Pilot Exercise: Generating the Illumina SampleSheet and sharing data via BaseSpace
Ruth Timme, Maria Balkey, Tunc Kayikcioglu, Candace Bias, Cameron Boerner, James Pettengill
Abstract
This protocol describes the required steps to generate Illumina SampleSheet and provides guidelines for sharing data via BaseSpace with FDA for the 2021 Pilot Exercise
Steps
Generating Sequencing SampleSheet
Create your SampleSheet using Excel or a text editor. Name your sample sheet according your internal protocols, and use a *.csv extension. SampleSheet.xlsx The sample sheet is organized in sections titled Header, Reads, Settings and Data. Section headings are case-sensitive and shown in brackets [ ]. Details for each section are described below.
Header Section.
| A | B |
|---|---|
| Parameter | Description |
| Investigator Name | First Name and Last Name for laboratory scientist |
| Experiment Name | Pilot Exercise SARS-Cov-2 |
| Date | YYYY-MM-DD |
| Workflow | GenerateFASTQ |
| Application | FASTQ Only |
| Instrument Type | MiSeq |
| Library Prep Kit | Swift Normalase Amplicon NGS Panels (SNAP) |
| Index Kit | Swift Normalase Amplicon NGS Panels (SNAP) |
| Description | FDA Wastewater Protocol Pilot Exercise |
| Chemistry | Amplicon |
Table 1: Header Section in Sequencing SampleSheet
Reads Section
**Enter the length of reads you are generating (could be different from 151)
| A | B |
|---|---|
| Parameter | Description |
| Number of cylces in Read 1 | 151 |
| Number of cycles in Read 2 | 151 |
Table 2: Reader Section in Sequencing SampleSheet
Settings Section
| A | B |
|---|---|
| ReverseComplement | 0 |
Table 3: Settings Section in Sequencing SampleSheet
Data Section: Include the machine ID in the Sample_Name column. e.g. sample_B01_M01234 where M01234 corresponds to the machine ID. This is the ID that will get populated to the fastq file names.
| A | B | C | D | E | F | G | H | I | J |
|---|---|---|---|---|---|---|---|---|---|
| Sample_ID | Sample_Name | Sample_Plate | Sample_Well | I7_Index_ID | index | I5_Index_ID | index2 | Sample_Project | Description |
| WPP_sample_B.01 | WPP_sample_B.01_M01234 | 1 | A1 | SU001 | TTGTTCCTTG | I5_1 | CCGAACAACA | LabAbreviation-WPP | |
| WPP_sample_C.01 | WPP_sample_C.01_WPP-sample_B.01 | 1 | B1 | SU002 | AGGTTGTGTT | I5_2 | AAGAATCGGC | LabAbreviation-WPP | |
| WPP_sample_SA-1.01 | WPP_sample_SA-1.01_WPP-sample_B.01 | 1 | C1 | SU003 | TATTGGTTGG | I5_3 | CACTTCGCTT | LabAbreviation-WPP | |
| WPP_sample_SA-2.01 | WPP_sample_SA-2.01_WPP-sample_B.01 | 1 | D1 | SU004 | CACTTCGCTT | I5_4 | TATTGGTTGG | LabAbreviation-WPP |
Table 4: Data Section in Sequencing SampleSheet (dashes converted to underscores for Illumina requirements)
If a new library or sequencing protocol is utilized for processing the pilot exercise samples, make the changes in the corresponding parameters for the sequencing sample sheet.
Starting your MiSeq run and connecting to BaseSpace:
When you set up the run on the MiSeq, select the option to log in to BaseSpace.
