Exploring Non-Coding RNAs in RNAcentral
Robert D. Finn, Robert D. Finn, Blake A. Sweeney, Blake A. Sweeney, Arina A. Tagmazian, Arina A. Tagmazian, Carlos E. Ribas, Carlos E. Ribas, Alex Bateman, Alex Bateman, Anton I. Petrov, Anton I. Petrov
Abstract
Non-coding RNAs are essential for all life and carry out a wide range of functions. Information about these molecules is distributed across dozens of specialized resources. RNAcentral is a database of non-coding RNA sequences that provides a unified access point to non-coding RNA annotations from >40 member databases and helps provide insight into the function of these RNAs. This article describes different ways of accessing the data, including searching the website and retrieving the data programmatically over web APIs and a public database. We also demonstrate an example Galaxy workflow for using RNAcentral for RNA-seq differential expression analysis. RNAcentral is available at https://rnacentral.org. © 2020 The Authors.
Basic Protocol 1 : Viewing RNAcentral sequence reports
Basic Protocol 2 : Using RNAcentral text search to explore ncRNA sequences
Basic Protocol 3 : Using RNAcentral sequence search
Basic Protocol 4 : Using RNAcentral FTP archive
Support Protocol 1 : Using web APIs for programmatic data access
Support Protocol 2 : Using public Postgres database to export large datasets
Support Protocol 3 : Analyze non-coding RNA in RNA-seq datasets using RNAcentral and Galaxy
INTRODUCTION
RNAcentral (https://rnacentral.org) is a database of non-coding RNA (ncRNA) sequences that aggregates ncRNA data from >40 member resources known as Expert Databases (Bateman et al., 2011; The RNAcentral Consortium, 2019). RNAcentral is designed as a single-entry point for biologists and bioinformaticians interested in ncRNAs, where they can find a high-level overview of ncRNA content in different species or taxonomic groups, as well as functional information about individual ncRNAs. This includes RNA secondary structure, genome locations, Rfam annotations (see Current Protocols article: Kalvari et al., 2017), orthologs and paralogs, miRNA targets, RNA modifications, and more. In addition to the data from member resources, RNAcentral generates additional annotations, such as comprehensive genome mapping for >350 reference genomes (The RNAcentral Consortium, 2019) and template-based RNA secondary structure diagrams.
RNAcentral provides four key functionalities:
- Viewing information about individual ncRNA sequences;
- Text search that enables exploration of ncRNA sequences from different sources;
- Sequence search for performing sequence similarity queries against a comprehensive set of ncRNA sequences;
- FTP archive with downloadable files, including genome annotations in BED and GFF3 formats.
The following four basic protocols describe how to use the main RNAcentral features, focusing on advanced methods for data access. In addition, three support protocols discuss programmatic data retrieval using web APIs or a public Postgres database, and describe an example Galaxy workflow (Afgan et al., 2018) for analyzing RNA-seq data using RNAcentral.
As the RNAcentral database and the website are under active development, the most recent RNAcentral version may contain new types of data or other functionality not covered here. This article is based on RNAcentral release 14.
Basic Protocol 1: VIEWING RNAcentral SEQUENCE REPORTS
At the time of this writing, RNAcentral contains >16 million non-redundant sequences from a wide range of species. Each sequence has a dedicated report page that always includes the following information: ncRNA sequence, cross-reference(s) to the databases where this sequence is annotated, and its unique accession number (see Guidelines for Understanding Results for more information about RNAcentral identifiers).
Depending on the ncRNA type, organism, and source database, the report pages may include one or more additional sections: RNA secondary structure, an embedded genome browser, Rfam classification (see Current Protocols article: Kalvari et al., 2017), Gene Ontology (GO) terms (Huntley et al., 2014), orthologs and paralogs from Ensembl Compara (Pignatelli et al., 2016), microRNA targets from TarBase (Karagkouni et al., 2018) and LncBase (Paraskevopoulou et al., 2016), modifications from Modomics (Machnicka et al., 2013), literature references, sequence feature viewer, and more.
In this protocol, we show the general structure of a sequence report page and give suggestions for identifying the most relevant information.
Necessary Resources
Hardware
- Any device with Internet access
Software
- An up-to-date Web browser, such as Chrome, Safari, or Firefox
View basic information about ncRNA sequence
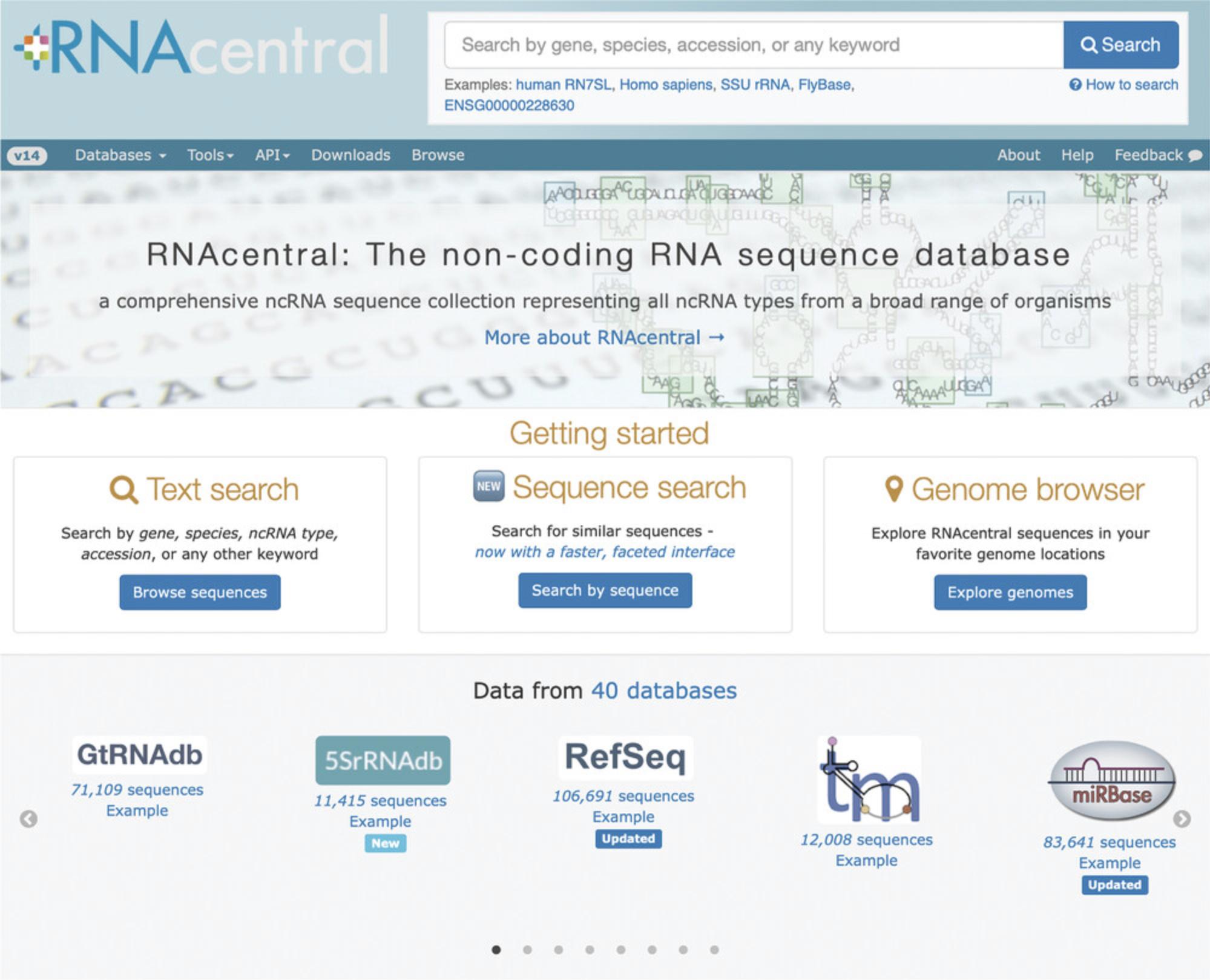
1.Start at the RNAcentral homepage (Fig. 1) and search for human miR-181b-1 stem-loop microRNA by entering URS0000530EBF_9606 in the search box located at the top of the page (more information about searching RNAcentral is found in Basic Protocol 2).
2.Click on the single search result to view the sequence report.
3.Explore the report page (Fig. 2), focusing on the RNA type, source databases, and genomic neighborhood. Most sections of the webpage are interactive. For example, you can click on the RNAcentral transcripts in the genome browser (Fig. 2C) to find what database these sequences come from, or you can visualize the GO term hierarchy by clicking on the tree icons (Fig. 2E).
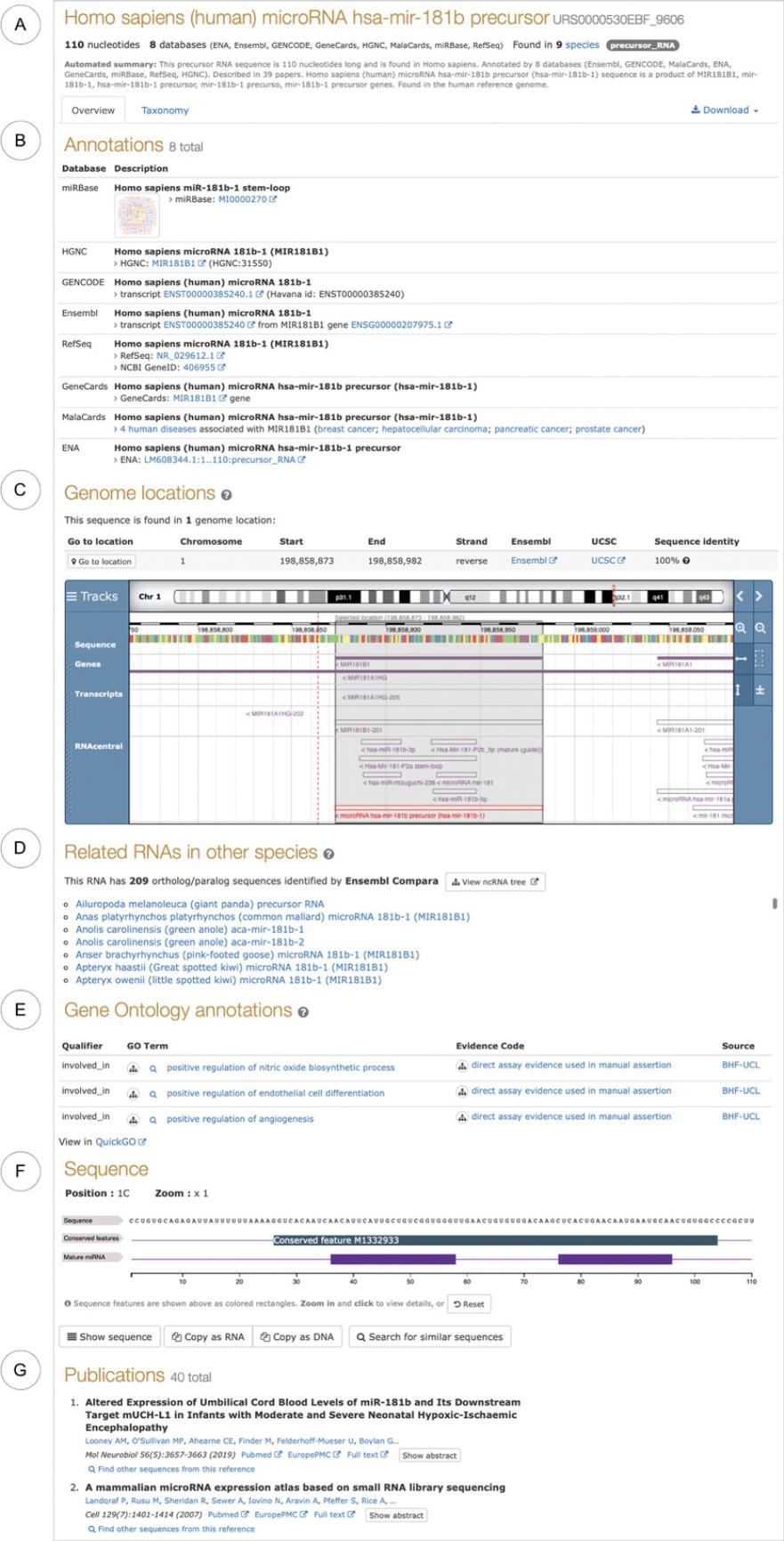
4.Follow the links to the source databases to find additional information about this RNA. For example, in miRBase (Kozomara, Birgaoanu, & Griffiths-Jones, 2019) you can view deep sequencing data supporting this microRNA as well as >370 papers that mention this RNA, while in MalaCards (see Current Protocols article: Rappaport et al., 2014) you can find details about the association of this microRNA with breast cancer and hepatocellular carcinoma, as well as pancreatic and prostate cancers.
