Mime-seq 2.0: a method to sequence microRNAs from specific mouse cell types
Ariane Mandlbauer, Qiong Sun, Niko Popitsch, Tanja Schwickert, Miroslava Spanova, Jingkui Wang, Stefan Ameres, Meinrad Busslinger, Luisa Cochella
Abstract
The description of precise miRNA expression patterns is crucial to understand what these small RNAs contribute to animal development and physiology. High-throughput sequencing of miRNAs from individual developmental stages has provided insight into temporal regulation but often lacks the cellular resolution to link miRNA-function to the biology of distinct cell types within complex tissues. Here, we provide a protocol for microRNome by methylation-dependent sequencing for mice (mime-seq 2.0). Mime-seq takes advantage of a chemical small RNA-tagging approach followed by chemo-selective, high-throughput sequencing that enables the identification of tissue- and cell type-specific miRNA profiles in animals in a sensitive and reproducible manner.
Steps
Total RNA isolation
Extract total RNA from frozen cells or tissue using TRIzol, depending on sample type follow manufacturer’s instructions for proper lysis.
Add 200 μl of chloroform per 1 ml of TRIzol, agitate samples vigorously and incubate for 5 min at room temperature (RT).
Centrifuge at full speed (12,000 x g) for 15 min at 4°C. The mixture separates into lower, red, phenol-chloroform phase, an interphase, and a colorless aqueous phase.
Carefully transfer the upper aqueous phase to a fresh tube.
Precipitate RNA by adding 1 μl of glycogen (20 mg/ml) and 500 μl of isopropyl alcohol, vortex.
Incubate at 4˚C for 10 minutes.
Centrifuge at full speed for 10 minutes at 4˚C; RNA will form a gel-like pellet on the side and bottom of the tube; for low input samples, extend centrifugation time to 60 min.
Remove supernatant without losing the RNA pellet.
Wash pellet
Add 300 μl of 80% cold ethanol, vortex.
Centrifuge 10 minutes at full speed at 4°C.
Remove supernatant, collect remaining ethanol by quick centrifugation and pipette all remaining liquid using a 10 μl pipette. Air dry pellet for approx. 5 min or until it becomes translucent.
Air dry pellet for approx. 5 min or until it becomes translucent.
Dissolve RNA pellet in 20-50 μl of DEPC-treated or other RNase free water, to obtain a target RNA concentration above 1 μg/μl.
Measure concentration (Qubit BR RNA kit); ideally, at least 3.3 μg of total RNA are used for small RNA library preparation per sample (will be split in ox/unox).
NaIO4-mediated oxidation
Prepare 3.3 μg of total RNA with a mix of methylated and unmethylated spike-ins in 44 μl RNase-free H2O (includes 10 % extra).
For Oxidation reaction, split each sample in half and incubate using following reaction mix 30 min at RT.
| A | B |
|---|---|
| RNA/Spike-in mix | 20 ul |
| NaIO4 (50mM) or H2O | 5 ul |
| 5x borate buffer | 8 ul |
| H2O | 7 ul |
Prepare a master mix +/- NaIO4 containing everything except for the respective sample. Transfer samples for oxidation and control reaction in 8 strips, distribute master mix last.
Add immediately 229 ul H2O, 30 ul NaOAc (pH 5.2) and 1 ul glycogen (20 mg/ml), mix well.
EtOH precipitation
Precipitate by adding 900 ul ice-cold EtOH to each sample.
Incubate for > 1h at -20˚C (this can be overnight).
Centrifuge for 1 h at 4˚C and remove supernatant without disturbing the pellet.
Wash
Add 500 μl of 80% cold ethanol.
Centrifuge 10 minutes at full speed at 4°C.
Remove supernatant, collect remaining ethanol by quick centrifugation and pipette all remaining liquid using a 10 μl pipette.
Air dry pellet for approx. 5 min or until it becomes translucent.
Resuspend each pellet in 6 ul H2O (DEPC-treated or RNase free).
3' Ligation
Add to each sample 1 μl srBC 3’ adapter (10 μM) and note used barcodes.
Note – oxidized samples should not be run on the same gel as unoxidized samples, due to big differences in ligatable small RNAs. Each sample run on the same gel should have a different srBC adapter.
Set up in parallel ligation reaction for 18mer and 30mer as later reference for PAGE purification. Mix 2.5 μl 18mer (10 μM) with 2.5 μl 30mer (10 μM) and 0.5 μl 100 μM 3’ linker and 1.5 μl H2O. 2O.
Thaw 50% PEG8000 at 37˚C, this makes pipetting of viscous solution easier.
Incubate RNA + srBC 3’ 5 min at 70˚C, snap-cool on ice.
