A Platform for Medium-Throughput Cell-Free Analyses of Microtubule-Interacting Proteins Using Mammalian Cell Lysates
A. S. Jijumon, A. S. Jijumon, Arya Krishnan, Arya Krishnan, Carsten Janke, Carsten Janke
Abstract
The microtubule (MT) cytoskeleton performs a variety of functions in cell division, cell architecture, neuronal differentiation, and ciliary beating. These functions are controlled by proteins that directly interact with MTs, commonly referred to as microtubule-associated proteins (MAPs). Out of the many proteins reported interact with MTs, only a some have been biochemically and functionally characterized so far. One of the limitations of classical in vitro assays and single-MT reconstitution approaches is that they are typically performed with purified proteins. As purification of proteins can be difficult and time-consuming, many previous studies have only focused on a few proteins, while systematic analyses of many different proteins by in vitro reconstitution assays were not possible. Here we present a detailed protocol using lysates of mammalian cells instead of purified proteins that overcomes this limitation. Those lysates contain all molecular components required for in vitro MT reconstitution including the endogenous tubulin and the recombinant MAPs, which form MT assemblies upon the injection of the lysates into a microscopy chamber. This allows to directly observe the dynamic behavior of growing MTs, as well as the fluorescently labeled associated proteins by total internal reflection fluorescence (TIRF) microscopy. Strikingly, all proteins tested so far were functional in our approach, thus providing the possibility to test virtually any protein of interest. This also opens the possibility to screen the impact of patient mutations on the MT binding behavior of MAPs in a medium-throughput manner. In addition, the lysate approach can easily be adapted to other applications that have predominantly been performed with purified proteins so far, such as investigating other cytoskeletal systems and cytoskeletal crosstalk, or to study structures of MAPs bound to MTs by cryo-electron microscopy. Our approach is thus a versatile, expandable, and easy-to-use method to characterize the impact of a broad spectrum of proteins on cytoskeletal behavior and function. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Preparation of lysates of human cells for TIRF reconstitution assays
Basic Protocol 2 : Quantification of GFP-tagged MAP concentration in cell lysates
Support Protocol 1 : Purification of KIF5B(N555/T92A) (dead kinesin) protein for TIRF reconstitution assays
Support Protocol 2 : Preparation of GMPCPP MT seeds for TIRF reconstitution assays
Basic Protocol 3 : TIRF-based MT-MAP reconstitution assays using cell lysates
INTRODUCTION
The microtubule (MT) cytoskeleton plays key roles in many cellular processes in every eukaryotic cell. MTs control cell shape, cell locomotion, intracellular transport, chromosome segregation, and are essential for neuronal development and function. MTs do not function in isolation; they interact with a wide variety of proteins including molecular motors, MT-severing, and MT-stabilizing proteins. MT interactors, often referred to as MT-associated proteins (MAPs), control dynamic properties, structure, as well as interactions of MTs with membranes, organelles, and other protein complexes. Although some MAPs have been investigated in detail using in vitro reconstitution approaches, many of them remain insufficiently characterized. In addition, the number of newly discovered MAPs or MAP candidates is constantly growing (Bodakuntla et al., 2019), underpinning the need for a broader approach to characterize MAPs biochemically and biophysically. Current approaches are typically based on purification of MAPs in their functional form, which can be technically challenging and, in some cases, even impossible (Dogterom & Surrey, 2013). The lack of molecular tools to rapidly characterize any given MAP limits the understanding of their biological roles and the identification of novel links to human diseases.
To overcome these limitations, we developed an approach that allowed us to study the interactions of a variety of MAPs with MTs at single-MT resolution. Our method uses lysates prepared from mammalian cells overexpressing fluorescently tagged MAPs, in which we can directly polymerize MTs from endogenous tubulin and observe the behavior of the overexpressed MAP of interest. Similar approaches have been used in previous studies, e.g., mammalian cell lysates overexpressing kinesins to investigate motor processivity (Soppina et al., 2014), budding yeast lysates to study MT dynamics (Bergman et al., 2019), or Xenopus laevis egg extracts to explore MT network organization (Alfaro-Aco & Petry, 2015; Field et al., 2017). These studies, and our recent work (Jijumon et al., 2022), demonstrate that the lysate approach that lies between cellular studies and in vitro assays with purified components, enables in-depth structural and functional characterization of a wide range of MT-MAP interactions.
The protocols described here have been developed mainly for determining the impact of a variety of MAPs on MT assembly using total internal reflection fluorescence (TIRF) microscopy. We showed that it can also be used for structural studies of MAP-bound MTs using cryo-electron microscopy to determine the crosstalk of multiple MAPs on the MT lattice, to measure the affinity of MAPs to different types of MTs, or to screen the impact of mutations found in human patients on the interactions of MAPs with MTs.
Basic Protocol 1 describes the preparation of cell lysate of MAPs from HEK 293 cells. Basic Protocol 2 describes the quantification of the protein of interest in cell lysates by immunoblot. Support Protocol 1 provides a detailed description of purification of dead kinesin (KIF5B-N555/T92A) protein. Support Protocol 2 describes the preparation of guanylyl-(alpha, beta)-methylene-diphosphonate (GMPCPP) seeds, and Basic Protocol 3 describes TIRF reconstitution assays using cell lysates.
Basic Protocol 1: PREPARATION OF LYSATES OF HUMAN CELLS FOR TIRF RECONSTITUTION ASSAYS
To study the function of specific MAPs, the first requirement is the generation of fluorescently tagged expression constructs for the MAPs of interest. If not available, we clone the open reading frames of MAPs into any mammalian expression vector in frame with a fluorescent protein of choice. While this can be done with any cloning method, we use the simple and fast sequence and ligation-independent cloning (SLIC) method (Jeong et al., 2012).
Basic Protocol 1 describes the preparation of cell lysates. Expression plasmids were first transfected into HEK 293 cells to overexpress fluorescently tagged MAPs of interest. Cells were lysed and the extracts were cleared by ultracentrifugation. The cleared lysates are then directly injected into a microscopy flow chamber to perform in vitro reconstitution assays and TIRF microscopy. Alternatively, the cell lysates can be snap-frozen in liquid nitrogen and stored at −80°C.
Before collecting the transfected cells for lysate preparation, cells should be inspected under a fluorescence microscope to determine the expression levels. MAPs with strong expression levels (intense fluorescence) can be collected after 24 hr, while weakly expressed MAPs should be collected after 48 hr. All steps from collecting cells in Dulbecco's modified Eagle medium (DMEM) to the actual lysis are performed at 4°C or on ice to assure an efficient depolymerization of the MTs. The lysate preparation pipeline, described here, is schematically outlined in Figure 1.
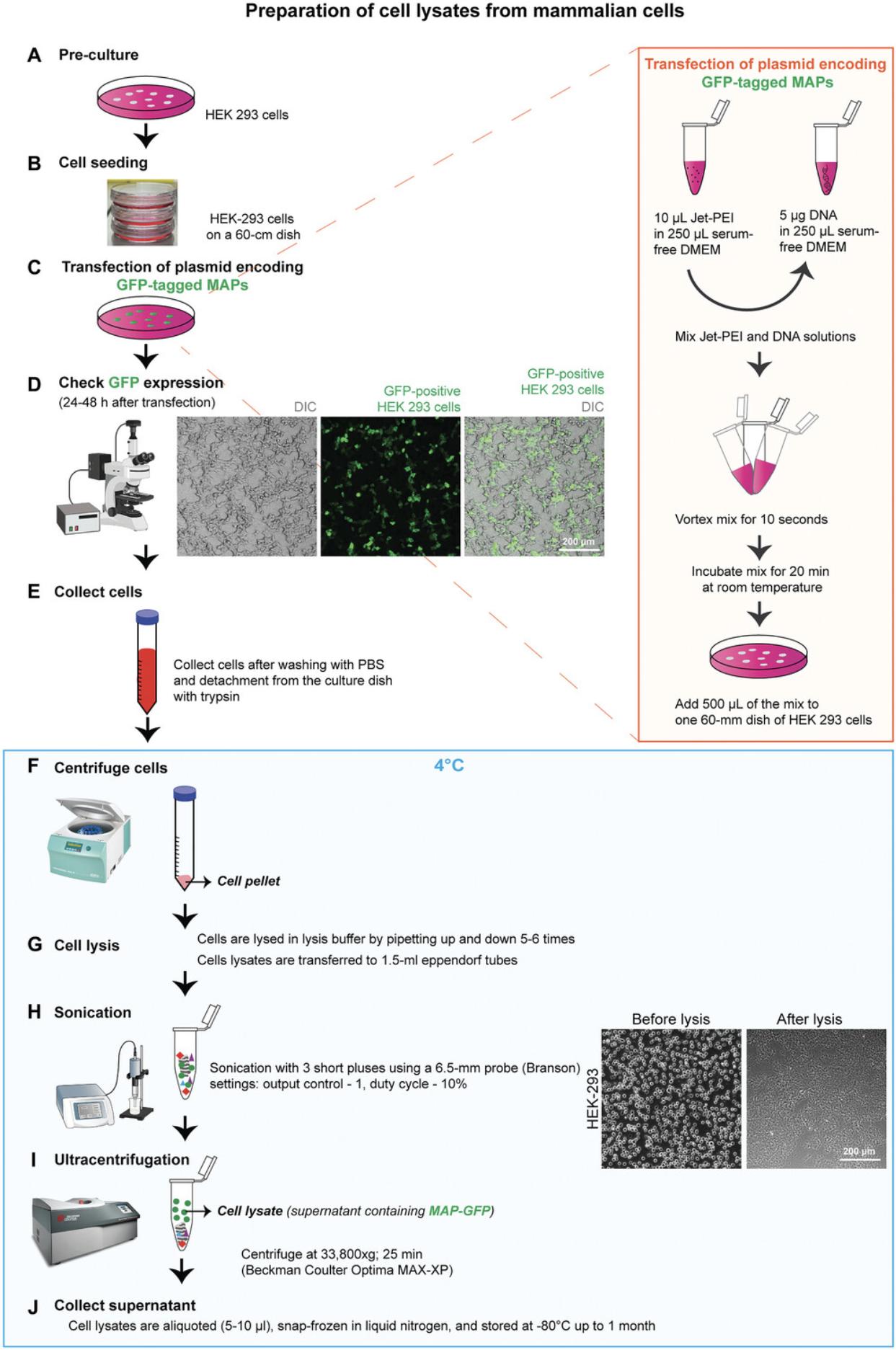
Materials
-
HEK 293 cells (ATCC, cat. no. CRL-1573)
-
DMEM (Life Technologies, cat. no. 41965062)
-
Fetal bovine serum (FBS) (Sigma-Aldrich, cat. no. F7524)
-
L-glutamine (Life Technologies, cat. no. 25030123)
-
Penicillin-streptomycin (Life Technologies, cat. no. 15140130)
-
Phosphate-buffered saline (PBS) (Life Technologies, cat. no. 14190169)
-
Trypsin-EDTA (Life Technologies, cat. no. 25200056)
-
Trypan blue solution (Sigma-Aldrich, cat. no. T8154)
-
jetPEI (Polyplus, cat. no. 101)
-
DNA (MAP-expressing plasmids)
-
Lysis buffer #1 (see recipe)
-
T75 TPP tissue culture flasks (Sigma-Aldrich, cat. no. Z707546)
-
Cell culture hood
-
Water bath (Julabo, cat. no. 9352508.33)
-
Micropipettes (p2.5, p10, p20, p100, p200, and p1000 and corresponding tips)
-
Pipette controller (Drummond Pipet-Aid XP, model no. 4-000-101)
-