5.Identify other sequences in the same genomic locations. The embedded genome browser (Fig. 2C) shows the sequence in the context of the reference genome, including other ncRNAs as well as protein-coding genes and pseudogenes found in the genomic neighborhood.
6.Click the “Taxonomy” tab to view a list of species where the RNA sequence also occurs. Note that this tab only shows the entries with the 100% identical sequence. For other related sequences, explore the “Related RNAs” section of the report page that shows ortholog and paralog sequences retrieved from Ensembl Compara.
7.Click the “Download” button to retrieve the ncRNA sequence in FASTA format or get the sequences and the annotations in JSON format.
Basic Protocol 2: USING RNAcentral TEXT SEARCH TO EXPLORE ncRNA SEQUENCES
The text search enables users to query RNAcentral by species, gene name, RNA type, or any other keyword. The search can be used for exploring the data from >40 databases on the RNAcentral website as shown in this protocol, or it can be used programmatically via an API (see Support Protocol 1).
Necessary Resources
Hardware
- Any device with Internet access
Software
- An up-to-date Web browser, such as Chrome, Safari, or Firefox
Browse all RNAcentral sequences
1.Start at the RNAcentral homepage (https://rnacentral.org) and click “Browse sequences” (Fig. 1).
Browse RNA sequences in a species or taxonomic group of interest
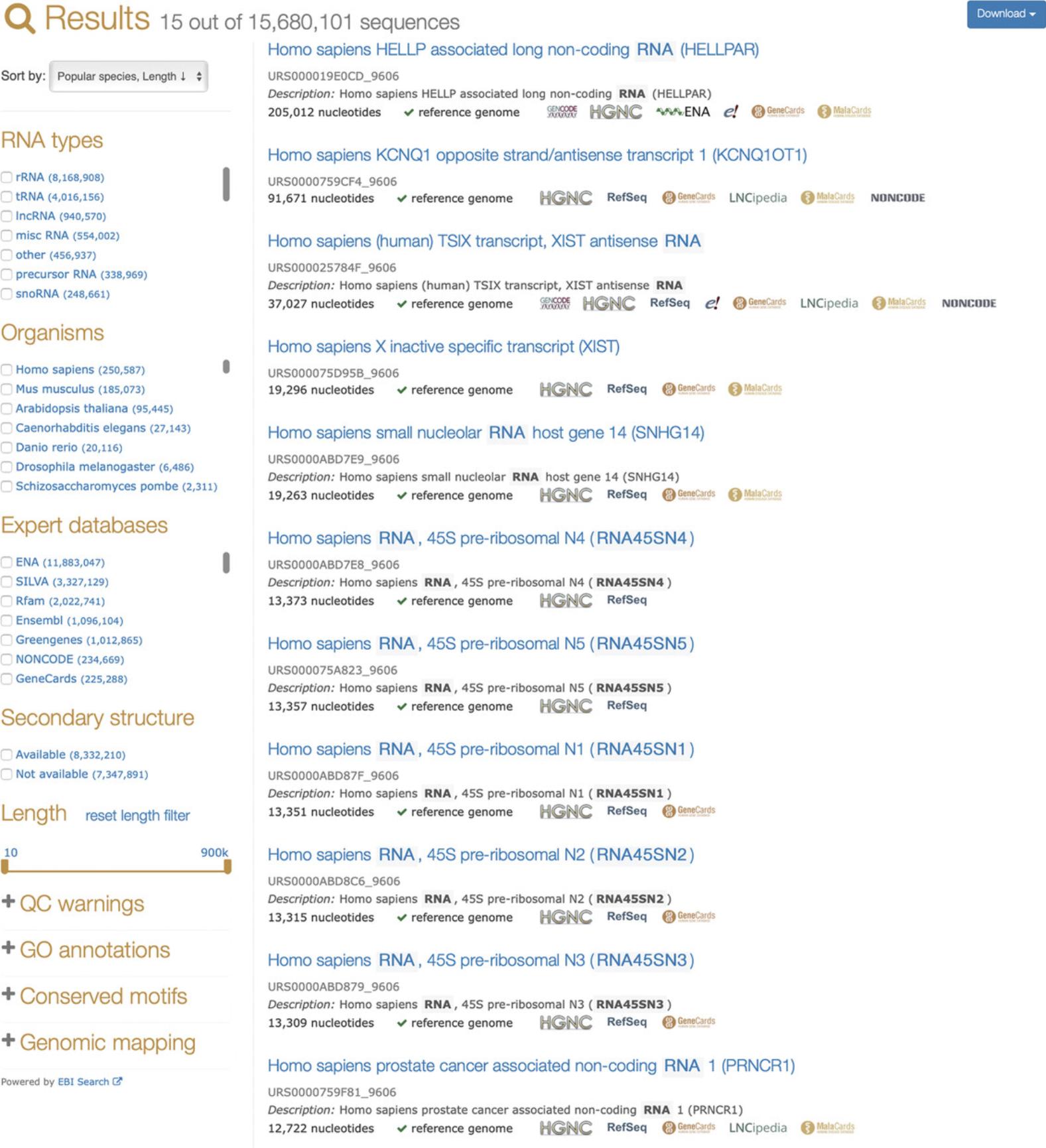
2.To list all ncRNAs in a particular species, type the species name in the search bar and select the species in the Organism facet.
- Alternatively, you can use the syntax:taxonomy: "NCBI_TAXID"whereNCBI_TAXIDis the NCBI taxonomy identifier (taxid) (Federhen,2012). For example,taxonomy: "9606"will return all sequences fromHomo sapiens, which has the assigned NCBI taxid of 9606.
For the most accurate results, we recommend using the Organism facet or entering NCBI taxids, because keyword-based queries can be ambiguous (for example, a search for Bacillus matches both Bacteria and Eukaryotes, seehttps://www.ncbi.nlm.nih.gov/taxonomy/?term=bacillus).
- It is also possible to search for ncRNA sequences from any taxon using the following syntax:tax_string: "mammalia"
wheretax_stringis the taxonomic lineage for a species. Note the use of the Latin taxon name (“mammalia,” not “mammals”).
Use advanced text search syntax
3.As shown above, it is possible to construct advanced queries using the Lucene query syntax (see https://www.ebi.ac.uk/ebisearch/documentation.ebi for more details). The following section provides examples for common use cases.
-
Use double quotes.
To make sure that the results contain the exact query string, surround the query with double quotes. For example, if you are looking for a specific microRNA like hsa-mir-126, run the following search:
- "hsa-mir-126"
A search without double quotes will also match hsa-mir-1261 , hsa-mir-1262 , and other sequences.
- b.Use logic operators.
The search supports logic operators, such as AND, OR, and NOT. For example, one can identify microRNAs from miRBase that are not found in RefSeq or Ensembl (note the use of parentheses for grouping the search terms):
-
rna_type: "miRNA" AND expert_db: "miRBase" NOT (expert_db: "RefSeq" OR expert_db: "Ensembl")
-
c.Limit sequence length.
One can restrict the length of the sequences; for example, the following search returns tRNAs between 60 and 100 nucleotides long (it is also possible to use the sequence length slider shown in Fig. 3).
- rna_type: "tRNA" length:[60 TO 100]
Download search results
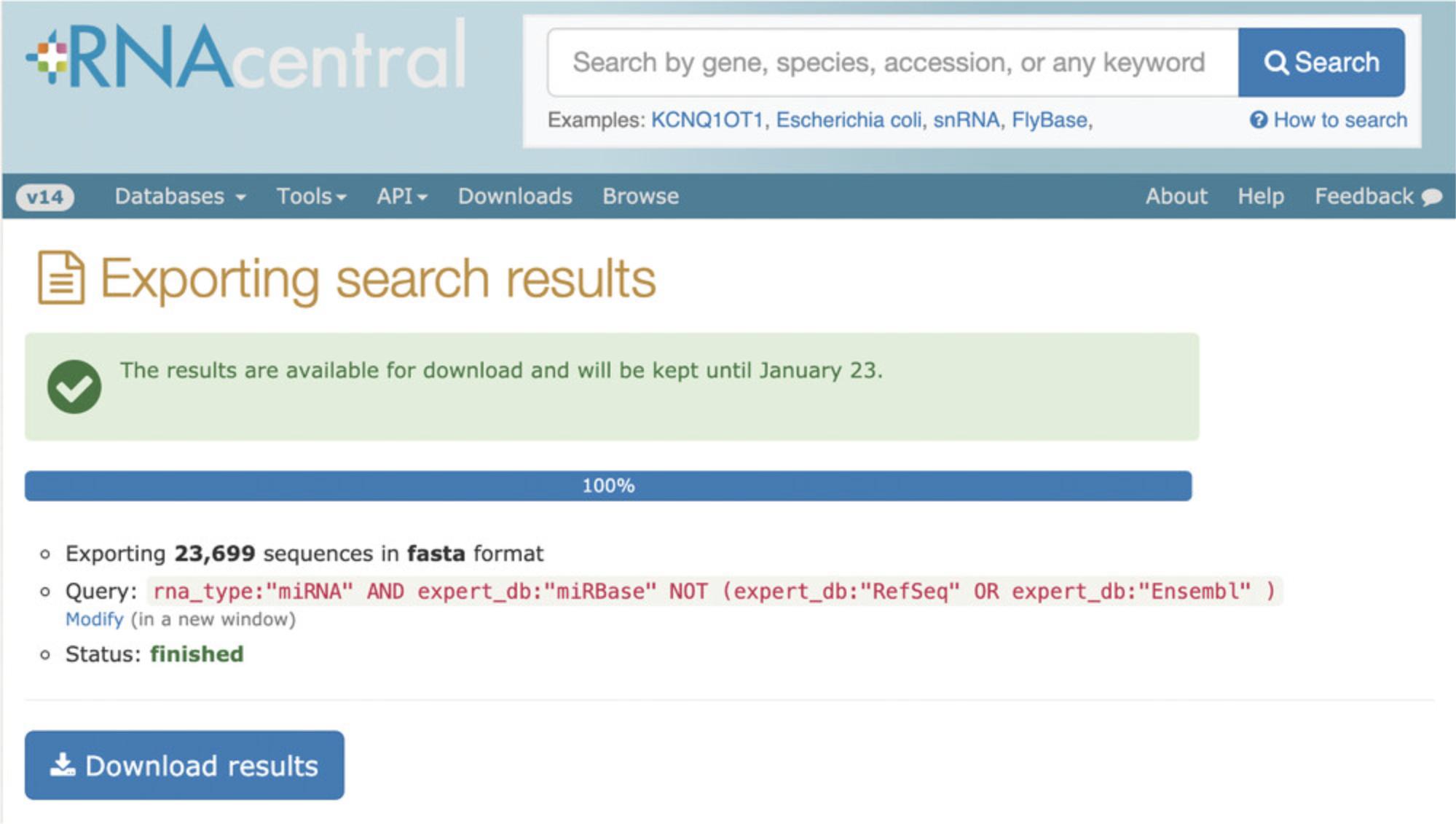
4.The search results can be downloaded for further local processing by clicking the Download button (Fig. 3, top right). You will be redirected to a page with a link to the downloadable file (Fig. 4). The results are kept for 7 days, and the URL can be bookmarked or shared.