Set up master mix at room temperature:
| A | B |
|---|---|
| PEG8000 50% | 10 ul |
| 10x 3' Ligation buffer | 2 ul |
| T4 RNA ligase trunc | 2 ul |
Add 13 µl MM to each sample and incubate o/n at 4˚C.
Note – we normally keep the ligation reactions in a metal cooling rack in the fridge or cold room.
Size selection (PAGE)
Prepare a 15% denaturing urea polyacrylamide gel (SequaGel; 20 cm × 16 cm × 1 mm, length × width × thickness).
Note – pre-treat glass plates (one with anti-fog, the other with water repellent) to avoid gel sticking to the plates.
Pre-run the gel in 0.5× TBE buffer Tat a constant power of 35 W for 30-60 min until the surface temperature of the gel reaches approximately 45–55°C.
In the meantime add an equal volume of form amide loading buffer to the samples and denature by incubating 5 min at 95ºC, snap-cool on ice.
Note – include ligated 18 and 30mers, as well as unligated 60mer as size reference.
Load samples into pre-washed wells.
Note – IMPORTANT rinse all wells before loading with a syringe to remove all accumulated urea and gel fragments. Rinse again before loading of each sample.
Do not use lanes with bubbles and leave at least one empty lane between each sample for easier handling. Do not run oxidized and unoxidized samples on the same gel.
Run gel first at constant power of 10 W until samples are migrated out of their wells. Run at a constant power of 35 – 45 W until the bromophenol blue is about halfway down the gel (1-1.5 h).
Remove the top glass plate and with the gel still on the bottom plate, pour 10-15 ml of SYBR gold in 0.5x TBE (for 50 ml use 5 μl of SYBR gold) and stain for 3-4 minutes.
Carefully transfer the gel onto a transparent sheet protector .
Note – open sheet protector on 3 sides, hold gel on one end and transfer between sheets. We normally use tape on the outside to label the individual sample lanes for orientation.
Visualize RNA on blue transilluminator (take a photograph if desired) and label area between ligated 18mer and 30mer on each lane on the sheet protector with a marker. Remove from transilluminator and cut out using a razor blade small RNAs between markers, collect in 1.5 ml LoBind tubes.
Note – cutting the gel piece in half and laying them on top of each other makes it easier to transfer gel piece to a tube.
Add 800 µl 0.3 M NaCl + 0.1% SDS and allow RNA to elute overnight at room temperature, while rotating.
EtOh precipitation
Precipitate each sample by transferring to two fresh tubes (without transferring gel pieces) and adding to each 400 µl aliquot, 1 µl glycogen (20 mg/mL) and 1 ml of 100% EtOH; incubate samples for 1.5-2 hours at -20˚C.
Centrifuge 40 minutes at full speed (12,000 x g) at 4˚C, decant supernatant and wash the pellet with 300 µl of 80% cold ethanol as before. Centrifuge again 10 minutes at 4˚C, remove the supernatant completely, perform an extra round of centrifugation for 2 minutes and pipette out all residual EtOH.
Without air-drying the pellet, dissolve small RNAs in 6 µl H2O + 1 µl 5’ adapter (10 µM).
5’ linker ligation
Incubate RNA + adapter mix 5 min at 70˚C, snap-cool on ice.
Prepare ligation master mix (volumes are calculated for one sample):
| A | B |
|---|---|
| PEG8000 50% | 10 ul |
| 10x RNA ligae buffer | 2 ul |
| T4 RNA Ligase 1 | 2 ul |
Add to each sample 13 µl of master mix, mix and give it a quick spin.
Incubate o/n at 4˚C.
Note – we normally keep the ligation reactions in a metal cooling rack in the fridge or cold room.
Purification using RNA Clean & Concentrator 5 (Zymo)
Add 80 µl H2O and mix by pipetting.
Purify using the RNA Clean & Concentrator 5 (RCC5) kit
Add 200 µl RNA binding buffer and mix.
Add 300 µl EtOH absolute and mix.
Continue according to manufacturer's instructions and elute in 12 µl H2O.
Reverse transcription
Transfer 11.5 µl sample into a PCR strip and add 1 µl RT primer (10 µM).
Incubate 5 min at 65˚C, snap-cool on ice.
Note – pre-heat PCR block to 50˚C.
RT reaction master mix:
| A | B |
|---|---|
| 5x FS buffer | 4 ul |
| dNTP mix (10mM) | 1 ul |
| 0.1 M DTT | 1 ul |
| RNase out | 0.5 ul |
| Superscript II or III | 1 ul |
Add on ice 7.5 µl RT master mix, mix well, spin down and immediately transfer to pre-heated PCR block at 50˚C, incubate 30 min.
Note – IMPORTANT, no heat inactivation step after RT.
Add 8 µl ExoSAP-IT and incubate 15 min at 37˚C followed by heat inactivation at 80˚C for 15 min.