5-, 10-, and 25-ml sterile pipettes (Corning, cat. nos. 4487, 4488, and 4489)
-
15- and 50-ml centrifuge tubes (Falcon, cat. nos. 352095 and 352070)
-
Laboratory centrifuge for 50-ml tubes (Sigma, cat. no. 4-16 K)
-
KOVA Glasstic Slide 10 (Thermo Fisher, cat. no. 11917967)
-
Microscope for cell counting
-
60-mm TPP tissue culture dish (Sigma-Aldrich, cat. no. Z707978)
-
Cell culture incubator, 37°C, 5% CO2 (Eppendorf, Galaxy 170S CO2 incubator)
-
1.5- and 2-ml tubes (Eppendorf, cat. nos. 0030125150 and 0030120094, respectively)
-
Vortex mixer (Scientific Industries, cat. no. SI-0236)
-
Inverted fluorescence microscope (to verify cell transfection)
-
1.5-ml ultracentrifuge tubes (Beckman Coulter, cat. no. 357448)
-
Sonicator (Branson Sonifier 450, model no. B450-WP)
-
6.5-mm diameter sonicator probe (Branson, cat. no. 101-148-070)
-
Ultracentrifuge (Beckman Coulter, model no. 393315 Optima MAX-XP)
-
TLA-55 rotor (Beckman Coulter, cat. no. 366725), 4°C
-
5-ml syringes without needles (Terumo, cat. no. SS-05S)
-
18G needles, 1.5-in., 1.2-mm × 38-mm (Terumo, cat. no. AN1838R1)
NOTE : All equipment and solutions coming in contact with cells must be sterile.
Cell seeding
1.Grow HEK 293 cells up to 60% to 70% confluency in DMEM supplemented with 10% (v/v) FBS, 2 mM L-glutamine, and 1× penicillin-streptomycin in a T75 flask.
2.Remove the medium from the flask and wash with 1× PBS.
3.Detach the cells from the flask by adding warm Trypsin-EDTA and collect them in pre-warmed DMEM. HEK 293 cells are easy to detach; either tap the flask after adding the trypsin or pipette out the DMEM with some force to detach them.
4.Transfer the cells into a conical centrifuge tube and centrifuge the collected cells 4 min at 134 × g , room temperature.
5.Remove the supernatant and resuspend the cells in DMEM.
6.Mix 10 µl resuspended cells and 10 µl Trypan blue dye for cell counting.
7.Load 10 µl of this mix into a KOVA Glasstic Slide 10 hemocytometer and count cells per quadrant under a microscope.
8.Seed 106 cells on a 22-cm2 dish (60-mm TPP tissue culture dish) for transfection and grow them at 37°C and 5% CO2 in a cell culture incubator for 16 to 24 hr.
Transfection
9.Before transfection, remove the medium from the dish and add 1.5 ml fresh serum-containing medium to each dish along the walls without disturbing the cells.
10.For tube 1, dilute 10 µl jetPEI in DMEM supplemented with 2 mM L-glutamine and 1× penicillin-streptomycin to a final volume of 250 µl (for one 22-cm2 dish).
11.For tube 2, dilute 5 µg DNA (MAP-expressing plasmids) in DMEM supplemented with 2 mM L-glutamine and 1× penicillin-streptomycin to a final volume of 250 µl (for one dish).
12.Add the 250 µl jetPEI mixture from tube 1 to the DNA-containing solution in tube 2.
13.Gently vortex the transfection mix at medium speed for 10 s.
14.Incubate the mix for 20 min at room temperature.
15.Per dish, add 500 µl jetPEI/DNA mix to 1.5 ml serum-containing medium and homogenize by gently shaking the dish in a cross shape (avoid swirling).
16.Incubate 24 to 48 hr at 37°C and 5% CO2 in a cell culture incubator.
Cell lysate preparation
Before lysate preparation, all transfected dishes need to be strictly checked with a fluorescence microscope to ensure transfection efficiency and to know the GFP expression levels (Fig. 1D). Dishes with low transfection efficiency or massive cell lethality should be discarded and must be repeated with a new set of cells.
17.Remove DMEM from the cells.
18.Gently wash the cells with 1.5 ml warm 1× PBS (per dish).
19.Detach the cells by adding 500 µl warm Trypsin-EDTA to the dish and incubate for 30 to 45 s at room temperature.
20.Add 2 ml warm DMEM containing 10% (v/v) FBS to inactivate trypsin and collect the detached cells.
21.Transfer the medium with detached cells to a 15- or 50-ml falcon tube (size chosen based on the number of dishes) and keep on ice.
22.Centrifuge the cells 10 min at 209 × g , 4°C. Prepare lysis buffer #1 and make sure it is ice-cold (incubate it on ice for 5 to 10 min).
23.Remove the supernatant and resuspend the cell pellets in freshly prepared, ice-cold lysis buffer #1.To obtain lysates with high protein concentrations, do not use more than 20 to 30 µl buffer for cells collected from one dish (for 5 dishes, use 100 to 150 µl).
24.Transfer the resuspended cells from the falcon tube into a 1.5-ml Beckman Coulter ultracentrifuge tube.
25.Pipette the cells up and down on ice for 2 to 3 min with short interruptions, e.g., 2 to 3 min incubations on ice. Repeat this process for 10 min total.
26.Sonicate the samples with three short pulses using the following probe settings:
- 6.5-mm probe diameter
- “Output control” set at 1
- “Duty cycle” set at 10%.
27.After cell lysis, ultra-centrifuge the sample 25 min at 38,500 × g , 4°C, with a precooled TLA-55 centrifuge rotor.
28.Collect the supernatant.
29.Lysates are aliquoted (5 to 10 µl) on ice, snap-frozen and stored at −80°C, or in liquid nitrogen for further use.
Basic Protocol 2: QUANTIFICATION OF GFP-TAGGED MAP CONCENTRATION IN CELL LYSATES
One application of the cell lysate approach is to rapidly screen and directly compare the MT-binding capacity of several proteins. It also allows to show to what extent different proteins induce MT polymerization in the lysates. Moreover, in our previous experiments we found that some of the 45 MAPs we characterized induced unique MT structures (Jijumon et al., 2022). We further demonstrated for some proteins that this is a concentration-dependent phenomenon. For example, a truncated version of the Microtubule-Actin Crosslinking Factor 1 [MACF1(C1023)-GFP] induced the formation of hooks at growing MT tips in cell lysates. This happened efficiently, that is, at almost all growing MTs at 520 nM of MACF1(C1023)-GFP, but quickly declined when the MAP was diluted in the lysates (Jijumon et al., 2022). Therefore, the quantification of GFP-MAPs in lysate is important for a more detailed characterization of their impact on MTs.
Techniques such as fluorescence spectroscopy, if available, can be used to determine the concentrations of GFP-tagged MAPs in lysates. Here, we used the immunoblot with an anti-GFP antibody (Fig. 2). To estimate the concentrations of GFP-MAPs in cell lysates, we calibrated the blots with a dilution series of a purified GFP-tagged protein of known concentration (Fig. 2A). The intensity of the GFP-positive bands was measured using FIJI software and GFP concentrations determined using GraphPad Prism or Microsoft Excel (Fig. 2C).
![Details are in the caption following the image Quantification of the concentration of GFP-tagged MAPs in cell lysates using an immunoblot. (A) A purified GFP-tagged protein [here: StrepII-GFP-Tau (0N 3R)] with known concentration was loaded at different dilutions alongside with a range of dilutions of the cell lysate expressing a GFP-tagged MAP of unknown concentration [here: MACF1 (N1023)-GFP]. GFP was labeled by immunoblot. (B) The intensity of each GFP-positive band was quantified using FIJI software. (C) The known protein concentrations of purified StrepII-GFP-Tau (0N 3R) were plotted against the measured intensities of the GFP-signals and a linear regression was applied. The GFP-intensity values of MACF1 (C1023)-GFP were then used to calculate the amount of GFP from this calibration curve, taking into consideration the dilution factors and the load volume of 10 µl. After averaging the results from 3 different dilutions, we obtained an average value of 1.24 pmol/µl (1.24 µM) for MACF1 (C1023)-GFP in undiluted lysate.](https://static.yanyin.tech/literature_test/cpz11070-fig-0002-m.jpg)
Materials
-
Purified GFP-tagged protein (Genova et al., 2023)
-
2× Laemmli sample buffer (see recipe)
-
Blocking buffer (see recipe)
-
Anti-GFP rabbit antibody (Torrey Pines Biolabs, cat. no. TP401)
-
PBST (see recipe)
-
Anti-rabbit HRP-conjugated antibody (Bethyl, cat.no. A120-201P)
-
Clarity Western ECL substrate (Bio-rad, cat. no. 1705061)
-
Trizma base (Sigma-Aldrich, cat. no. T1503)
-
Glycine (Sigma-Aldrich, cat. no. G8898)
-
Heating block (Stuart, cat. no. SBH130D)
-
SDS-PAGE electrophoresis equipment (Bio-Rad, cat. no. 1658001FC)
-
Trans-Blot Turbo transfer starter system, midi nitrocellulose (Bio-Rad, cat. no. 17001915)
-
Shaker (Heidolph Promax, cat. no. HEID_14009)
-
Cling wrap (Thermo fisher, cat. no. 11785873)
-
Chemidoc gel imager (VILBER, Fusion FX)
-
FIJI software
-
Prism software
1.Prepare dilution series of a purified GFP-tagged protein (Genova et al., 2023) of known concentrations as standard along with dilution series of the cell lysates (Fig. 2A).