Basic Protocol 3: USING RNAcentral SEQUENCE SEARCH
RNAcentral hosts a sequence-similarity search powered by nhmmer (Wheeler & Eddy, 2013) that enables users to compare any sequence against a collection of ncRNA sequences available in RNAcentral. The RNAcentral sequence search can be used to find similar sequences, check if the exact sequence has been observed before, or confirm that a sequence does not match any known ncRNAs.
Necessary Resources
Hardware
- Any device with Internet access
Software
- An up-to-date Web browser, such as Chrome, Safari, or Firefox
1.Find similar sequences. Go to the sequence search page (https://rnacentral.org/sequence-search) and enter the following microRNA sequence:
- GGGAUGAGGUAGUAGGUUGUAUAGUUUUAGGGUCACACCCACCACUGGGAGAUAACUAUACAAUCUACUGUCUUUC
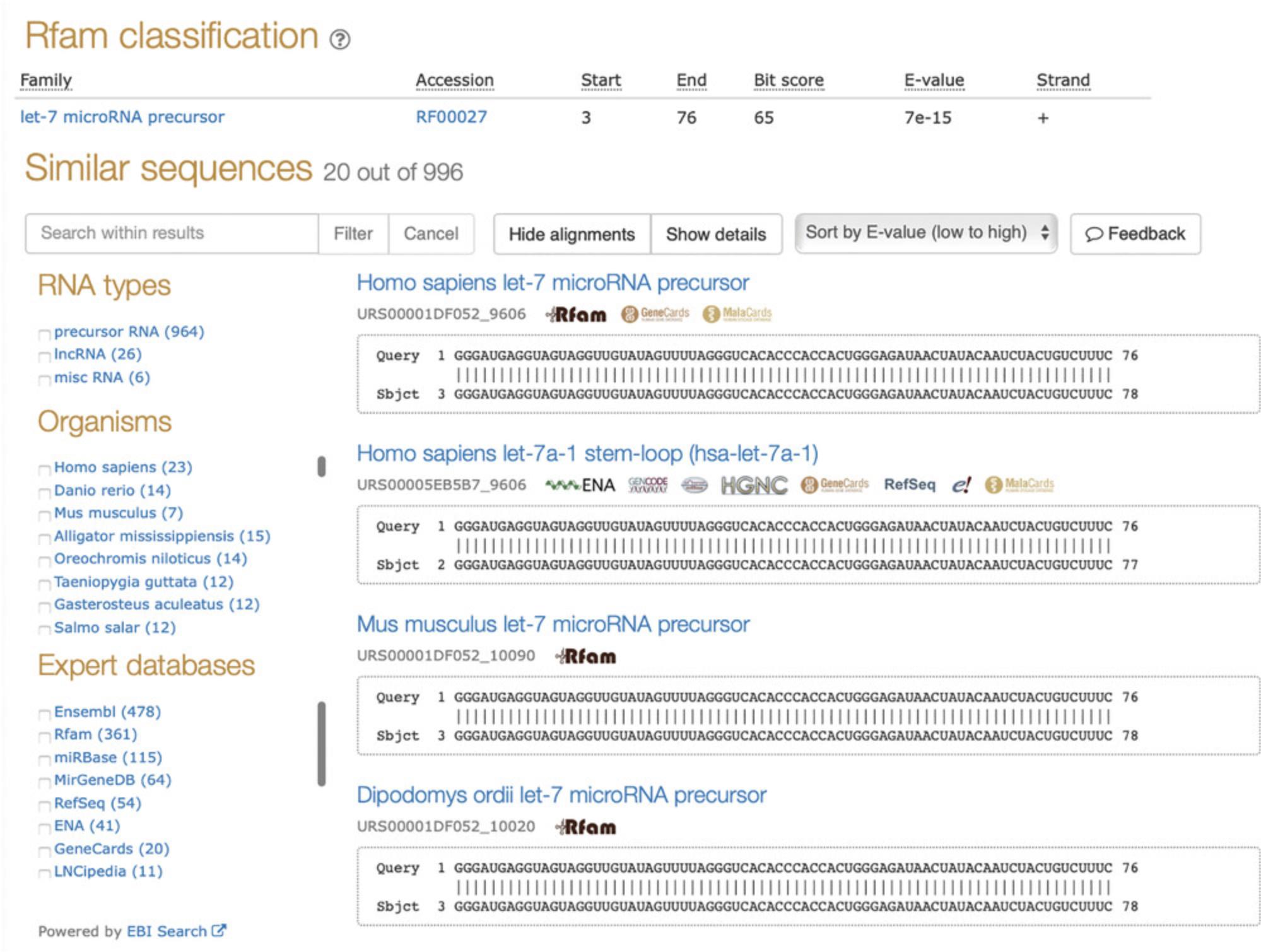
2.Click the “Show details” button to show information about the match such as e-value, sequence identity, and numbers of matching bases. By default, the results are sorted by e-value, but the “Sort by” drop-down menu allows alternative orderings.
3.Explore the results using the same facets as in the text search (Basic Protocol 2). For example, the query sequence matched the human let-7 precursor microRNA as the top hit; however, one can view hits in other species listed under the Organisms facet. Each result links out to the sequence report page (see Basic Protocol 1).
The results can also be filtered by any keyword using the “Search within results” field (Fig. 5). See Basic Protocol 2 for more information on facets and different search strategies.

Basic Protocol 4: USING RNAcentral FTP ARCHIVE
The FTP archive provides users with an easy way to fetch large amounts of data from RNAcentral. The archive contains downloadable files, including sequences in FASTA format, identifier mapping files that can be used to convert between RNAcentral and external identifiers, Rfam annotations, GO annotations, and genome annotations in GFF3 and BED formats. In addition, the archive enables access to previous RNAcentral releases.
In the following protocol, we demonstrate how to compare a set of genomic coordinates with a comprehensive collection of ncRNAs from RNAcentral by intersecting an RNAcentral BED file with an example GFF3 file (the same steps work with BED, VCF, or BAM files). This protocol can be used to find if a genomic region of interest overlaps with RNAcentral sequences as part of RNA-seq data analysis (see Support Protocol 3 for an alternative workflow using Galaxy).
Necessary Resources
Hardware
- A computer with access to UNIX terminal and the Internet
Software
- BedTools (see Current Protocols article: Quinlan, 2014); curl; gzip; grep
Compare a set of genome coordinates with RNAcentral genome annotations
1.Go to the RNAcentral FTP archive and select a BED file for your species of interest (in this example we use Drosophila melanogaster). Download and uncompress the file:
- curl
- `ftp://ftp.ebi.ac.uk/pub/databases/RNAcentral/releases/14.0/genome_coordinates/bed/drosophila_melanogaster.BDGP6.22.bed.gz' | gzip -d > rnacentral.bed
2.Use the Supporting Information file example.gff3 to intersect it with the RNAcentral BED file using bedtools (this step also works with BED, VCF, or BAM files):
- bedtools intersect -a rnacentral.bed -b example.gff3 > output.bed
3.Filter results using RNAcentral-specific metadata BED fields. In addition to the standard fields, the RNAcentral BED files contain two additional columns (RNA type and a list of source databases) that enable selecting a subset of sequences using a command-line tool like grep (Fig. 7). For example, it is possible to filter the genomic regions by RNA type, member database, and other criteria using grep commands, as in the following.
-
Filter by RNA type (for example, exclude tRNA):
-
grep -v tRNA output.bed > output-no-trna.bed
-
b.Filter by RNAcentral member database (for example, FlyBase):
-
grep FlyBase output.bed > output-flybase.bed

Support Protocol 1: USING WEB APIs FOR PROGRAMMATIC DATA ACCESS
In addition to accessing the RNAcentral data through the website, it is possible to use it programmatically by taking advantage of two web APIs:
- 1.The RNAcentral API can be used to retrieve information about individual ncRNA entries;
- 2.The text search API can be used to search and retrieve information about RNAcentral entries as described in Basic Protocol 1.
These APIs can be used independently or cooperatively, depending on the use case, as explained below.
Necessary Resources
Hardware
- A computer with access to UNIX terminal and the Internet
Software
- To interact with the API, you will need to run custom programs in your preferred programming language. The following example is written in Python and requires the requests package.
Using text search API
The RNAcentral text search (see Basic Protocol 2) is powered by the EMBL-EBI Search engine and has a REST API that can be used from any programming language that supports retrieving data over the Internet, such as Python or JavaScript (Madeira et al., 2019). This protocol covers basic usage of the API; for more detailed information about programmatic access, please refer to the EMBL-EBI Search documentation (https://www.ebi.ac.uk/ebisearch) and the RNAcentral API help page (https://rnacentral.org/api). Here we demonstrate how to programmatically perform a search for has-mir-126 sequences from miRBase (Kozomara et al., 2019) using the text search, and then retrieve the description and RNA type of the results.
1a. Use a programming language to access the following URL:
This URL is equivalent to searching "hsa-mir-126" AND expert_db: "mirbase" on the RNAcentral website (see Basic Protocol 2). In Python this is done with:
- import requests
- response = requests.get("https://www.ebi.ac.uk/ebisearch/ws/rest/rnacentral?query=%22hsa-mir-126%22%20AND%20expert_db:%22mirbase%22&fields=description,rna_type&format=json")
2a. Display the data from the text search API. In Python, this is done with:
- data = response.json()
- print(data)

Combining text search and RNAcentral APIs
The RNAcentral text search and the RNAcentral API can be used together to search and access RNAcentral data. For example, in the previous section we retrieved several fields directly from the text search (description and rna_type, see Fig. 8). However, not all of the metadata about the sequence is available in the text search. Most importantly, the nucleotide sequence is not part of the text search index, but it can be accessed via the RNAcentral API.
1b. Use a programming language to lookup the sequence for the results from above. In python this can be done with:
- for entry in data[ʹentriesʹ]:
- urs = entry[ʹidʹ].split(ʹ_ʹ)[0]
- response = requests.get(ʹhttps://rnacentral.org/api/v1/rna/%s.fastaʹ%urs)
- sequence = response.text
- print(sequence)

Support Protocol 2: USING PUBLIC POSTGRES DATABASE TO EXPORT LARGE DATASETS
A public copy of the RNAcentral Postgres database is made available in order to enable users to query RNAcentral in any programming language with database connectivity. This functionality can be used to automate data export or to export large datasets that cannot be downloaded from the RNAcentral website. The database is updated with each RNAcentral release and contains a copy of the data available through the RNAcentral website.