On ice, add 2 µl H2O for a total volume of 30 µl.
qPCR amplification
Transfer ½ of each sample (15 µl) to strip tube. Add 10 µl i5/i7 primer mix and 25 µl Kapa HiFi Library Amp Kit mix.
Prepare also in duplicates 50 µl of fluorescent standard (STD) 3 and 4.
Close strip with an optical flat-cap, vortex, and spin down.
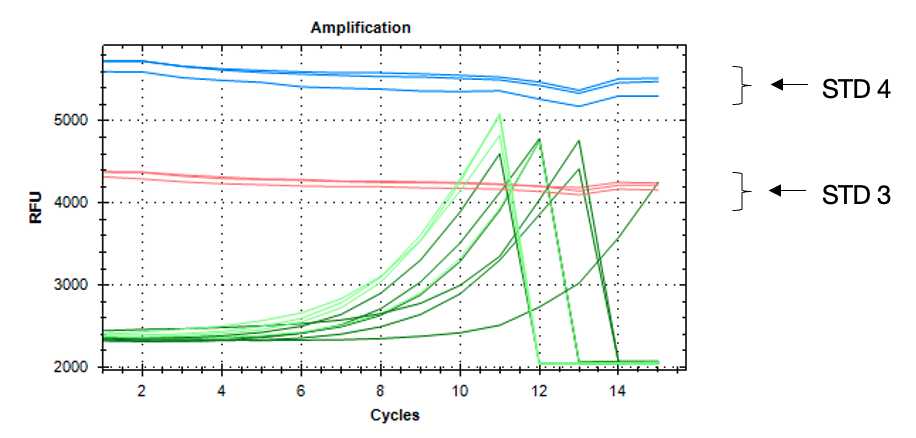
Run the following program until samples reach the fluorescence level of standard 3 (assessed from the image acquired by the PCR machine in each cycle).
Note - use qPCR machine (i.e. BioRad CFX96) to monitor library amplification in real-time. Base-line subtraction hould be turned off.
| A | B | C |
|---|---|---|
| 95˚C | 45 sec | |
| 95˚C | 15 sec | n cycles |
| 65˚C | 15 sec | |
| 72˚C | 20 sec | |
| plate read | ||
| 72˚C | 10 sec |
When a sample reaches signal between standard 3 and 4, pause the program during the 10 sec step after the plate read and remove carefully the respective tubes from the PCR machine. Repeat until all samples are amplified sufficiently.
Gel purification of amplified cDNA
Prepare a 3% Low-Range Ultra agarose gel according to the manufacturer’s instructions (swirl the flask often during heating procedure), with 1.5 µl SYBR Safe per 50 ml of gel.
Load 6 µl of GeneRuler 50 bp DNA Ladder and the amplified cDNA after addition of 10 µl of Orange G loading dye 6x (total volume 60 µl)
Run the gel at constant V (depending on chamber size between 80 and 150V) until the Orange G loading dye band is at 2/3 of the gel length.
Visualize on a UV transilluminator and excise upper DNA band between 160-200 bp with a clean scalpel or razor blade and place in a clean tube, as shown in Figure 2 below.
Note - IMPORTANT in case 2 bands are visible – avoid cutting into lower band as this corresponds to adapter dimers; this is especially common with oxidized library samples.
Dissolve gel by adding 3 volumes (~800-900 µl) of ADB buffer (ZymocleanTM Gel DNA Recovery Kit) and incubating samples at 55ºC for ~10 minutes or until the gel slice is completely dissolved; mix and collect by centrifugation.
Transfer the melted agarose solution to a Zymo-SpinTM Column in a collection tube, centrifuge 1 minute at 10,000 rpm and discard the flow-through.
Add 200 µl of DNA Wash Buffer to the column, centrifuge 1 minute and discard the flow-through; repeat the wash step once more.
Add 20 µl of water directly to the column matrix, place the column into a 1.5 ml tube and centrifuge for 1 minute to elute DNA; purified DNA is now ready for quality control and deep sequencing.
Note - we routinely run libraries on fragment analyzer or tape station to make sure adapter dimers are sufficiently removed.
Bioinformatics and data analysis
Libraries are sequenced using an Illumina short read sequencer with a minimum read length of 50 nucleotides as for example NovaSeq SP in SR100 XP mode.
Custom NextFlow pipelines for processing can be found on GitHub under h https://github.com/popitsch/pysrna. In short:
Sequencing quality control with fastqc v 0.11.8.
Raw reads are demultiplexed, parsed and filtered.
Spike-in reads are filtered and counted allowing for alignment 1 mismatch.
Remaining small RNA reads are mapped to a miRNA transcriptome obtained from MirGeneDB v2.0 or miRbase annotations.
Small RNAs were counted and normalized to reads obtained from methylated spike-ins.