2.Add 2× Laemmli sample buffer to all the samples.
3.Heat the mixture at 95°C for 5 min.
4.Run the protein gel at 100 V for 10 to 15 min and increase the voltage to 180 V for the rest of the run (∼45 min).
5.Gently transfer the gel onto a nitrocellulose membrane with Bio-Rad Trans-Blot Turbo system using Trans-Blot Turbo midi nitrocellulose transfer packs.
6.Transfer for 8 to 10 min and remove the membrane from the transfer device.
7.Block the membrane with blocking buffer (5% skimmed milk in PBST) for 1 hr at room temperature on a shaker.
8.Dilute the primary anti-GFP rabbit antibody 1:2000 in blocking buffer.
9.Incubate the membrane in primary antibody for 2 hr at room temperature on a shaker.
10.Wash the membrane with PBST for 5 min at room temperature on a shaker.
11.Repeat this step 3 times.
12.Dilute the secondary anti-rabbit HRP-conjugated antibody 1:10,000 in PBST.
13.Incubate the membrane in secondary antibody for 45 min at room temperature on a shaker.
14.Wash the membrane with PBST for 5 min at room temperature on a shaker.
15.Repeat this step 3 times.
16.Prepare the chemiluminescence substrate by mixing 1:1 reagent A (luminol/enhancer solution) to reagent B (peroxide solution).
17.Incubate the membrane for 1 min in the chemiluminescence substrate at room temperature.
18.Cover the membrane with cling wrap.
19.Use a gel imager with chemiluminescence detection to image the bands.
20.To quantify the immunoblot by FIJI, use the rectangular selection tool to define the left most lane.
21.Select Analyze >Gels>Select First Lane.
22.Move the rectangular selection right to the next lane.
23.Select Analyze>Gels>Select Next Lane.
24.Move the rectangular selection right to the next lane.
25.Repeat this step for each lane.
26.Generate lane profile plots by selecting Analyze>Gels>Plot Lanes.
27.Select the straight-line tool and draw base lines to form a closed area for each peak.
28.Select the wand tool from the FIJI menu and measure the area of each plot by clicking inside the peak.
29.Measure the area of each lane by selecting Analyze>Gels>Label Peaks.
30.To calculate the unknown concentrations using standard curve in Prism, enter the known concentration values into the X column and the corresponding quantified results of detection intensity into the Y column.
31.Enter the detection intensity for the unknown samples into the Y column, leaving the corresponding X cells as blank.
32.Click on the “Analyze” button and select “Simple Linear regression” option.
33.In the dialogue box, check the box labeled interpolate unknown from the standard curve and click OK.
34.To interpolate the X values to the graph, click on the “Change” button and select “Add Data sets”.
35.Click on the “Add” button in the dialog box that appears.
Support Protocol 1: PURIFICATION OF KIF5B(N555/T92A) (DEAD KINESIN) PROTEIN FOR TIRF RECONSTITUTION ASSAYS
This Support Protocol describes the purification of the recombinant 6×His-mKIF5B(N555/T92A) protein that we used to attach MTs to the cover slip in the TIRF reconstitution assays. The KIF5B(N555/T92A) mutant lacks the motor activity, while still binding MTs (Nakata & Hirokawa, 1995). This “dead kinesin” is flushed into the TIRF chamber to allow a proper attachment of GMPCPP seeds to the glass surface of the TIRF chamber, and to also bind the dynamically polymerizing MTs from the cell extracts to the surface.
Additional Materials (also see Basic Protocols 1 and 2)
-
Escherichia coli Rosetta (DE3) cells (Sigma-Aldrich, cat. no. 70954)
-
Plasmid DNA expression vector
-
LB medium (see recipe)
-
LB agar dishes (see recipe)
-
50 mg/ml kanamycin, 1000× (see recipe)
-
0.5 M isopropyl β-D-thiogalactopyranoside (IPTG) (see recipe)
-
Lysis buffer #2 (see recipe)
-
Ni-NTA agarose (Qiagen, cat. no. 30210)
-
Wash buffer (see recipe)
-
Elution buffer (see recipe)
-
Ice box and ice
-
Incubator (Memmert, cat. no. 03953)
-
Shaker (Infors Multitron Standard Shaker, cat. no. 29530)
-
Benchtop centrifuge (Sigma 4-16K, cat. no. 97262)
-
L-rod spreader (Sigma-Aldrich, cat. no. S4522)
-
Inoculation loop
-
100-ml conical flask
-
4-L conical flask
-
Disposable plastic cuvette (VWR, cat. no. 97000-586)
-
BioPhotometer (Eppendorf biophotometer, cat. no. P230609057)
-
1-L bottles for centrifugation (Beckman Coulter, cat. no. C31597)
-
Avanti J-26S centrifuge (Beckman Coulter, cat. no. B22985), 4°C
-
Ultracentrifuge (Beckman Coulter, Optima XPN-80, cat. no. B10051), 4°C
-
Rotator (Labinco, cat. no. 76000)
Transformation
For the purification of dead kinesin (6×His-mKIF5B-N555/T92A), first transform the expression vector into E. coli Rosetta (DE3) cells.
1.Thaw the competent E. coli cells from −80°C and keep the thawed cells on ice.
2.Add 100 ng of plasmid DNA to 100 µl competent cells, and gently mix by pipetting up and down without vortexing.
3.Incubate the mix on ice for 30 min.
4.Heat-shock at 42°C in a water bath for 45 s.
5.Incubate the mix on ice for 5 min.
6.Add 1 ml LB medium at room temperature to the cells.
7.Keep the cells in a 37°C incubator with shaking (250 rpm) for 1 hr.
8.In a benchtop centrifuge, centrifuge the tube 3 min at 197 × g , room temperature.
9.Discard the supernatant and resuspend the cells in 100 µl LB medium.
10.Spread 100 µl bacteria suspension onto a LB agar dish supplemented with 1× kanamycin antibiotic and incubate at 37°C for 12 hr.
Protein expression and purification
11.For the starter culture, inoculate 25 ml LB medium containing kanamycin with a single colony in a 100-ml conical flask.
12.Incubate the culture in a 37°C shaker (250 rpm) overnight.
13.For the secondary culture, in a 4-L conical flask, inoculate 1 L of LB medium supplemented with 1× kanamycin with 10 ml starter culture.
14.Incubate in a 37°C shaker (150 rpm) until the OD600nm measurement of bacterial culture reaches 0.6.Use a disposable plastic cuvette and the BioPhotometer to measure OD600nm.
15.Induce the protein expression by adding 1 ml of 0.5 M IPTG (for 1 L culture).
16.Incubate the IPTG-induced culture at 20°C with shaking (150 rpm) for 12 hr.
17.Using 1-L bottles and the Avanti J-26S, centrifuge the culture 15 min at 3993 × g , 4°C.
18.Discard the supernatant and collect the cells.
19.Add 10 ml cold lysis buffer #2 to a cell pellet from 1 L bacterial culture and resuspend the cells in lysis buffer by pipetting up and down.
20.Place the cell slurry in an ice-water bath for sonication and put the sonication probe inside the cell suspension.
21.Sonicate the sample at 50 kHz, 40% duty cycle with 20 s on, 30 s pause settings and repeat 10 times.
22.Ultracentrifuge the lysed cells 15 min at 14,822 × g , 4°C.
23.Transfer the supernatant into a fresh tube and discard the pellet.
24.Incubate the supernatant with 200 µl Ni-NTA agarose for 2 hr at 4°C on a rotating wheel at 5 rpm.
25.Using a benchtop centrifuge, centrifuge 2 min at 303 × g , 4°C, and remove the supernatant.
26.Wash the beads with 2 ml wash buffer, followed by centrifugation for 2 min at 303 × g , 4°C.
27.Repeat the washing step 3 times.
28.Elute the His-tagged protein by subsequently resuspending the beads in different volumes of elution buffer containing 350 mM imidazole (2 × 100 µl, 1 × 150 µl, 1 × 250 µl, and 1 × 300 µl). For each elution step, the beads were sedimented by centrifuging 2 min at 303 × g , 4°C, to collect the supernatants.
29.Collect each eluate (supernatant) in a separate tube and determine protein concentrations.
30.Prepare 10 to 20 µl aliquots of each eluted fractions separately, snap-freeze in liquid nitrogen, and store at −80°C or in liquid nitrogen.
Support Protocol 2: PREPARATION OF GMPCPP MT SEEDS FOR TIRF RECONSTITUTION ASSAYS
This Support Protocol describes the preparation of fluorescently labeled, GMPCPP-stabilized MT seeds that are used as nucleators of MT growth in our cell-lysate assays. GMPCPP MT seeds are prepared by polymerizing purified tubulin in the presence of GMPCPP, a slowly hydrolysable analogue of GTP (Hyman et al., 1992). Here we performed two rounds of tubulin polymerization with GMPCPP to increase the stability of the MT seeds, which also allows one to keep functional seeds for longer times at −80°C (Caplow & Shanks, 1996; Gell et al., 2010). The unlabeled tubulin used to prepare MT seeds was purified from mouse brain, using cycles of polymerization and depolymerization (Bodakuntla et al., 2020; Souphron et al., 2019), and was supplemented with commercially available rhodamine-labeled brain tubulin.