The database connection details can be found in Table 1.We recommend using a Postgres client like DBeaver or PgAdmin for exploring the schema and testing SQL queries, but for exporting large volumes of data, it is best to use a command-line client.
| Hostname | hh-pgsql-public.ebi.ac.uk |
|---|---|
| Port | 5432 |
| Database | Pfmegrnargs |
| User | Reader |
| Password | NWDMCE5xdipIjRrp |
- a
See https://rnacentral.org/help/public-database for the most up-to-date version.
Necessary Resources
Hardware
- A computer with access to UNIX terminal and the Internet
Software
- A Postgres database management application, such as DBeaver (https://dbeaver.io/) or PgAdmin (https://www.pgadmin.org/); psql for connecting to the database on command line or Docker for using Docker containers; seqkit (Shen, Le, Li, & Hu, 2016) for extracting sequences
Example workflow to extract all bacterial rRNA sequences
The RNAcentral text search can export up to 1 million search results, but you can extract a larger number of sequences using the Postgres database, as described below. Connection details for the database are in Table 1.
1.Connect to the RNAcentral Postgres database. If you have psql installed on your machine, use the following command:
- psql postgres://reader:NWDMCE5xdipIjRrp@hh-pgsql-public.ebi.ac.uk:5432/pfmegrnargs
Alternatively, use docker to get started with a pre-configured Postgres image:
- docker pull postgres
- docker run -it postgres psql postgres://reader:NWDMCE5xdipIjRrp@hh-pgsql-public.ebi.ac.uk:5432/pfmegrnargs
2.Create a file query.sql (Fig. 10) with an SQL statement that retrieves RNAcentral identifiers of all bacterial rRNA sequences (matches 3,863,753 sequences in RNAcentral release 14).

3.Run the following command to execute the query if psql is installed on your machine:
- psql -t -A -f query.sql postgres://reader:NWDMCE5xdipIjRrp@hh-pgsql-public.ebi.ac.uk:5432/pfmegrnargs > ids.txt
If using Docker, run:
- docker run -v ∼pwd∼:/rnacentral -it postgres /bin/sh -c `cd /rnacentral && psql -t -A -f query.sql postgres://reader:NWDMCE5xdipIjRrp@hh-pgsql-public.ebi.ac.uk:5432/pfmegrnargs > ids.txt'
The command will create a file ids.txt with a list of RNAcentral identifiers, which can be used to fetch sequences from the FASTA files.
4.Download a file with RNAcentral sequences in FASTA format and place it into the same directory as ids.txt:
- ftp://ftp.ebi.ac.uk/pub/databases/RNAcentral/current_release/sequences/rnacentral_species_specific_ids.fasta.gz
5.Extract RNA sequences with the identifiers listed in file ids.txt using seqkit:
- seqkit grep -f ids.txt rnacentral_species_specific_ids.fasta.gz > output.fasta
In Docker this can be done with:
- docker pull pegi3s/seqkit
- docker run -v ∼pwd∼:/rnacentral -it pegi3s/seqkit /bin/sh -c `cd /rnacentral && seqkit grep -f ids.txt rnacentral_species_specific_ids.fasta.gz > output.fasta'
Support Protocol 3: ANALYZE NON-CODING RNA IN RNA-seq DATASETS USING RNAcentral AND GALAXY
RNA-seq experiments can provide information about gene expression in the cells of interest. There are a wide range of RNA-seq technologies (Stark, Grzelak, & Hadfield, 2019) targeting different types of transcripts. For example, TGIRT-seq (Nottingham et al., 2016) uses the thermostable group II intron reverse transcriptase and can process highly structured, short RNAs, such as tRNAs and snoRNAs (Boivin et al., 2018, 2020).
In the following protocol, we demonstrate an example RNA-seq workflow using Galaxy, a web platform that enables users to perform computational workflows in the cloud (Afgan et al., 2018). We will analyze a single-end RNA-seq dataset based on Drosophila melanogaster S2 cells sequenced under normal conditions and amino-acid starvation (project PRJNA601750, https://www.ebi.ac.uk/ena/data/view/PRJNA601750). We will use the RNAcentral ncRNA annotations to compare ncRNA expression between the two conditions.
The workflow includes the following steps:
- 1.Importing the data.
- 2.Preprocessing the reads using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and aligning them to the reference genome with HISAT2 (Kim, Langmead, & Salzberg, 2015).
- 3.Counting reads using the FeatureCounts software (Liao, Smyth, & Shi, 2014) and ncRNA annotations from RNAcentral.
- 4.Performing differential expression (DE) analysis using DESeq2 (Love, Huber, & Anders, 2014).
Necessary Resources
Hardware
- Any device with Internet access
Software
- An up-to-date Web browser, such as Chrome, Safari, or Firefox
- Optional: IGV (Robinson et al., 2011).
Files
- RNA-seq dataset (FASTQ format)
- RNAcentral annotations (GFF3 format)
- Reference genome (FASTA format)
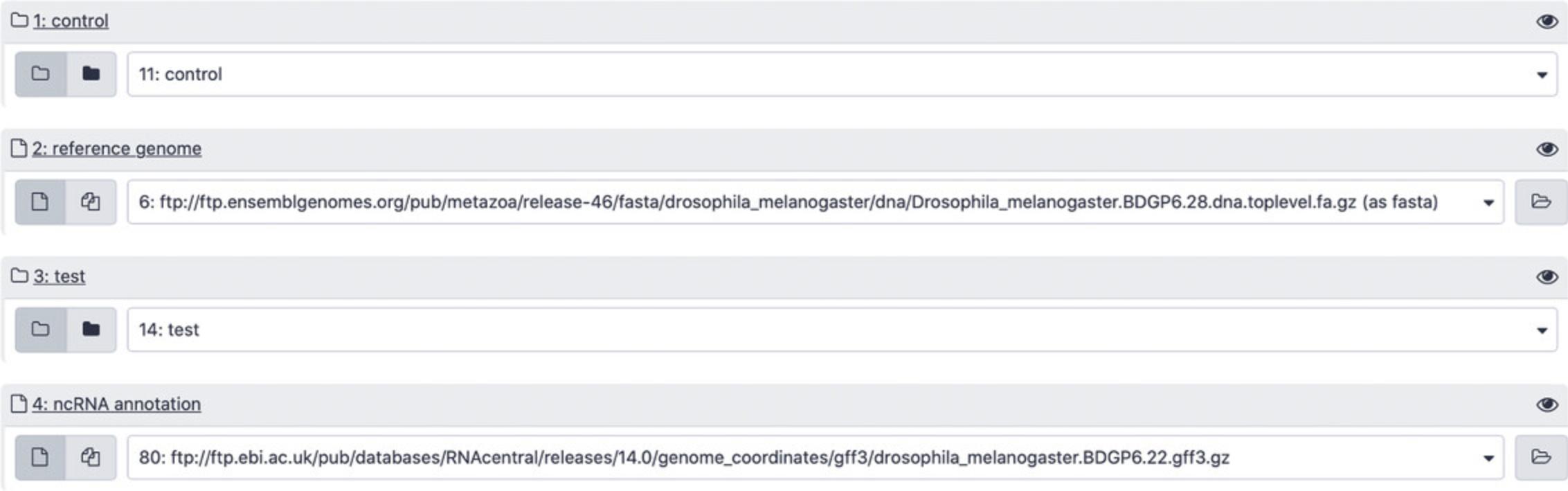
1.Import RNA-seq data into Galaxy.
An RNA-seq dataset can be uploaded to Galaxy from a local computer or imported directly from the biological databases, an FTP archive, or any public URL.
-
Begin at the Galaxy homepage (https://usegalaxy.org/) and log in to your Galaxy account. Click the “+” icon in the History panel to create a new history and keep track of input and output files (Fig.11, right).
-
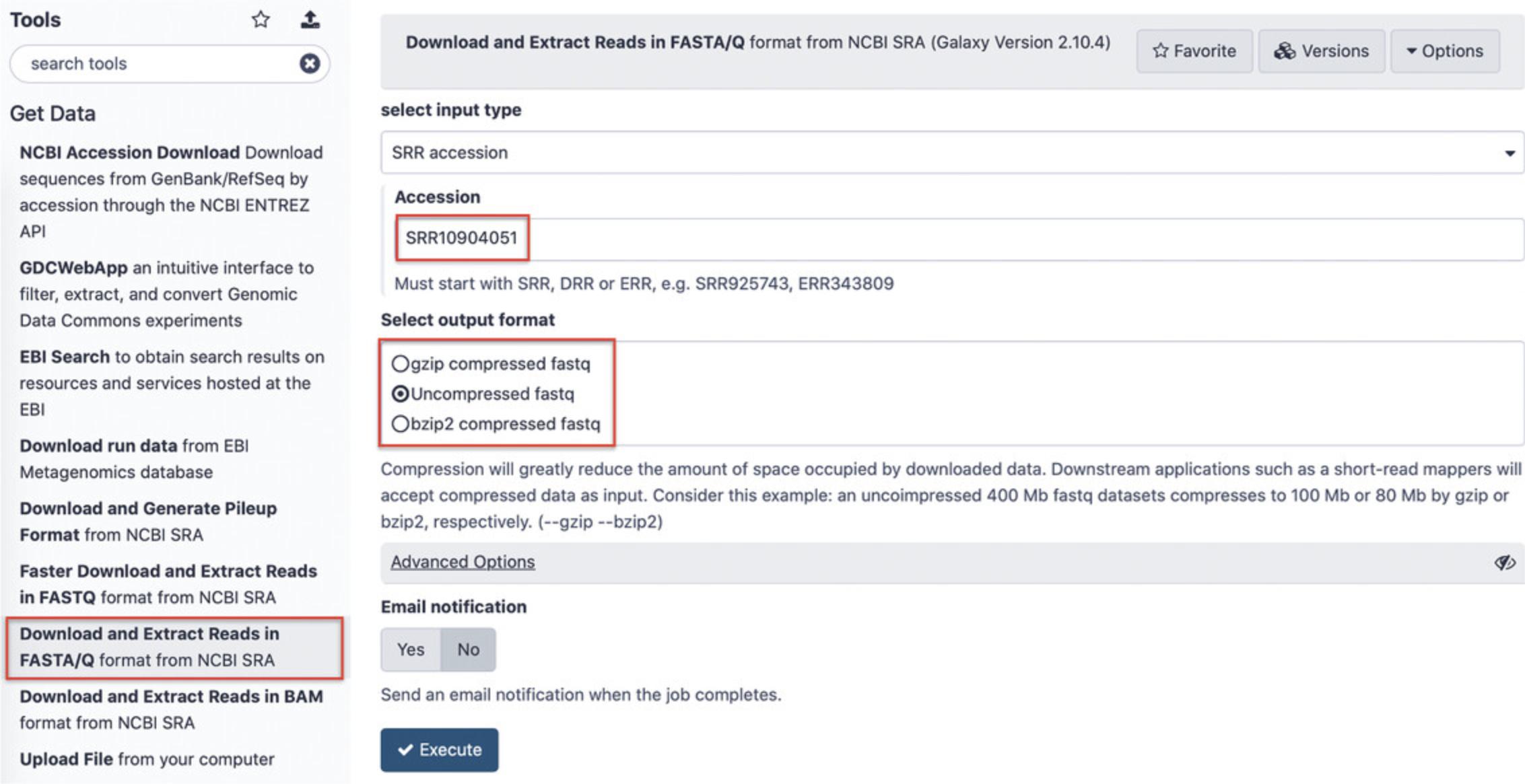
Find the section “Get data” in the Tools panel (Fig.11, left). Click “Download and Extract Reads in FASTA/Q format from NCBI SRA” in order to import data directly from the NCBI SRA database (Amid et al.,2020). Enter the four SRR accessions from Table2as shown in Figure12(each accession needs to be imported separately).
