cell , 16(4), 597–607. https://doi.org/10.1016/j.molcel.2004.10.031
- Zhou, Z., & Fu, X. D. (2013). Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma , 122(3), 191–207. https://doi.org/10.1007/s00412-013-0407-z
- Zillmann, M., Rose, S. D., & Berget, S. M. (1987). U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Molecular and Cellular Biology , 7(8), 2877–2883. https://doi.org/10.1128/mcb.7.8.2877-2883.1987