Additional Materials (also see Basic Protocol 1)
-
Rhodamine-labeled brain tubulin (Cytoskeleton Inc, cat. no. TL590M-A)
-
Unlabeled brain tubulin, purified as described in Souphron et al. (2019)
-
BRB80 (see recipe)
-
GMPCPP (Jena Biosciences, cat. no. NU-405S)
-
Glycerol (VWR Chemicals, cat. no. 24388.295)
-
Tabletop centrifuge for 1.5-ml tubes (Eppendorf, cat. no. 5417R)
1.Add 15 µM unlabeled tubulin and 5 µM labeled brain tubulin in cold BRB80 buffer (60 µl total volume).
2.In an Optima MAX-XP and TLA-55 rotor, ultracentrifuge 20 min at 135,800 × g , 4°C.
3.Carefully collect the supernatant and discard the pellet.
4.Add and mix 0.5 mM GMPCPP to the supernatant.
5.Keep the sample on ice for 5 min.
6.Incubate at 37°C for 1 hr.
7.Centrifuge 10 min at 9180 × g , 37°C.
8.Remove the supernatant.
9.Wash the pellet by adding 30 µl of warm BRB80.
10.Centrifuge 3 min at 9180 × g , 37°C.
11.Remove the supernatant.
12.Resuspend the pellet in 55 µl of cold BRB80 on ice.
13.Incubate the sample on ice for 15 min, and vigorously pipette up and down every 3 min.
14.Add 0.5 mM GMPCPP to the resuspended sample.
15.Incubate on ice for 5 min.
16.Incubate at 37°C for 1 hr.
17.Centrifuge 10 min at 9180 × g , 37°C.
18.Remove the supernatant.
19.Wash the pellet by adding 30 µl of warm BRB80 to the pellet.
20.Centrifuge 3 min at 9180 × g , 37°C.
21.Remove the supernatant.
22.Resuspend the pellet in 75 µl of warm BRB80 supplemented with 10% (v/v) glycerol.
23.Aliquot in small volumes (2 to 5 µl) and snap-freeze in liquid nitrogen.
24.Store at −80°C for up to 1 month.
Basic Protocol 3: TIRF-BASED MT-MAP RECONSTITUTION ASSAYS USING CELL LYSATES
This Basic Protocol describes the use of cell lysates prepared in Basic Protocol 1 for the reconstitution of MT-MAP interactions. This method was used to screen the MT-binding capacity of proteins of interest, as well as to determine MAP-induced MT phenotypes. It is recommended to quantify the GFP-MAP candidate in cell lysate (as described in Basic Protocol 2) before performing TIRF assays.
The first part of the TIRF experiment is the preparation of the flow chambers. For that, glass slides and coverslips were cleaned by sonication in 100% acetone followed by rinsing with ultra-pure water and sonication in 100% ethanol. These glass slides and coverslips can be stored in fresh 100% ethanol for up to 1 month. To assemble TIRF chambers, four strips of double-sided adhesive tape were affixed to each glass slide, creating three chambers. Fresh coverslips, stored in ethanol, were then gently pressed onto the sticky tape strips to complete the chamber assembly. The TIRF chamber volume is ∼10 µl. To start the experiment, we sequentially introduce different solutions into the flow chamber: β-casein to block glass surfaces to reduce any non-specific protein binding, dead kinesin, GMPCPP seeds, and finally the cell lysates. The binding of the fluorescently labeled MAPs on growing MTs is followed using a TIRF microscopy. At the end of each experiment, to visualize the MTs and actin filaments, the chamber can be flushed with the antibody YL1/2 to detect MTs, and with SiR-actin to visualize actin filaments. A schematic representation of each step in Basic Protocol 3 is shown in Figure 3.
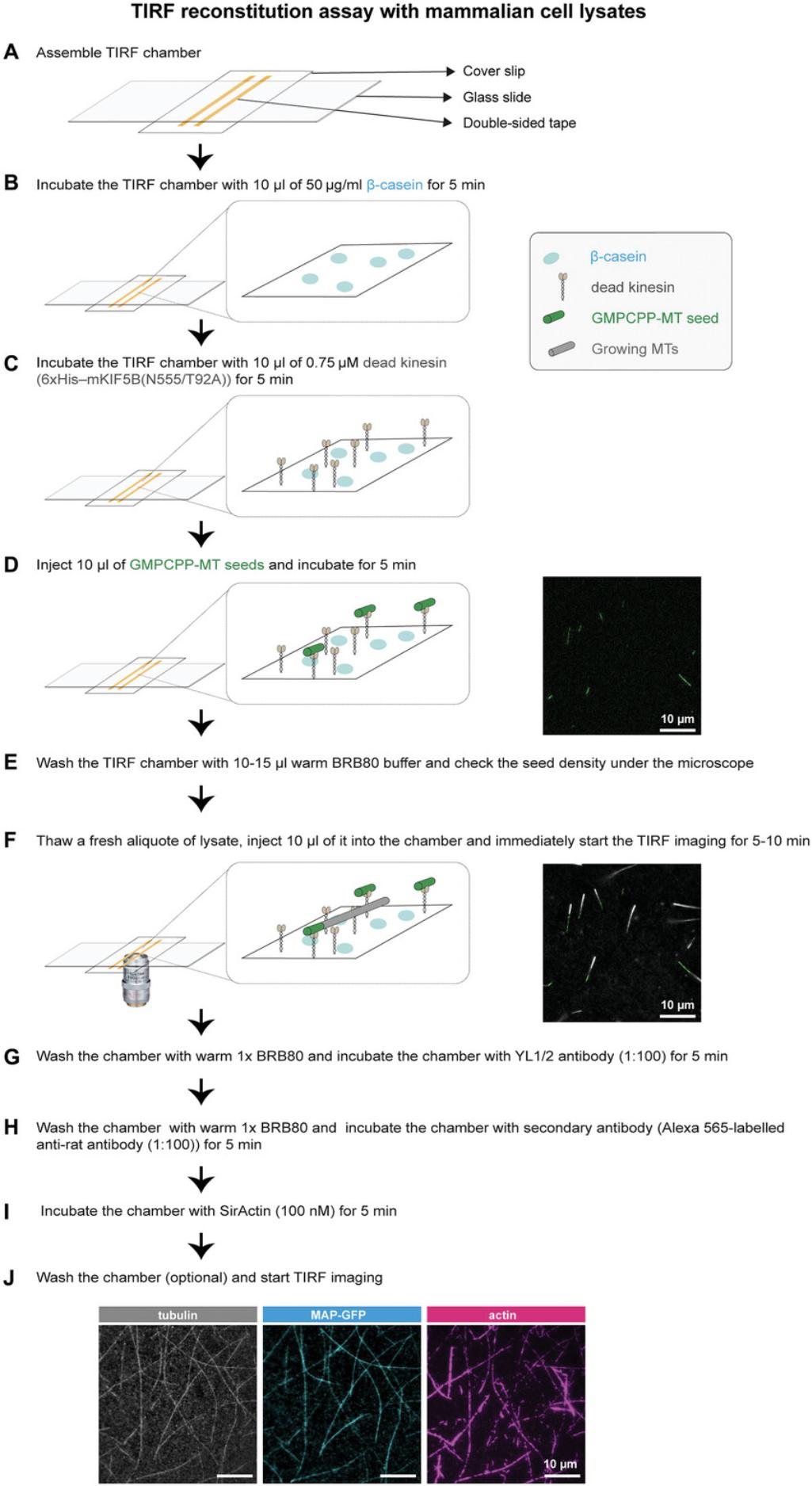
Materials
-
Dead kinesin (from Support Protocol 1)
-
GMPCPP MT seeds (from Support Protocol 2)
-
β-casein (see recipe)
-
BRB80 (see recipe)
-
Frozen and cleared HEK 293 cell lysates with overexpressed MAPs (from Basic Protocol 1)
-
YL1/2 primary antibody (see recipe)
-
Secondary antibody (see recipe)
-
SiR-actin (see recipe)
-
Latrunculin A (see recipe)
-
76-mm × 26-mm glass slides (Fisher Scientific, cat. no. 12-550-15) or 24-mm × 50-mm coverslip, thickness no. 1.5 (VWR, cat. no. 48393-241)
-
22-mm × 22-mm coverslips, thickness no. 1.5, (VWR, cat. no. 631-0125)
-
tesa double-sided tape, 10-m × 15-mm (tesa)
-
Dry heat block (Eppendorf Thermomixer C Model 5382)
-
Azimuthal TIRF controlled by Modular software (Gataca Systems)
-
Evolve EMCCD camera (16-µm pixel size), Photometrics Evolve 512 EMCCD (Photometrics)
-
Nikon 100× TIRF oil objective, NA 1.49 (Nikon)
-
Ice
-
Liquid nitrogen
-
TIRF chambers
-
Whatman filter paper (GE Healthcare, cat. no. 3017-915)
1.Switch on the heat block (37°C) and TIRF microscope 10 to 15 min in advance.
2.Thaw a fresh vial of GMPCPP MT seeds, dead kinesin, and β-casein. Keep the thawed kinesin and β-casein on ice.
3.Warm the GMPCPP seeds and some BRB80 buffer in the 37°C heat block.
4.Pipette in 10 µl of 50 µg/ml β-casein (in 1× BRB80) into the TIRF chamber from one side and absorb the outgoing solution with a small piece of filter paper from the other side of the chamber. Incubate for 5 min.
5.Inject 10 µl of 0.75 µM 6×His-mKIF5B(N555/T92A) kinesin and incubate for 5 min at room temperature.
6.Introduce GMPCPP MT seeds into the chamber and incubate for 5 min at room temperature.
7.Wash the TIRF chamber with 10 µl of warm BRB80 to remove excess and unattached MT seeds.