| Condition | Accessions |
|---|---|
| Control (normal conditions) | SRR10904051, SRR10904052 |
| Test (amino-acid starvation) | SRR10904053, SRR10904054 |
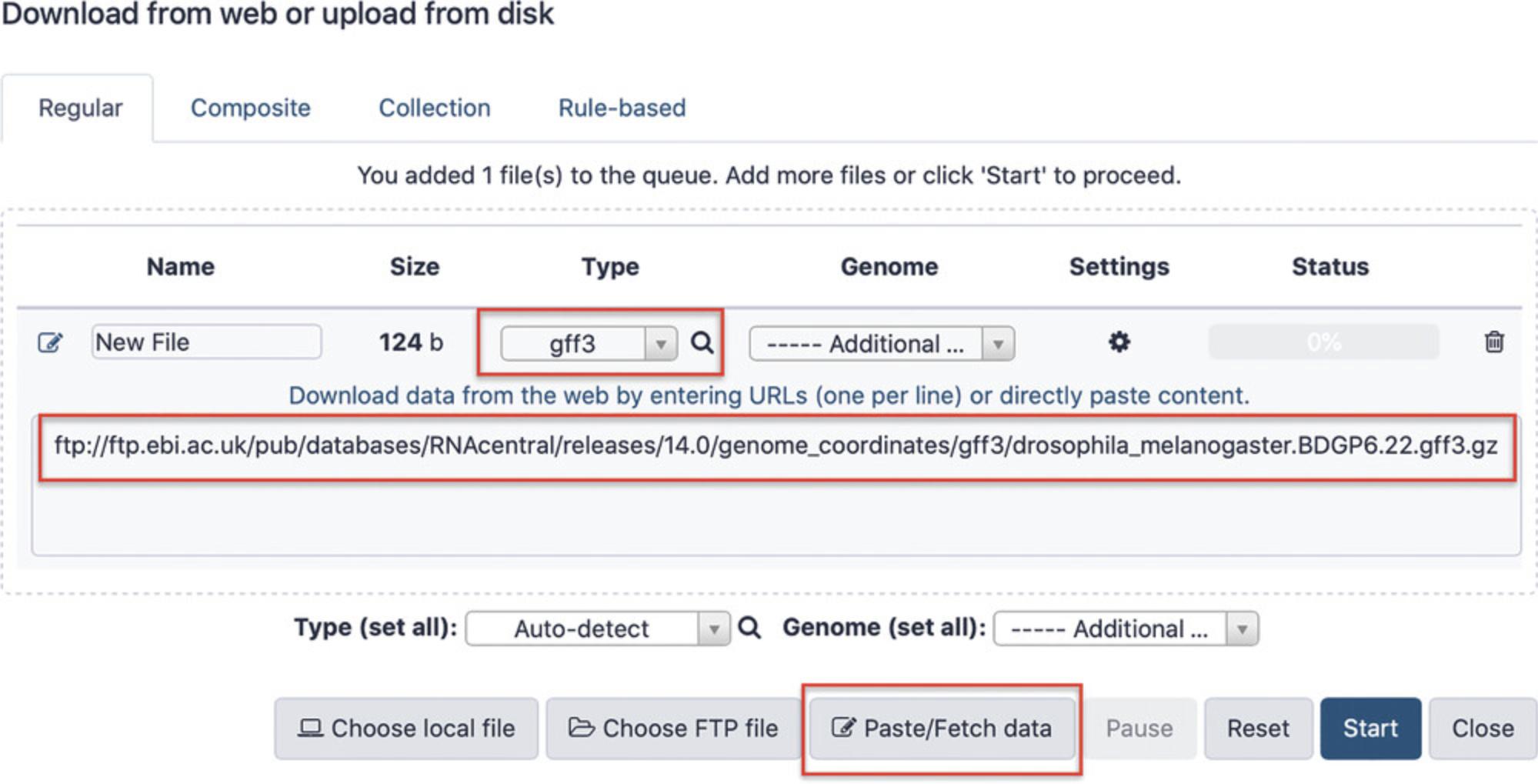
2.Import ncRNA annotations from RNAcentral.
-
Copy the following stable FTP address that points to a GFF3 file with RNAcentral annotations forDrosophila melanogaster:
- ftp://ftp.ebi.ac.uk/pub/databases/RNAcentral/releases/14.0/genome_coordinates/gff3/drosophila_melanogaster.BDGP6.22.gff3.gz.
For reproducibility purposes, the link above refers to RNAcentral release 14, but in general it is recommended to use the most recent RNAcentral version. The up-to-date GFF3 files can be found by navigating to the RNAcentral FTP archive (https://rnacentral.org/downloads) and browsing the genome_coordinates folder of the current release.
- b.Go to the Galaxy dashboard and click the arrow icon in the upper right of the Tools panel, select “Paste/Fetch data,” enter the GFF3 file address, and specify “gff3” for the file Type (Fig. 13).
3.Upload the reference genome.
-
Select the reference genome to use. Depending on the genome, you can download the FASTA format from Ensembl or the UCSC Genome browser (Kent et al.,2002).
-
Ensembl:ftp://ftp.ensembl.org/pub/current_fasta
-
UCSC Genome browser:ftp://hgdownload.soe.ucsc.edu/goldenPath/
Make sure that the genome assembly of the reference genome matches the assembly version available in RNAcentral.
-
Import the reference genome, as with the RNAcentral GFF3 file in the previous step. Click the arrow icon in the upper right of the Tools panel, select “Paste/Fetch data,” and enter the FASTA file address.
In this example, the following Ensembl file is used:
- ftp://ftp.ensemblgenomes.org/pub/metazoa/release-46/fasta/drosophila_melanogaster/dna/Drosophila_melanogaster.BDGP6.28.dna.toplevel.fa.gz
4.Import the Galaxy workflow.
-
At the top of the Galaxy page locate the “Workflow” tab that contains all available workflows.
-
Click the “Import” button to upload a Galaxy workflow from a local file (see Supporting Information fileRNAcentral-single-end-workflow.ga) or follow the URL below and click the “+” icon to add the workflow to your Galaxy account:https://usegalaxy.org/u/rnacentral/w/current-protocols-workflow.
-
Go to the “Workflow” tab again, click the newly imported workflow, and select “Edit” to view the steps (Fig.14).
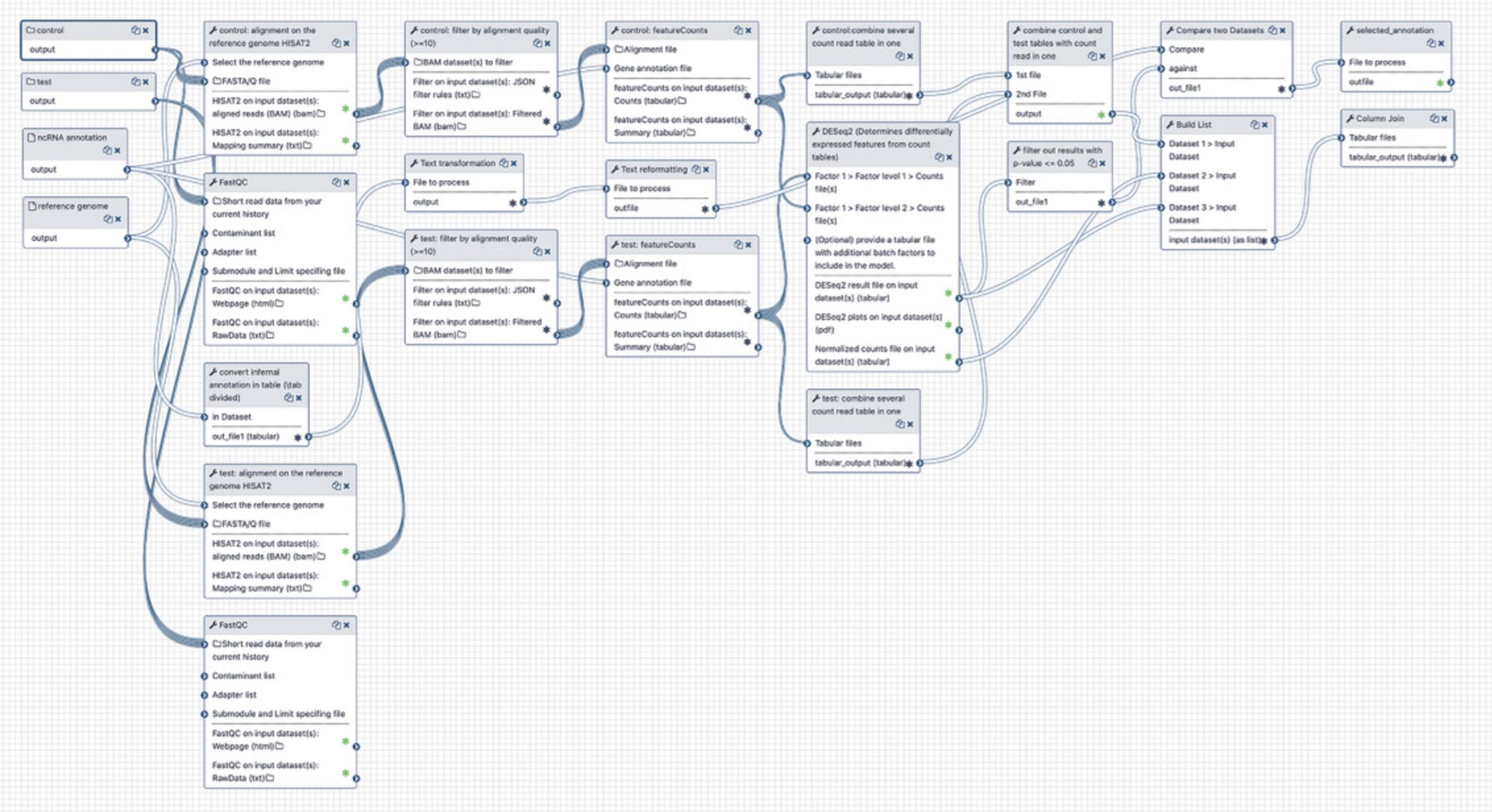
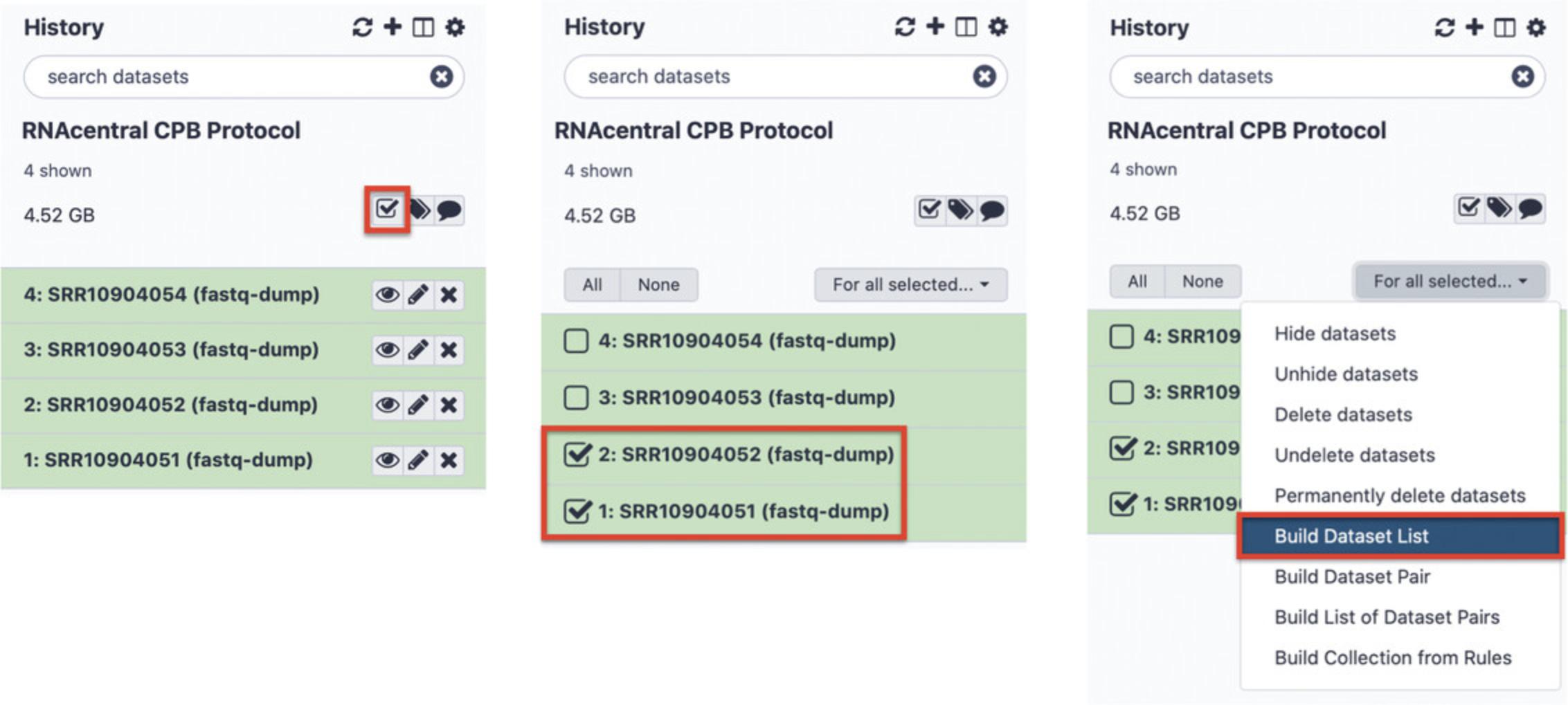
5.Prepare RNA-seq data for differential expression analysis.