8.Thaw a fresh aliquot of lysate and make appropriate dilutions with either cold BRB80 or freshly thawed control cell lysate (from non-transfected cells) if necessary. Flow 10 µl lysate into the TIRF chamber.
9.Once the lysates are injected:
-
Place the TIRF chamber immediately on a TIRF microscope stage.
-
Find a field of view with sparsely distributed seeds at the center of TIRF chamber.
-
Start the timelapse with preloaded acquisition settings.
The timelapses were acquired with a Nikon Ti-E azimuthal TIRF setup utilizing an iLas-2 system (Gataca Systems) on an inverted microscope featuring a 100× TIRF NA 1.49 oil objective and equipped with the Nikon perfect focus system (PFS). The imaging field measured 68.1 × 68.1–µm, and videos were recorded with an Evolve EMCCD camera and MetaMorph software. The total duration of the timelapse for each MAP candidate was 11.612 min with a frame interval of 1.98 frames/s. TIRF chambers were illuminated at 488 nm for GFP-tagged proteins, at 561 nm for GMPCPP MT-seeds (rhodamine-labeled brain tubulin), and at 642 nm for actin filaments labeled with Sir-Actin.
10.Once the acquisition is finished, remove the chamber from the stage and wash with warm BRB80.
11.To visualize MTs, inject YL1/2 (anti-tyr-tubulin) primary antibody (1:100) into the chambers and incubate for 5 min.
12.Wash the chamber with warm BRB80 to remove unbound antibody.
13.Flow secondary antibody (Alexa 565-labeled anti-rat antibody diluted 1:100) and SiR-actin (500 nM; to visualize F-actin) in warm BRB80 into the TIRF chamber and incubate for 5 min.
14.Place the TIRF chamber on a microscope stage and perform tricolor-TIRF microscopy.
REAGENTS AND SOLUTIONS
Blocking buffer
- 5 g skimmed milk powder (REGILAIT)
- 100 ml PBST (see recipe)
- Prepare fresh before use
BRB80
- 20 ml of 0.5 M K-PIPES (see recipe) (final concentration: 80 mM)
- 250 µl of 0.5 M K-EGTA (see recipe) (final concentration: 1 mM)
- 125 µl of 1 M MgCl2 (Sigma-Aldrich, cat. no. M1028) (final concentration: 1 mM MgCl2)
- Make up to 125 ml with MilliQ H2O
- Sterilize using a 0.2-µm filter
- Store up to 3 months at 4°C
β-casein
- 6 mg β-casein (Sigma-Aldrich, cat. no. C6905)
- 200 µl H2O
- Sterilize using a 0.2-µm filter
- Aliquot and store up to 6 months at −20°C
Elution buffer
- 50 mM Tris pH 8.0 (see recipe)
- 100 mM NaCl (see recipe)
- 350 mM imidazole (see recipe)
- 5% (v/v) glycerol (VWR Chemicals, cat. no. 24388.295)
- Prepare fresh before use
Imidazole, 1 M, pH 8.0
- 3.4 g imidazole (Sigma-Aldrich, cat. no. I2399)
- 40 ml MilliQ H2O
- Adjust to pH 8.0 with KOH
- Bring up to 50 ml with MilliQ H2O
- Sterilize using a 0.2-µm filter
- Store up to 3 months at 4°C
IPTG, 0.5 M
- 1.19 g IPTG (Sigma-Aldrich, cat. no. I6758)
- 10 ml MilliQ H2O
- Bring up volume to 500 ml with MilliQ H2O
- Sterilize using a 0.2-µm filter
- Store up to 6 months at −20°C
Kanamycin, 1000×, 50 mg/ml
- 5 g kanamycin (Sigma-Aldrich, cat. no. K0129)
- 100 ml H2O, distilled
- Vortex the solution to dissolve
- Sterilize using a 0.2-µm filter
- Store up to 6 months at −20°C
K-EGTA, 0.5 M
- 47.5 g EGTA (Sigma-Aldrich, cat. no. E3889)
- 150 ml MilliQ H2O
- Adjust to pH 7.7 with KOH
- Bring up volume to 250 ml with MilliQ H2O
- Sterilize using a 0.2-µm filter
- Store indefinitely at room temperature
K-PIPES, 0.5 M
- 75.5 g PIPES (1,4-piperazine ethanesulfonic acid) (Sigma-Aldrich, cat. no. P6757)
- 300 ml MilliQ H2O
- Adjust to pH 6.8 with KOH while PIPES is dissolving, as it will not dissolve at low pH
- Bring up volume to 250 ml with MilliQ H2O
- Sterilize using a 0.2-µm filter
- Store up to 3 months at 4°C
Laemmli sample buffer, 2×
- 4% sodium dodecyl sulphate (SDS) (VWR, cat. no. 442444H)
- 160 mM Tris·HCl, pH 6.8 (Current Protocols, 2006)
- 180 mM dithiothreitol (DTT) (Sigma-Aldrich, cat. no. D9779)
- 20% (v/v) glycerol (VWR Chemicals, cat. no. 24388.295)
- 100 mg bromophenol blue (Sigma-Aldrich, cat. no. 1.08122)
- Store up to 6 months at −20°C
Latrunculin A
- 11 µM latrunculin A (Sigma-Aldrich, cat. no. L5163)
- Store up to 3 months at −20°C
LB agar dishes
- 25 mg LB broth powder (Sigma-Aldrich, cat. no. L3522)
- 15 mg agar (Sigma-Aldrich, cat. no. A1296)
- 1 L ddH2O
- Swirl to mix and autoclave
- Let cool down to ∼60° to 70°C
- Add antibiotic to the mix
- Pour LB agar into dishes under a laminar flow hood
- Keep the dishes in the hood until it solidifies (∼30 min)
- Store up to 3 weeks at 4°C
Pour the medium while still liquid but cool enough for safe handling. LB agar bottle should be opened, and preparation of dish should be conducted only in laminar flow hood to avoid contamination.
LB medium
- 25 mg LB broth powder (Sigma-Aldrich, cat. no. L3522)
- 1 L ddH2O
- Swirl to mix and autoclave
- Store up to 3 months at room temperature
Lysis buffer #1
- 990 µl BRB80 (see recipe)
- 5 µl of 200× protease inhibitor cocktail (see recipe) (final concentration: 1×)
- 5 µl of 10% (v/v) Triton X-100 (see recipe) (final concentration: 0.05%)
- Prepare fresh before use
Lysis buffer #2
- 50 mM Tris pH 8.0 (see recipe)
- 100 mM NaCl (see recipe)
- 30 mM imidazole (see recipe)
- 1× protease inhibitor (see recipe)
- 1 mM PMSF (see recipe)
- 0.1% Triton X-100 (see recipe)
- Prepare fresh before use
NaCl, 5 M
- 146.1 g NaCl (Sigma-Aldrich, cat. no. S5886)
- Make final volume to 500 ml with MilliQ H2O
- Sterilize using a 0.2-µm filter
- Store up to 3 months at room temperature
PBST
- 1 ml TWEEN 20 (Sigma-Aldrich, cat. no. P1379)
- 999 ml 1× PBS (Life Technologies, cat. no. 14190169)
- Store up to 1 month at room temperature
PMSF, 0.1 M
- 435.5 mg PMSF (phenylmethanesulfonyl fluoride) (Sigma-Aldrich, cat. no. P7626)
- 25 ml isopropanol (VWR, cat. no. 20842.298)
- Store up to 6 months at −20°C
Protease inhibitor cocktail, 200×
- 10 mg aprotinin (Sigma-Aldrich, cat. no. A1153)
- 10 mg leupeptin (Sigma-Aldrich, cat. no. L2884)
- 10 mg 4-(2-aminoethyl)-benzenesulfonyl fluoride (Sigma-Aldrich, cat. no. A8456)
- 2.5 ml MilliQ H2O
- Aliquot and store up to 6 months at −20°C
Secondary antibody
Dilute Alexa 565-labeled anti-rat antibody (Thermo Fisher Scientific, cat. no. A-11077) 1:100 in BRB80 (see recipe). Prepare fresh before use.
SiR-actin
- 500 nM SiR-actin (Spirochrome, cat. no. SC001)
- Store up to 3 months at −20°C
Tris, 1 M, pH 8
- 60.57 g Tris (Sigma-Aldrich, cat. no. T1503)
- 400 ml ddH2O
- Adjust to pH 8.0 with HCl
- Bring up to 500 ml with water
- Sterilize using a 0.2-µm filter
- Store up to 3 months at room temperature
Triton X-100, 10% (v/v)
- 5 ml Triton X-100 (Sigma-Aldrich, cat. no. T9284)
- 45 ml ddH2O
- Stir vigorously until completely mixed
- Sterilize using a 0.2-µm filter
- Store indefinitely at room temperature
Wash buffer
- 50 mM Tris pH 8.0 (see recipe)
- 100 mM NaCl (see recipe)
- 30 mM imidazole (see recipe)
- Prepare fresh before use
YL1/2 primary antibody
Dilute YL1/2 antibody (Sigma-Aldrich, cat. no. MAB1864-I) 1:100 in BRB80 (see recipe). Prepare fresh before use.
COMMENTARY
Background Information
In the past decades, in vitro reconstitution studies with purified MAPs, as well as cell-based studies have provided a large body of knowledge on selected MAPs and their interactions with MTs. However, both approaches have their own limitations. In vitro reconstitution assays typically require protein purification, which is cumbersome and often requires time-consuming optimization steps, and some proteins might never be purified in a functional state. In cell-based studies, exogenous expression of MAPs often leads to overexpression artefacts such as uncontrolled MT bundling, which we indeed observed for many MAPs (Jijumon et al., 2022). These limitations have restricted in-depth molecular characterizations to only few MAPs, while to date, many proteins have been proposed to be MAPs without solid experimental evidence so far.