The data files must be grouped by experimental condition into the Galaxy “Dataset Lists” (for example, healthy and cancerous samples, or different stages of embryonic development). All replicates/samples for each condition must be located in one Dataset List. Make sure that your FASTQ files are unzipped.
-
Go the Galaxy dashboard and click the checkbox icon in the History panel. Select all FASTQ replicates/samples that should be in one list (Fig.15, left and center).
-
Click “For all selected…” and select “Build Dataset List” to create a folder with the selected samples (Fig.15, right). In this example, SRR10904051 and SRR10904052 should be grouped and called “control” while SRR10904053 and SRR10904054 should be grouped and called “test” or similar.
If a sample is subdivided into several FASTQ files (which does not apply to this example), then before creating the Dataset List, the FASTQ files from the same sample should be combined using the “Collapse collection” function (available under “Collection operations” in the Tools section).
Detailed instructions on combining multiple samples in collections can be found in a Galaxy tutorial (https://galaxyproject.org/tutorials/collections/).
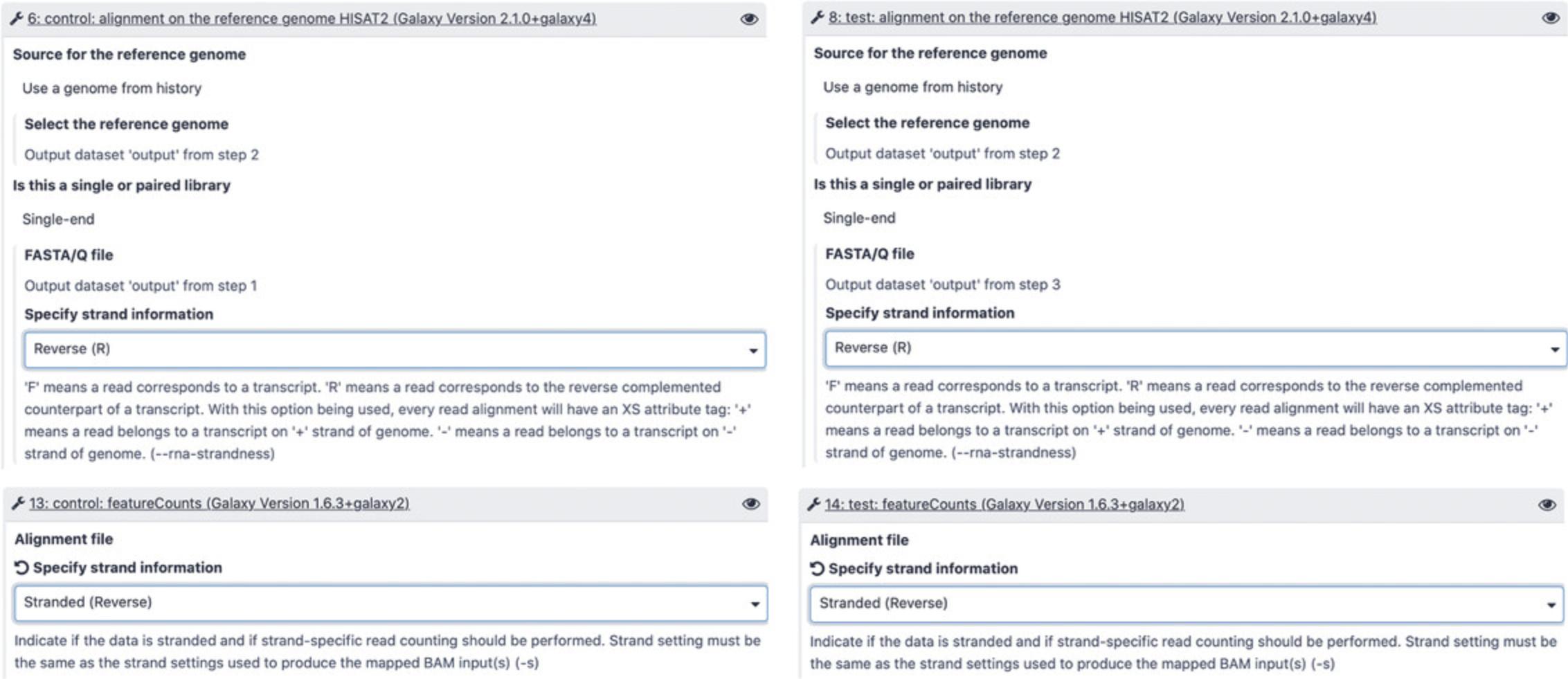
6.Run Galaxy workflow.
The main purpose of this workflow is the DE analysis of ncRNAs under different conditions. The workflow aligns the FASTQ files onto the reference genome to produce BAM files, analyses the alignments, counts reads using the RNAcentral annotations, normalizes count matrices, and performs the DE analysis using DESeq2.
-
Click “Workflows” at the top of the webpage, select the imported workflow and choose “Run.”
-
Select workflow inputs (Fig.16):
control : a Dataset List containing the control samples. In this example, it is the dataset list containing files SRR10904051 and SRR10904052 where cells were grown under normal conditions
test : a Dataset List containing samples subjected to amino-acid starvation (files SRR10904053 and SRR10904054)
reference genome : the Drosophila melanogaster reference genome
ncRNA annotation : RNAcentral ncRNA annotations or any other annotations in GFF3 format
- c.Change individual tool settings. Depending on the sequencing technology, the strand-specificity options need to be specified for the HISAT2 and featureCounts tools. Since in this example the sample was prepared using the Illumina TruSeq Stranded Total RNA library kit, the “Reversed” option should be selected in test and control steps for HISAT2 and featureCounts (Fig. 17). When following this protocol for analyzing different datasets, make sure to specify the correct strandedness depending on your specific use case.
- d.Click “Run workflow.”
7.Examine workflow outputs.
When all tasks in the History panel turn green, the results are ready and you should see the following outputs:
- A selected_annotation GFF3 file containing ncRNAs with statistically significant changes in expression levels between the control and test conditions. Click the eye icon (Fig. 18, left) to view the data (Fig. 18, right).
- DEseq2 output files (DESeq2_results, a table containing p -values and other statistics for each ncRNA, DESeq2_plots with DESeq2 graphical outputs, and DESeq2_normalized_table, a table with normalized read counts).
In addition, the History panel will contain quality control reports from FASTQC, alignments of the reads to the reference genome (BAM format), and Count read matrix for all samples.
The precomputed results are stored in a public Galaxy history and can be viewed at https://usegalaxy.org/u/rnacentral/h/rnacentral-cpb-protocol.
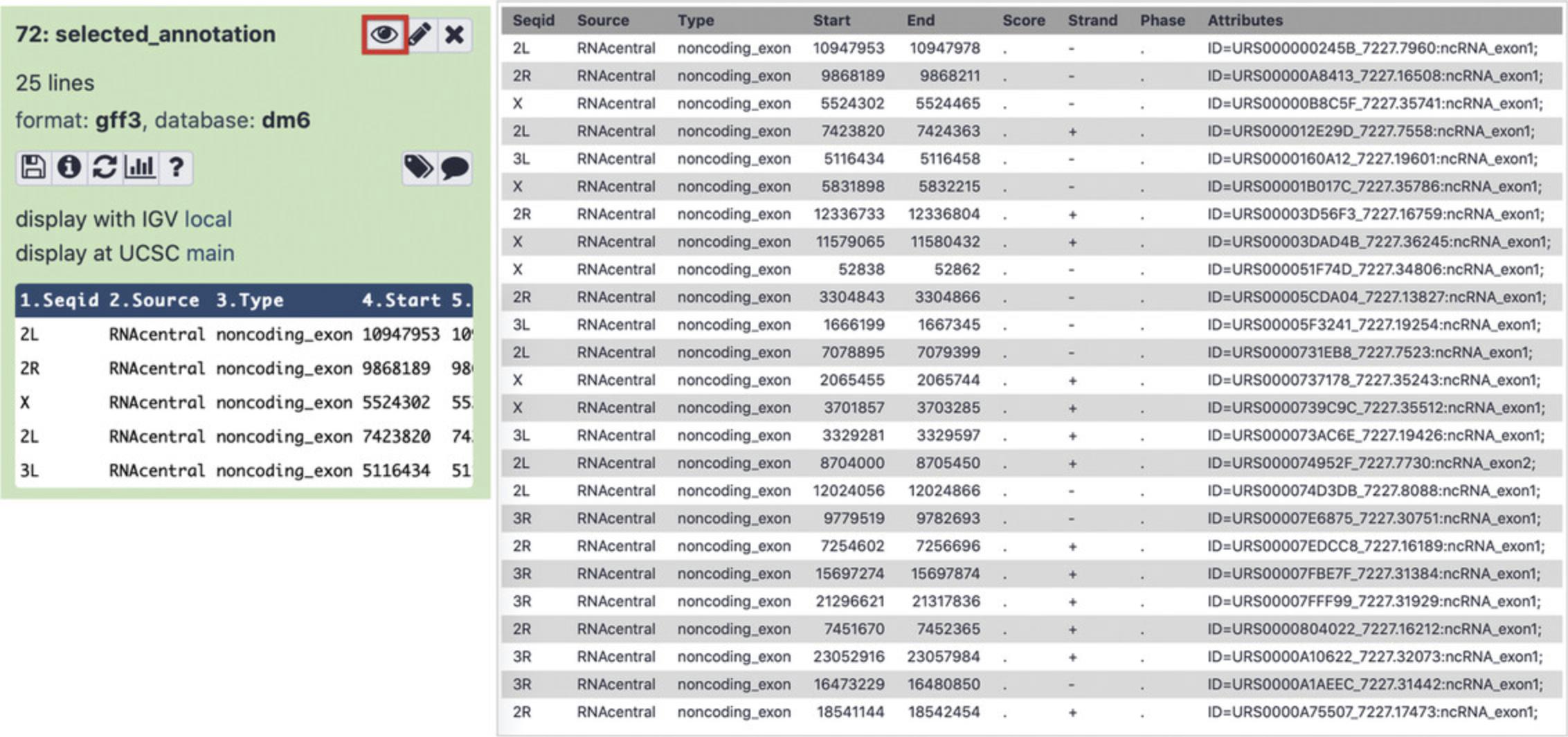
8.Visualize results locally using IGV.