To overcome these limitations, we took advantage of the simplicity to express functional proteins in mammalian cells, which we then lysed to directly use them for in vitro reconstitution experiments. The use of cell lysates for in vitro studies is not entirely new. Xenopus egg extracts (Belmont et al., 1990), mammalian cell lysates (Saredi et al., 1997), Drosophila embryo extracts (Telley et al., 2013), or yeast cell lysates (Bergman et al., 2019) were previously employed to study MT functions. Experiments using Xenopus egg extracts have contributed greatly to our understanding of the function of the mitotic/meiotic spindle, and in the investigation of other subcellular structures (reviewed in Milunović-Jevtić et al., 2016). Similarly, extracts from single Drosophila embryos were used to study the mitotic machinery (Telley et al., 2013), and yeast extracts were used to study MT dynamics (Bergman et al., 2019). First studies using mammalian cell extracts allowed to identify and visualize proteins decorating MT-based structures such as MT asters. These experiments helped in the discovery and characterization of MAPs, such as NuMa (nuclear protein that associates with the mitotic apparatus), ch-TOGp and protein 4.1R, and later to conduct more detailed experiments to study the role of these proteins in cell division (Dionne et al., 2000; Gaglio et al., 1995; Huang et al., 2004). Another study used highly diluted lysates of mammalian Cos7 cells as a source of kinesin motor proteins to perform single-molecule experiments (Soppina et al., 2014).
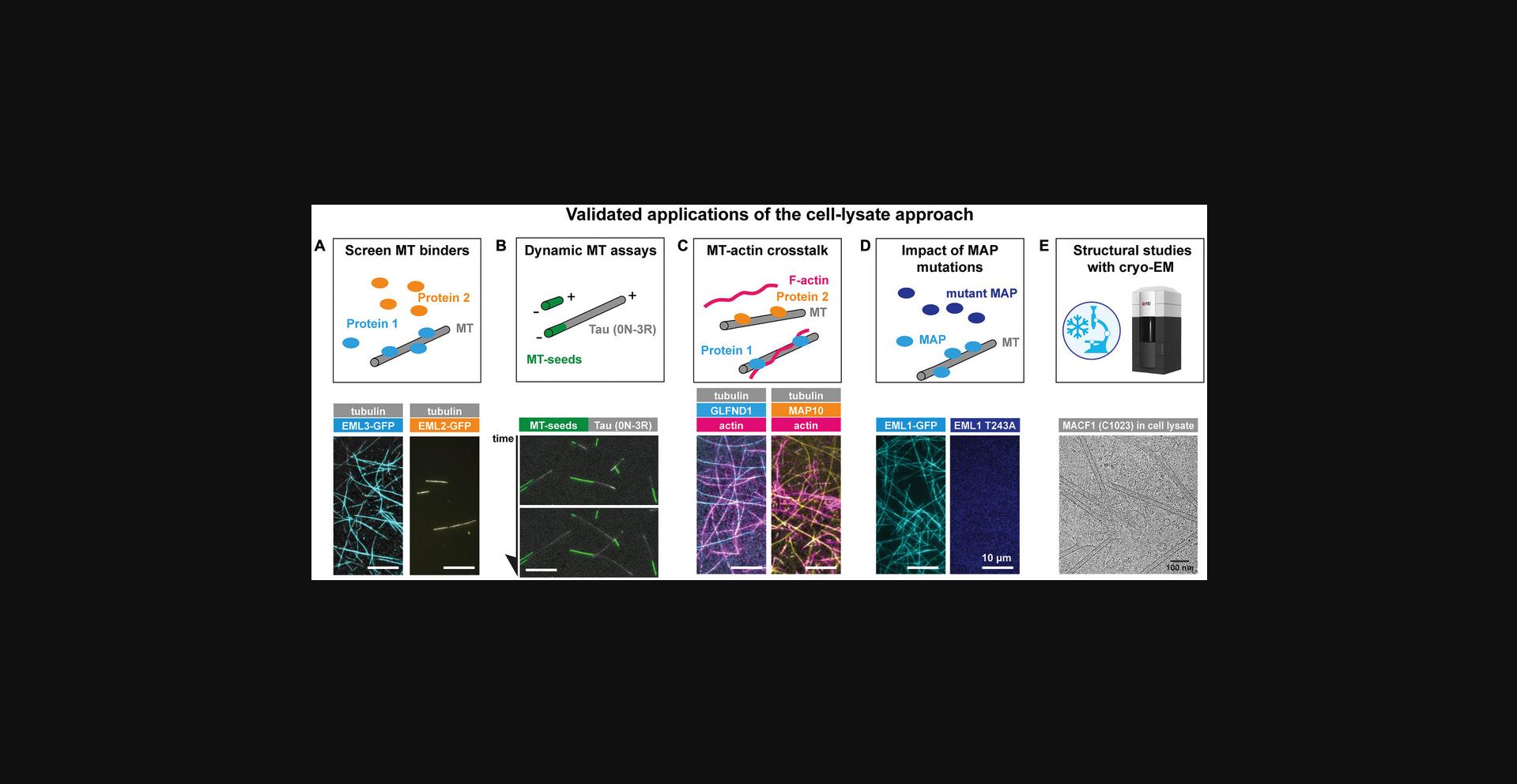
The method presented herein uses lysates prepared from mammalian HEK 293 cells. Our method uses highly concentrated cell lysates, which has the great advantage that MTs dynamically assemble from endogenous tubulin in the absence of crowding agents, or other agents inducing MT polymerization. Our method allowed us to identify new MAPs, to discover novel properties of known MAPs, to perform structural studies of MAPs bound to MTs by cryo-electron microscopy, and to study the role of MAPs in actin-MT crosstalk, as well as the impact of patient mutations in MAP-MT interactions (Jijumon et al., 2022; Krajka et al., 2022) (Fig. 4).
Given our observation that >50 randomly chosen proteins, MAPs and potential MAPs were soluble and functional in our assay, it is likely that in vitro reconstitution assays that were previously limited by the availability of functional, purified MAPs, can now be conducted using our lysate approach. Therefore, the potential applications of our lysate method are vast. Novel applications could include, but are not restricted to, testing the impact of drugs on MAP-MT interactions. Preparing lysates from different cell types would allow for the identification of cell-type specific cofactors regulating MAP-MT interactions. Similarly, using cell lysates prepared from synchronized cells could allow for observing the behavior of MAPs in different stages of cell division.
Taken together, the use of mammalian cell lysates for in vitro reconstitution assays opens a new perspective for medium-throughput in vitro analyses of MT interactors and regulators. Given that proteins can subsequently be purified from the cell lysates, it could be directly coupled to follow-up analyses with purified components, thus overcoming the potential drawback of the lysate-based method: the presence of other proteins in the cell extracts.
Critical Parameters and Troubleshooting
The cell lysate approach is a powerful tool to rapidly test the impact of any protein of interest on MT assembly, MT structure, and cytoskeletal crosstalk. While the technique has already proven its utility (Jijumon et al., 2022; Krajka et al., 2022), there are several critical measures to follow while preparing the lysates and performing the TIRF experiments to get reproducible and reliable results.
One of the most critical elements is the need of sufficiently concentrated lysates in order to allow for MT polymerization. First, it is important to use healthy cells with low number of passages. Unhealthy cells might die upon overexpression of MAPs, which could lower the overall concentration of the MAP in the lysate, thereby affecting the reproducibility of experiments. Second, use of the smallest-possible volume of BRB80 buffer at the cell lysis step is crucial to obtain concentrated lysates. Dilution of lysate can affect the critical tubulin concentration to polymerize, which in turn can lead to poor or no MT growth. Overall, it is essential to ensure the application of similar conditions in independent lysate preparations.
Some of the MAP-induced MT phenotypes we observed were concentration-dependent, therefore we recommended to conduct TIRF assays with different MAP concentrations in the lysate. To dilute the overexpressed MAP by keeping the concentration of all other components constant, cell lysates containing the overexpressed MAP were diluted with lysates from non-transfected cells. In cases where the concentration is found to be an important factor, it becomes essential to estimate the concentrations of the overexpressed MAP, endogenous tubulin, and total protein concentrations in the lysate to be able to reproduce the observed MT phenotype with new batches of lysates (Jijumon et al., 2022).
Commonly observed problems, their possible causes, and troubleshooting suggestions are listed in Table 1.
| Step | Problem | Possible reasons | Solution |
|---|---|---|---|
| Basic Protocol 1, step 9 | Non-uniform distribution of cells on a culture dish in the seeding step | Seeding and direct incubation of cells without any shaking, or improper shaking | Shake the culture dish 2-3 times in a cross-shape fashion by moving on a flat surface; do not swirl |
| Basic Protocol 1, step 17 | Cell death/floating cells after transfection | Unhealthy cells; overexpression of some MAPs can be toxic to cells; addition of transfection mix to only one region of the culture dish (Basic Protocol 1, step 15) | Make sure to use well-maintained healthy cells with low passage number (8-10 passages); reduce the time after transfection; if you are trying a new protein candidate, observe the cells frequently under a microscope and find the time point before cells starts dying; disperse the transfection mix as small volumes at different spots in the culture dish, and homogeneously distribute the mix by gentle shaking (Basic Protocol 1, step 15) |
| Basic Protocol 1, step 17 | No GFP fluorescence signal after transfection | Expressions of some MAPs is lower compared to others; use of wrong plasmid or molecular cloning of incorrect DNA sequence | Wait longer before harvesting cells to check whether expression increases; given the large number of plasmids handled, it might happen that a wrong construct is used in an assay; we always include a positive control such as a GFP plasmid in all assays |
| Basic Protocol 1, step 17 | Low transfection efficiency | Low plasmid concentration; expired transfection reagents, or aliquots that have been opened multiple times and used for >1 month; unhealthy cells | Check the plasmid quality and concentration, repeat plasmid purification if required; repeat experiment with new transfection reagents; see “Cell death/floating cells after transfection” above |
| Basic Protocol 1, step 24-27 | Inefficient cell lysis, often represented as green pellet after ultracentrifugation (Basic Protocol 3, step 28) | Incomplete cell lysis | Make sure the lysis step (pipetting up and down and sonication steps) are done as described; to confirm lysis, it is possible to check the slurry of cells under a simple dissection microscope by putting a few drops of lysate on a glass coverslip; to confirm efficient cell lysis, check for indicators (shrunk cell membrane, absence of round cells, and presence of cell debris; Fig. 1H); increase the number of sonication pulses or pipetting if needed |
| Basic Protocol 1, step 29 | Floating lipid layer after ultracentrifugation | Unknown reason | It is normal to see this floating layer after ultracentrifugation step especially for HEK 293 cells; try to collect lysate using a fine pipette tip without disturbing the floating later; if the lysate is valuable and in limited supply, collect it in separate clean and dirty fractions and test them separately |
| Basic Protocol 2 | Low protein concentration, or total protein concentration in the lysate is <8 µg/µl | Use of excess buffer led to lysate dilution; low number of seeded cells; inefficient collection of cells from the culture dish; partial cell lysis | Repeat the lysate preparation, taking care to remove as much PBS and cell culture medium as possible (Basic Protocol 1, step 24), and add strictly 20-30 µl lysis buffer; make sure the cell counting is correct; make sure the number of dishes, cells, plasmid DNA concentration are same between experiments and are same as in the protocol; see “Inefficient cell lysis, often represented as green pellet after ultracentrifugation” above |
| Basic Protocol 3, step 6 and 7 | No or few MT seeds are attaching | Use of low kinesin concentration | Use a fresh aliquot of kinesin or re-quantify the kinesin concentration |
| Basic Protocol 3, step 7 | High MT-seed density | High concentration of kinesin; MT seeds are highly concentrated | Decrease kinesin concentration; dilute the MT seeds |
| Basic Protocol 3, step 7 | Long MT seeds | Old MT GMPCPP seeds | Prolonged incubation of MT seeds at warm conditions can induce MT annealing, and therefore forms long MT seeds; annealing can be reversed by gently shatter the seeds with a p20 pipette tip; alternatively, use a fresh aliquot of MT seeds |
| Basic Protocol 3, step 9 | No MT polymerization events/visible MAP decoration | Non-optimal MAP concentration; lysate is too diluted; tested protein is possibly not a MT binder or can be a MT-depolymerize; use of old lysates (frozen lysates older than 2-4 weeks) | Some MAPs might not induce MT polymerization at higher concentrations in lysates (to confirm, perform the TIRF assays with different lysate dilutions; in these assays we use MAP binding as a proxy to visualize MTs, therefore, no MAP decoration does not necessarily mean there is no MT polymerization, thus, at the end of TIRF assay, stain the MTs with YL1/2 antibody (Basic Protocol 3, step 10) to visualize MTs and thereby confirm the MT formations); verify protein concentration, if the concentration is too low, prepare a new lysate for that particular MAP or MAP candidate; for each set of TIRF experiments, include a positive and a negative control (a positive control that can be used is any known MAP that decorates MTs; a negative control can be lysate either with overexpressed GFP or from non-transfected cells); perform the reconstitution experiment with same conditions and stain MTs with YL1/2 tubulin antibody; prepare fresh cell lysates |
| Basic Protocol 3, step 9 | Rapid MT polymerization events | Use of highly concentrated cell lysate; high MT-seed density; use of warm lysate; tested protein can be a potential MT-polymerization enhancer | Dilute the lysate with cold BRB80 and repeat the TIRF experiment; reduce the MT-seed concentration; add only freshly thawed cold lysate into the TIRF flow chamber to prevent rapid MT polymerization, however, rest of the solutions (GMPCPP seeds, antibodies, SirActin and all BRB80 washes) are pre-warmed before introducing into the TIRF chamber; test this possibility by sequential dilutions of the MAP |
| Basic Protocol 3, step 9 | Many MT gliding events | Some MAPs could recruit motor proteins from the cell lysate | Add non-hydrolysable ATP (not tested by us but suggested as potential solution) |
| Basic Protocol 3, step 9 | Not able to reproduce a MT phenotype that was observed before | Non-optimal MAP/ tubulin concentration | The concentration of MAPs and tubulin can vary from one lysate preparation to the other; for testing simple MAP-MT binding, such variations are not important; nonetheless, to reproduce particular MT phenotypes induced by specific MAPs the concentration can be critical; therefore, for any newly observed MT phenotype, save an aliquot of the lysate for quantifying tubulin and MAP concentrations by immunoblot (Basic Protocol 2); this will allow to adjust these concentrations in experiments with new lysates |
| Basic Protocol 3, step 9 | High fluorescence background | Using TIRF chambers that are >1 week old, or dirty glass slide or coverslip; protein/MAP degradation in lysates; contamination with floating layer while collecting lysate (Basic Protocol 3, step 29); low binding affinity of MAPs to growing MTs | Use a newly assembled TIRF chamber with clean glass slide and coverslip and perform experiments in a dust-free environment; use a standard glass treatment for TIRF-based MT in vitro reconstitution assays; increase the concentration of protease inhibitors in the lysate or use a fresh aliquot of protease inhibitors; perform a short cold ultracentrifugation of lysate just before the TIRF assay; perform a western blot with GFP antibody (in case of GFP-tagged proteins) to verify protein degradation (Basic Protocol 2); use a syringe with long needle (18G) or a long SDS-PAGE gel-loading pipette tip to collect the lysate without disturbing the upper floating layer (Basic Protocol 3, step 29); after the polymerization step, wash the TIRF chamber with fresh BRB80 (Basic Protocol 3, step 9), this replaces the lysate inside the TIRF chamber leaving only MAP bound MTs, thereby reducing the background fluorescence signal and improving the visualization of weaker-bound MAPs |
| Basic Protocol 3, step 9 | Missed initial time frames of MT polymerization during TIRF imaging | The use of pre-warmed lysate could lead to immediate MT assembly in the TIRF chamber | Instead of injecting warm lysates into the TIRF chamber, add cold lysate freshly thawed on ice; for dilution of lysates, always use cold BRB80 buffer, or cold non-transfected cell lysate; alternatively, use TIRF chambers with cross design, that way there is no need to take the TIRF chamber out of the microscope stage to add the lysate (Fig. 3) |
| Basic Protocol 3, step 9 | MT polymerization does happen spontaneously, and not from seeds | Use of highly concentrated lysates; MAPs inducing spontaneous MT polymerization | Further dilute lysates; since many MAPs induce spontaneous MT nucleation, rapid MT polymerization is inevitable and part of the MAP-induced phenotype |
| Basic Protocol 3, step 14 | Bleed-through between different fluorophore channels | Wavelength spectrum of filter-cube in the TIRF microscope | Use correct filter cubes assuring a clean separation of the emission spectra of each fluorophore used in the experiment |
Understanding Results
Our lysate-based approach was originally developed with an intention to rapidly screen any protein of interest for their ability to bind to MTs. This can be tested in two ways:
- 1.MAP binding to stable MTs. In this case, we immobilize pre-polymerized, fluorescently labeled MTs stabilized with Taxol or GMPCPP in a TIRF chamber and introduce the diluted lysate (Fig. 4A).
- 2.MAP binding to dynamic MTs. In this approach, we introduce concentrated cell lysates into a TIRF chamber containing immobilized MT seeds, from which MTs grow, and will be decorated with the fluorescently labeled MAP (Fig. 4B).
The interpretation of these experiments is straightforward and provides a qualitative readout: a rapid MT decoration by a candidate protein confirms it as a MAP. However, because we work with cell lysates, it cannot be excluded that a candidate protein interacts with MTs via the intermediate of an endogenous MAPs, in which case it could be misinterpreted as a MAP (similar to what can happen in cell-based studies). While this possibility can only be excluded by reconstituting MT interactions with the purified protein of interest, we would argue that the high levels of overexpression of exogenous proteins in HEK 293 cells makes it rather unlikely that potential adapter proteins are present at sufficiently high concentrations to efficiently link the protein of interest to MTs.
In addition to the screening of MT binders, our protocol can be adapted to other applications, such as studying the role of MAPs in actin-MT crosstalk (Fig. 4C), the impact of MAP mutations found in human pathologies on MT binding (Fig. 4D), and for structural studies using cryo-electron microscopy (Fig. 4E). Those applications are reported and discussed in detail in Jijumon et al. (2022).
Because we visualized the growing MTs via the GFP-tagged MAPs binding to them, there was no need of additional MT staining during the dynamic assays. However, to confirm the presence of MTs, we performed an immunostaining at the end of each experiment using YL1/2 antibody (Basic Protocol 3 step 10). This antibody is specific to the tyrosinated form of α-tubulin, which is predominant in HEK 293 cells (Barisic et al., 2015). The choice of the antibody was made based on the highly efficient labeling; however, other antibodies could potentially be used in our assay.
If required, our assay can also be performed with direct visualization of growing MTs. This can be done by addition of purified, fluorescently labeled tubulin into the cell lysate. However, given that the addition of labeled tubulin can affect MT behavior, one should aim for the addition of minimal amounts to avoid artefacts. Alternatively, cell lysates expressing the MAP of interest can be prepared from cell lines stably expressing fluorescently labeled tubulin. We successfully tested HeLa cells expressing EGFP-α-tubulin in our lysate approach (Jijumon et al., 2022).
Our goal in developing this approach was to create a rapid and simple method for testing many MAPs and MAP candidates. Therefore, we chose a straightforward and simple technique to treat the glass surfaces of the TIRF chambers. For assays with lower fluorescent signals, such as those with weaker-binding MAPs or single-molecule studies with cell lysates, a more intricate glass surface treatment (Bieling et al., 2010) is recommended to reduce background fluorescence and improve signal-to-noise ratios.