To visualize the genome location of the differentially expressed ncRNAs, you can use the IGV software (Robinson et al., 2011).
-
To specify the species and the genome assembly of the output GFF3 file, find theselected_annotationfile in the History panel and click “Edit attributes” pencil icon. Next, choose thedm6genome assembly in the “Database/Build” drop-down list.
-
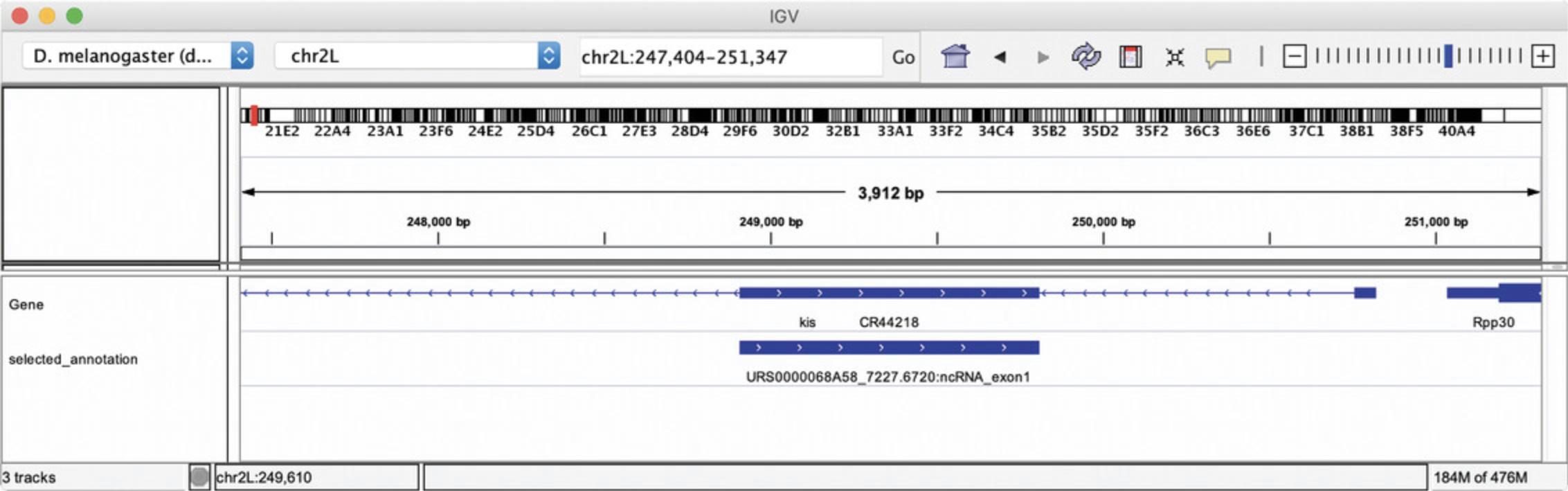
Open the IGV application on your computer and click “display with IGV local” in the Galaxy History panel. You can navigate the genome to zoom in to the differentially expressed RNAs shown in theselected_annotationIGV track (Fig.19).
GUIDELINES FOR UNDERSTANDING RESULTS
RNAcentral identifiers
In RNAcentral, each distinct ncRNA sequence is assigned a Unique RNA Sequence identifier (URS ID), which is stable across releases. As the same sequence can be observed in multiple species, RNAcentral also supports species-specific identifiers (The RNAcentral Consortium, 2017), which consist of the URS ID joined with the NCBI taxid (Federhen, 2012) for the species where the sequence occurs.
For example, URS00004BFD1E_9606 refers to the human hsa-let-7f-1 microRNA, while URS00004BFD1E_9544 refers to the same sequence in rhesus macaque. Note that the URS ID is the same in both cases while 9606 and 9544 are the NCBI taxids for human and rhesus macaque, respectively.
The URS IDs can be marked inactive if there are no current cross references. For example, at the time of this writing, URS000063488A is inactive. This tRNA sequence was present in the Rfam database version 12.0; however, later Rfam versions did not include this sequence. Since there are no other cross references to this sequence, it is marked as inactive.
The URS IDs are never deleted or reassigned, and they can always be accessed on the RNAcentral website using direct URLs in the following format: https://rnacentral.org/rna/
Genome mapping
RNAcentral genome annotations are based on a comprehensive mapping procedure that aligns all sequences without genome coordinates to the corresponding reference genome (The RNAcentral Consortium, 2019). If a member database provides the genome locations to RNAcentral when submitting the data, these coordinates are used and no mapping is performed. The source of genomic coordinates is specified both in the genome browser (Fig. 2C) and the GFF3 files in the FTP archive.
RNAcentral maps the sequences onto the most recent reference genome assembly from Ensembl. This procedure finds coordinates for ncRNA sequences from a database without an explicit connection to a genome, such as the ENA (Amid et al., 2020). The RNAcentral genome mapping helped improve model organism database annotations. For example, ten D. melanogaster snoRNA genes from the ENA database were added to FlyBase after these sequences were mapped to the fly genome in RNAcentral (The RNAcentral Consortium, 2019).
Note that the current procedure reports all genome alignments, and the results may require additional filtering. For example, short sequences, such as piRNAs can be similar to multiple genome regions, including those outside the piRNA clusters. Depending on the use case, such sequences can be excluded using a strategy described in Basic Protocol 4.
Alternative Galaxy workflows
Note that the results of the workflow described in Support Protocol 3 require additional interpretation and analysis. The readers are referred to the protocols dedicated to RNA-seq analysis for further information (see Current Protocols article: Ji & Sadreyev, 2018; also see Yalamanchili, Wan, & Liu, 2017). The workflow described above may require modifications if newer software versions become available in Galaxy, or depending on the specific RNA-seq technology. For example, paired-end RNA-seq datasets should be processed using a different workflow (see https://rnacentral.org/help/galaxy for an example).
If ncRNA annotations for the genome of interest is not available in RNAcentral, the genome can be annotated with Rfam covariance models and Infernal (Nawrocki & Eddy, 2013) using the steps outlined in the Rfam protocol (see Current Protocols article: Kalvari et al., 2018). In addition, RNAcentral sequences from a related taxonomic group can be mapped onto the genome using BLAST, blat, or other software (for example, all Diptera sequences from RNAcentral can be aligned to a newly sequenced fly genome). These workflows are discussed in more detail at https://rnacentral.org/help/galaxy.
COMMENTARY
Background Information
Non-coding RNAs
Non-coding RNAs (ncRNAs) are transcribed from the DNA similar to messenger RNAs, but are not translated into proteins. ncRNAs are found in all organisms and have a broad range of functions. For example, tRNA and rRNA are required for protein synthesis and are essential for all life, while the functions of many lncRNAs are still unclear. In humans, ncRNA expression has been tied to a variety of diseases such as ovarian cancer (Huang et al., 2002), hearing impairment (Finnilä & Majamaa, 2003), and dermatomyositis (Eisenberg et al., 2007). Due to their importance to cellular function, it is important to consider not only protein-coding genes, but also ncRNAs when analyzing RNA-seq datasets, as shown in Support Protocol 3.
Critical Parameters and Troubleshooting
Sequence naming and RNA type
RNAcentral provides descriptions and RNA type for all sequences. These annotations are essential to understanding the function of any RNA sequence, but there are some important factors to consider. These annotations are computed automatically from the descriptions and RNA types provided by the member databases. No description or RNA type is assigned manually in RNAcentral, although sequences from certain member databases, such as GENCODE or HGNC, may be manually curated. Additionally, member databases may disagree on an annotation. In such cases, RNAcentral strives to pick the annotations that are most consistent with the available data.
Transcript-level organization
RNAcentral is currently organized at the level of individual transcripts. This means that if several databases provide different sequences for the same RNA gene, all of these sequences will be available in RNAcentral under separate URS IDs. In order to visualize these related sequences, you can use the embedded genome browser on sequence report pages, as similar sequences will be mapped to the same genomic region (see Basic Protocol 1).
For example, Figure 20 shows the details of the overlapping RNAcentraltranscripts corresponding to the human miR-181b-1 microRNA. There are two alternative versions of the precursor sequence, three alternative 5′ and two 3′ mature sequences. These entries come from different databases and have different types of annotations. At the time of writing, to get a complete picture of all the information available for this microRNA, it is recommended to view all seven sequence reports, as demonstrated in Basic Protocol 1. Work is underway to create gene-level entries that would aggregate and prioritize transcripts from the same gene.

Acknowledgments
We would like to thank the RNAcentral Consortium for contributing data to RNAcentral. We would like to thank Alexandra Martin-Geary (University of Manchester) for providing feedback about the Galaxy protocol. This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) [BB/N019199/1]. Funding for open access charge: Research Councils UK (RCUK). This work is also supported by core EMBL funds. Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Blake A. Sweeney : Investigation; methodology; software; supervision; visualization; writing-original draft; writing-review & editing. Arina A. Tagmazian : Data curation; investigation; methodology; software; visualization; writing-original draft; writing-review & editing. Carlos E. Ribas : Software; validation. Robert D. Finn : Funding acquisition; project administration; supervision; validation; writing-review & editing. Alex Bateman : Funding acquisition; project administration; supervision; writing-review & editing. Anton I. Petrov : Conceptualization; methodology; project administration; software; supervision; validation; visualization; writing-original draft; writing-review & editing.
Supporting Information
| Filename | Description |
|---|---|
| SupportingInformation.zip6.8 KB | Supplementary Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., … Blankenberg, D. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research , 46(W1), W537–W544. doi: 10.1093/nar/gky379.