Time Considerations
Once a protein of interest is cloned into a mammalian expression plasmid, the preparation of lysates can be performed in 3 days (Basic Protocol 1). After seeding HEK 293 cells, they require 12 to 16 hr growth before transfection. After transfection the cells are further grown for 24 to 48 hr based on the expression level of the protein of interest (GFP intensity). The lysate preparation from transfected cells takes ∼3 to 4 hr (the precise timing depends on the number of lysates to be prepared in parallel). We experienced that a single person can prepare up to 10 cell lysates (five 22-cm2 dishes for each MAP) in parallel. Going beyond this number risks to markedly slow down the lysate preparation and could affect lysate quality. For higher numbers, multiple experimenters should be involved, or the lysate preparation should be done in batches. The quantification of proteins in lysates takes time depending on the techniques used. Estimation of total protein concentration with a nanodrop takes only few min but immunoblots with an antibody against the proteins of interest or GFP can takes 1 to 2 days (Basic Protocol 2). The MT reconstitution assays and TIRF microscopy experiment takes 20 to 30 min for one MAP including post-staining with tubulin and actin markers (Basic Protocol 3).
Acknowledgments
This work was supported by the ANR-10-IDEX-0001-02, the LabEx Cell'n’Scale ANR-11-LBX-0038 and the Institut de convergence Q-life ANR-17-CONV-0005. C.J. is supported by the Institut Curie, the French National Research Agency (ANR) awards ANR-17-CE13-0021 and ANR-20-CE13-0011, and the Fondation pour la Recherche Medicale (FRM) grants DEQ20170336756 and MND202003011485. A.S.J. was supported by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 675737, and the FRM grant FDT201904008210. The authors acknowledge all members of C. Janke lab; T. Surrey; and M. Sirajuddin and his lab members for the technical advice, in vitro reconstitution training and cryo-EM experiments.
Author Contributions
A. S. Jijumon : Conceptualization; data curation; formal analysis; methodology; supervision; visualization; writing—original draft; writing—review and editing. Arya Krishnan : Data curation; writing—original draft; writing—review and editing. Carsten Janke : Conceptualization; funding acquisition; project administration; supervision; validation; writing—review and editing.
Conflict of Interest
The authors declare that they do not have any conflicts of interest.
Open Research
Data Availability Statement
All commercial reagents and antibodies used in this protocol are detailed in respective sessions. All data supporting the findings of this study are available from the corresponding author upon email requests.
Literature Cited
- Alfaro-Aco, R., & Petry, S. (2015). Building the microtubule cytoskeleton piece by piece. Journal of Biological Chemistry , 290(28), 17154–17162. https://doi.org/10.1074/jbc.R115.638452
- Barisic, M., Silva e Sousa, R., Tripathy, S. K., Magiera, M. M., Zaytsev, A. V., Pereira, A. L., Janke, C., Grishchuk, E. L., & Maiato, H. (2015). Microtubule detyrosination guides chromosomes during mitosis. Science , 348(6236), 799–803. https://doi.org/10.1126/science.aaa5175
- Belmont, L., Hyman, A., Sawin, K., & Mitchison, T. (1990). Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell , 62(3), 579–589. https://doi.org/10.1016/0092-8674(90)90022-7
- Bergman, Z. J., Wong, J., Drubin, D. G., & Barnes, G. (2019). Microtubule dynamics regulation reconstituted in budding yeast lysates. Journal of Cell Science , 132(4), jcs219386. https://doi.org/10.1242/jcs.219386
- Bieling, P., Telley, I. A., Hentrich, C., Piehler, J., & Surrey, T. (2010). Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and+ TIP dynamics. Methods in Cell Biology , 95, 555–580. https://doi.org/10.1016/S0091-679X(10)95028-0
- Bodakuntla, S., Jijumon, A., Janke, C., & Magiera, M. M. (2020). Purification of tubulin with controlled posttranslational modifications and isotypes from limited sources by polymerization-Depolymerization cycles. Journal of Visualized Experiments : JoVE , (165), https://doi.org/10.3791/61826
- Bodakuntla, S., Jijumon, A., Villablanca, C., Gonzalez-Billault, C., & Janke, C. (2019). Microtubule-associated proteins: Structuring the cytoskeleton. Trends in Cell Biology , 29(10), 804–819. https://doi.org/10.1016/j.tcb.2019.07.004
- Caplow, M., & Shanks, J. (1996). Evidence that a single monolayer tubulin-GTP cap is both necessary and sufficient to stabilize microtubules. Molecular Biology of the Cell , 7(4), 663–675. https://doi.org/10.1091/mbc.7.4.663
- Current Protocols. (2006). Commonly Used Reagents. Current Protocols in Microbiology , 00, A.2A.1–A.2A.15. https://doi.org/10.1002/9780471729259.mca02as00
- Dionne, M. A., Sanchez, A., & Compton, D. A. (2000). ch-TOGp is required for microtubule aster formation in a mammalian mitotic extract. Journal of Biological Chemistry , 275(16), 12346–12352. https://doi.org/10.1074/jbc.275.16.12346
- Dogterom, M., & Surrey, T. (2013). Microtubule organization in vitro. Current Opinion in Cell Biology , 25(1), 23–29. https://doi.org/10.1016/j.ceb.2012.12.002
- Field, C. M., Pelletier, J. F., & Mitchison, T. J. (2017). Xenopus extract approaches to studying microtubule organization and signaling in cytokinesis. Methods in Cell Biology , 137, 395–435. https://doi.org/10.1016/bs.mcb.2016.04.014
- Gaglio, T., Saredi, A., & Compton, D. A. (1995). NuMA is required for the organization of microtubules into aster-like mitotic arrays. The Journal of Cell Biology , 131(3), 693–708. https://doi.org/10.1083/jcb.131.3.693
- Gell, C., Bormuth, V., Brouhard, G. J., Cohen, D. N., Diez, S., Friel, C. T., Helenius, J., Nitzsche, B., Petzold, H., Ribbe, J., Schäffer, E., Stear, J. H., Trushko, A., Varga, V., Widlund, P. O., Zanic, M., & Ribbe, J. (2010). Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods in Cell Biology , 95, 221–245. https://doi.org/10.1016/S0091-679X(10)95013-9
- Genova, M., Grycova, L., Puttrich, V., Magiera, M. M., Lansky, Z., Janke, C., & Braun, M. (2023). Tubulin polyglutamylation differentially regulates microtubule-interacting proteins. The EMBO Journal , 42(5), e112101. https://doi.org/10.15252/embj.2022112101
- Huang, S. C., Jagadeeswaran, R., Liu, E. S., & Benz, E. J. (2004). Protein 4.1 R, a microtubule-associated protein involved in microtubule aster assembly in mammalian mitotic extract. Journal of Biological Chemistry , 279(33), 34595–34602. https://doi.org/10.1074/jbc.M404051200
- Hyman, A. A., Salser, S., Drechsel, D., Unwin, N., & Mitchison, T. J. (1992). Role of GTP hydrolysis in microtubule dynamics: Information from a slowly hydrolyzable analogue, GMPCPP. Molecular Biology of the Cell , 3(10), 1155–1167. https://doi.org/10.1091/mbc.3.10.1155
- Jeong, J. Y., Yim, H. S., Ryu, J. Y., Lee, H. S., Lee, J. H., Seen, D. S., & Kang, S. G. (2012). One-step sequence-and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Applied and Environmental Microbiology , 78(15), 5440–5443. https://doi.org/10.1128/AEM.00844-12
- Jijumon, A., Bodakuntla, S., Genova, M., Bangera, M., Sackett, V., Besse, L., Maksut, F., Henriot, V., Magiera, M. M., Sirajuddin, M., & Janke, C. (2022). Lysate-based pipeline to characterize microtubule-associated proteins uncovers unique microtubule behaviours. Nature Cell Biology , 24(2), 253–267. https://doi.org/10.1038/s41556-021-00825-4
- Kielar, M., Tuy, F. P. D., Bizzotto, S., Lebrand, C., de Juan Romero, C., Poirier, K., Oegema, R., Mancini, G. M., Bahi-Buisson, N., Olaso, R., Le Moing, A. G., Boutourlinsky, K., Boucher, D., Carpentier, W., Berquin, P., Deleuze, J. F., Belvindrah, R., Borrell, V., Welker, E., & Francis, F. (2014). Mutations in Eml1 lead to ectopic progenitors and neuronal heterotopia in mouse and human. Nature Neuroscience , 17(7), 923–933. https://doi.org/10.1038/nn.3729
- Krajka, V., Vulinovic, F., Genova, M., Tanzer, K., Jijumon, A., Bodakuntla, S., Tennstedt, S., Mueller-Fielitz, H., Meier, B., Janke, C., Klein, C., & Rakovic, A. (2022). H-ABC–and dystonia-causing TUBB4A mutations show distinct pathogenic effects. Science Advances , 8(10), eabj9229. https://doi.org/10.1126/sciadv.abj9229
- Milunović-Jevtić, A., Mooney, P., Sulerud, T., Bisht, J., & Gatlin, J. (2016). Centrosomal clustering contributes to chromosomal instability and cancer. Current Opinion in Biotechnology , 40, 113–118. https://doi.org/10.1016/j.copbio.2016.03.011
- Nakata, T., & Hirokawa, N. (1995). Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. The Journal of Cell Biology , 131(4), 1039–1053. https://doi.org/10.1083/jcb.131.4.1039
- Saredi, A., Howard, L., & Compton, D. A. (1997). Phosphorylation regulates the assembly of NuMA in a mammalian mitotic extract. Journal of Cell Science , 110(11), 1287–1297. https://doi.org/10.1242/jcs.110.11.1287
- Soppina, V., Norris, S. R., Dizaji, A. S., Kortus, M., Veatch, S., Peckham, M., & Verhey, K. J. (2014). Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proceedings of the National Academy of Sciences , 111(15), 5562–5567. https://doi.org/10.1073/pnas.1400759111
- Souphron, J., Bodakuntla, S., Jijumon, A., Lakisic, G., Gautreau, A. M., Janke, C., & Magiera, M. M. (2019). Purification of tubulin with controlled post-translational modifications by polymerization–depolymerization cycles. Nature Protocols , 14(5), 1634–1660. https://doi.org/10.1038/s41596-019-0153-7
- Telley, I. A., Gáspár, I., Ephrussi, A., & Surrey, T. (2013). A single Drosophila embryo extract for the study of mitosis ex vivo. Nature Protocols , 8(2), 310–324. https://doi.org/10.1038/nprot.2013.003