- Amid, C., Alako, B. T. F., Balavenkataraman Kadhirvelu, V., Burdett, T., Burgin, J., Fan, J., … Cochrane, G. (2020). The European Nucleotide Archive in 2019. Nucleic Acids Research , 48(D1), D70–D76.
- Bateman, A., Agrawal, S., Birney, E., Bruford, E. A., Bujnicki, J. M., Cochrane, G., … Zwieb, C. (2011). RNAcentral: A vision for an international database of RNA sequences. RNA , 17(11), 1941–1946. doi: 10.1261/rna.2750811.
- Boivin, V., Deschamps-Francoeur, G., Couture, S., Nottingham, R. M., Bouchard-Bourelle, P., Lambowitz, A. M., … Abou-Elela, S. (2018). Simultaneous sequencing of coding and noncoding RNA reveals a human transcriptome dominated by a small number of highly expressed noncoding genes. RNA , 24(7), 950–965. doi: 10.1261/rna.064493.117.
- Boivin, V., Reulet, G., Boisvert, O., Couture, S., Elela, S. A., & Scott, M. S. (2020). Reducing the structure bias of RNA-Seq reveals a large number of non-annotated non-coding RNA. Nucleic Acids Research , 48, 2271-2286. doi: 10.1093/nar/gkaa028.
- Cunningham, F., Achuthan, P., Akanni, W., Allen, J., Amode, M. R., Armean, I. M., … Flicek, P. (2019). Ensembl 2019. Nucleic Acids Research , 47(D1), D745–D751. doi: 10.1093/nar/gky1113.
- Eisenberg, I., Eran, A., Nishino, I., Moggio, M., Lamperti, C., Amato, A. A., … Kunkel, L. M. (2007). Distinctive patterns of microRNA expression in primary muscular disorders. Proceedings of the National Academy of Sciences of the United States of America , 104(43), 17016–17021. doi: 10.1073/pnas.0708115104.
- Federhen, S. (2012). The NCBI Taxonomy database. Nucleic Acids Research , 40(Database issue), D136–D143. doi: 10.1093/nar/gkr1178.
- Finnilä, S., & Majamaa, K. (2003). Lack of a modulative factor in locus 8p23 in a Finnish family with nonsyndromic sensorineural hearing loss associated with the 1555A>G mitochondrial DNA mutation. European Journal of Human Genetics: EJHG , 11(9), 652–658.
- Fromm, B., Domanska, D., Høye, E., Ovchinnikov, V., Kang, W., Aparicio-Puerta, E., … Peterson, K. J. (2020). MirGeneDB 2.0: The metazoan microRNA complement. Nucleic Acids Research , 48(D1), D1172. doi: 10.1093/nar/gkz885.
- Huang, K.-C., Rao, P. H., Lau, C. C., Heard, E., Ng, S.-K., Brown, C., … Ng, S.-W. (2002). Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Molecular Cancer Therapeutics , 1(10), 769–776.
- Huntley, R. P., Sawford, T., Mutowo-Meullenet, P., Shypitsyna, A., Bonilla, C., Martin, M. J., & O'Donovan, C. (2014). The GOA database: Gene ontology annotation updates for 2015. Nucleic Acids Research , 43(D1), D1057–D1063. doi: 10.1093/nar/gku1113.
- Ji, F., & Sadreyev, R. I. (2018). RNA-seq: Basic bioinformatics analysis. Current Protocols in Molecular Biology , 124(1), e68. doi: 10.1002/cpmb.68.
- Kalvari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., … Petrov, A. I. (2017). Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Research , 41(D1), D335–342, doi: 10.1093/nar/gkx1038.
- Kalvari, I., Nawrocki, E. P., Argasinska, J., Quinones-Olvera, N., Finn, R. D., Bateman, A., & Petrov, A. I. (2018). Non-coding RNA analysis using the Rfam database. Current Protocols in Bioinformatics , 62, e51. doi: 10.1002/cpbi.51.
- Karagkouni, D., Paraskevopoulou, M. D., Chatzopoulos, S., Vlachos, I. S., Tastsoglou, S., Kanellos, I., … Hatzigeorgiou, A. G. (2018). DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA—gene interactions. Nucleic Acids Research , 46(D1), D239–D245. doi: 10.1093/nar/gkx1141.
- Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., & Haussler, D. (2002). The human genome browser at UCSC. Genome Research , 12(6), 996–1006. doi: 10.1101/gr.229102.
- Kim, D., Langmead, B., & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods , 12(4), 357–360. doi: 10.1038/nmeth.3317.
- Kozomara, A., Birgaoanu, M., & Griffiths-Jones, S. (2019). miRBase: From microRNA sequences to function. Nucleic Acids Research , 47(D1), D155–D162. doi: 10.1093/nar/gky1141.
- Liao, Y., Smyth, G. K., & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics , 30(7), 923–930. doi: 10.1093/bioinformatics/btt656.
- Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology , 15(12), 550. doi: 10.1186/s13059-014-0550-8.
- Machnicka, M. A., Milanowska, K., Osman Oglou, O., Purta, E., Kurkowska, M., Olchowik, A., … Grosjean, H. (2013). MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Research , 41(Database issue), D262–D267.
- Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., … Lopez, R. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research , 47(W1), W636–W641. doi: 10.1093/nar/gkz268.
- Nawrocki, E. P., & Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics , 29(22), 2933–2935. doi: 10.1093/bioinformatics/btt509.
- Nottingham, R. M., Wu, D. C., Qin, Y., Yao, J., Hunicke-Smith, S., & Lambowitz, A. M. (2016). RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA , 22(4), 597–613. doi: 10.1261/rna.055558.115.
- Paraskevopoulou, M. D., Vlachos, I. S., Karagkouni, D., Georgakilas, G., Kanellos, I., Vergoulis, T., … Hatzigeorgiou, A. G. (2016). DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Research , 44(D1), D231–D238. doi: 10.1093/nar/gkv1270.
- Pignatelli, M., Vilella, A. J., Muffato, M., Gordon, L., White, S., Flicek, P., & Herrero, J. (2016). ncRNA orthologies in the vertebrate lineage. Database—The Journal of Biological Databases and Curation , 2016, bav127. doi: 10.1093/database/bav127.
- Quinlan, A. R. (2014). BEDTools: The swiss-army tool for genome feature analysis. Current Protocols in Bioinformatics , 47, 11.12.1–34. doi: 10.1002/0471250953.bi1112s47.
- Rappaport, N., Twik, M., Nativ, N., Stelzer, G., Bahir, I., Stein, T. I., … Lancet, D. (2014). MalaCards: A comprehensive automatically-mined database of human diseases. Current Protocols in Bioinformatics , 47(1), 1.24.1–1.24.19. doi: 10.1002/0471250953.bi0124s47.
- Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., & Mesirov, J. P. (2011). Integrative genomics viewer. Nature Biotechnology , 29(1), 24–26. doi: 10.1038/nbt.1754.
- Shen, W., Le, S., Li, Y., & Hu, F. (2016). SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PloS One , 11(10), e0163962. doi: 10.1371/journal.pone.0163962.
- Stark, R., Grzelak, M., & Hadfield, J. (2019). RNA sequencing: The teenage years. Nature Reviews. Genetics , 20(11), 631–656. doi: 10.1038/s41576-019-0150-2.
- The RNAcentral Consortium. (2017). RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Research , 45(D1), D128–D134. doi: 10.1093/nar/gkw1008.
- The RNAcentral Consortium. (2019). RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Research , 47(D1), D221–D229. doi: 10.1093/nar/gky1034.
- Wheeler, T. J., & Eddy, S. R. (2013). nhmmer: DNA homology search with profile HMMs. Bioinformatics , 29(19), 2487–2489. doi: 10.1093/bioinformatics/btt403.
- Yalamanchili, H. K., Wan, Y.-W., & Liu, Z. (2017). Data analysis pipeline for RNA-seq experiments: From differential expression to cryptic splicing. Current Protocols in Bioinformatics , 59, 11.15.1–11.15.21. doi: 10.1002/cpbi.33.
Key References
- The RNAcentral Consortium. (2015). RNAcentral: An international database of ncRNA sequences. Nucleic Acids Research , 43(D1), D123–D129. doi: 10.1093/nar/gku991.
Introduces the RNAcentral website and describes Unique RNA Sequence identifiers.
- The RNAcentral Consortium. (2017). See above.
Describes RNAcentral species-specific identifiers.
- The RNAcentral Consortium. (2019). See above.
Describes the comprehensive genomic mapping pipeline, Rfam classification, and various types of functional annotations.
Internet Resources
RNAcentral database.
RNAcentral help and documentation.
RNAcentral FTP archive.
Galaxy portal.
Citing Literature
Number of times cited according to CrossRef: 5
- Liliia Nurullina, Salvatore Terrosu, Alexander G. Myasnikov, Lasse Bohl Jenner, Marat Yusupov, Cryo‐EM structure of the inactive ribosome complex accumulated in chick embryo cells in cold‐stress conditions, FEBS Letters, 10.1002/1873-3468.14831, 598 , 5, (537-547), (2024).
- Christophe Penno, Julien Tremblay, Mary O’Connell Motherway, Virginie Daburon, Abdelhak El Amrani, Analysis of Small Non-coding RNAs as Signaling Intermediates of Environmentally Integrated Responses to Abiotic Stress, Plant Abiotic Stress Signaling, 10.1007/978-1-0716-3044-0_22, (403-427), (2023).
- Baibhav Kumar, Animesh Kumar, Sarika Jaiswal, Mir Asif Iquebal, Ulavappa B. Angadi, Rukam S. Tomar, Anil Rai, Dinesh Kumar, Genome-Wide Identification of Long Non-Coding RNAs in Pearl Millet (Pennisetum glaucum (L.)) Genotype Subjected to Drought Stress, Agronomy, 10.3390/agronomy12081976, 12 , 8, (1976), (2022).
- Marios A. Diamantopoulos, Konstantina K. Georgoulia, Andreas Scorilas, Identification and expression analysis of ten novel small non-coding RNAs (sncRNAs) in cancer cells using a high-throughput sequencing approach, Gene, 10.1016/j.gene.2021.146025, 809 , (146025), (2022).
- Blake A Sweeney, Anton I Petrov, Carlos E Ribas, Robert D Finn, Alex Bateman, Maciej Szymanski, Wojciech M Karlowski, Stefan E Seemann, Jan Gorodkin, Jamie J Cannone, Robin R Gutell, Simon Kay, Steven Marygold, Gil dos Santos, Adam Frankish, Jonathan M Mudge, Ruth Barshir, Simon Fishilevich, Patricia P Chan, Todd M Lowe, Ruth Seal, Elspeth Bruford, Simona Panni, Pablo Porras, Dimitra Karagkouni, Artemis G Hatzigeorgiou, Lina Ma, Zhang Zhang, Pieter-Jan Volders, Pieter Mestdagh, Sam Griffiths-Jones, Bastian Fromm, Kevin J Peterson, Ioanna Kalvari, Eric P Nawrocki, Anton S Petrov, Shuai Weng, Philia Bouchard-Bourelle, Michelle Scott, Lauren M Lui, David Hoksza, Ruth C Lovering, Barbara Kramarz, Prita Mani, Sridhar Ramachandran, Zasha Weinberg, RNAcentral 2021: secondary structure integration, improved sequence search and new member databases, Nucleic Acids Research, 10.1093/nar/gkaa921, 49 , D1, (D212-D220), (2020).