Transfer sequencing data through BaseSpace
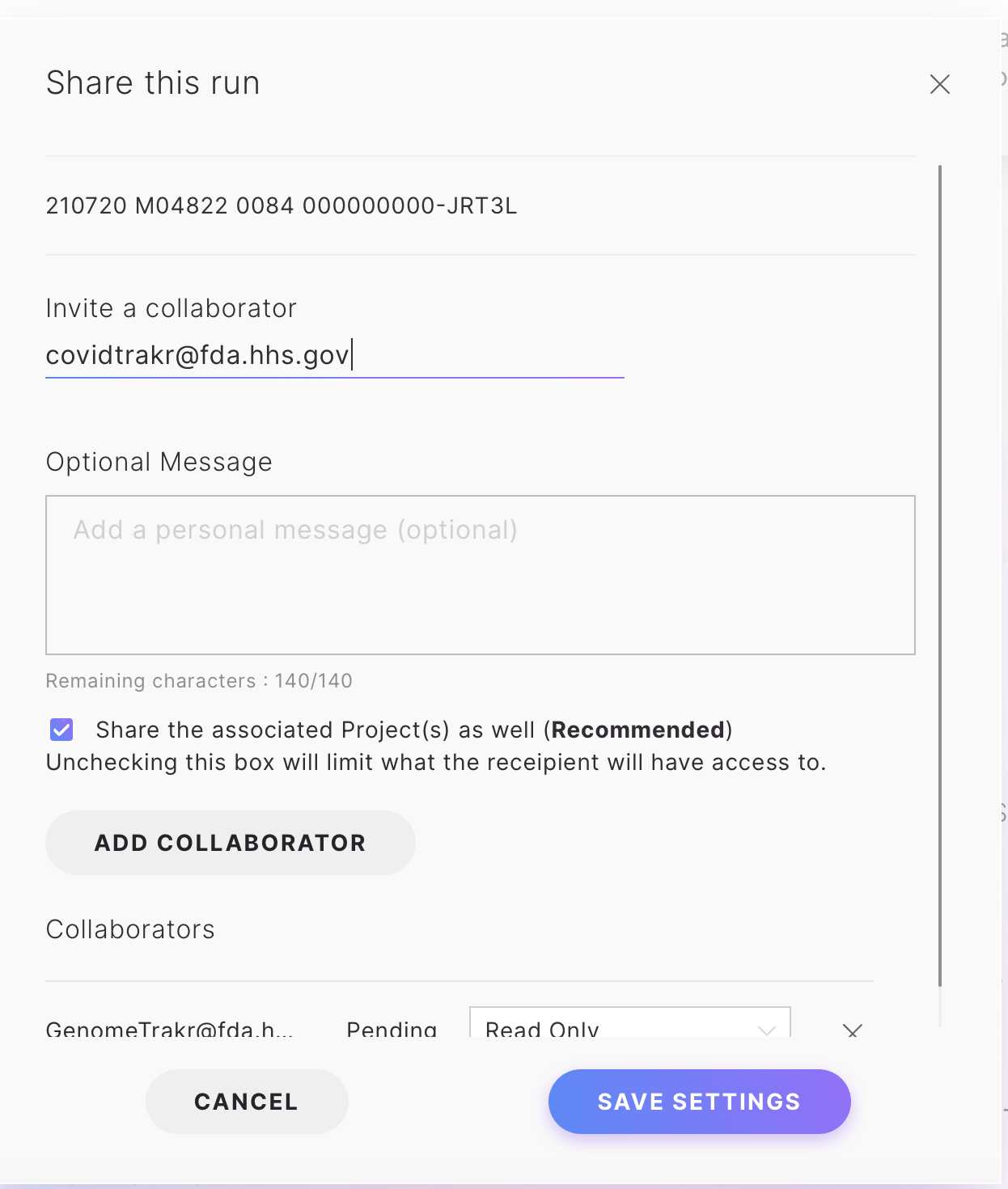
Click the "Share Project" option to share the run using an email address.
Click the Runs tab.
Select the run that you would like to share with the FDA LFFM wastewater project team.
Click Share.
Enter the email address for the FDA team ( covidtrakr@fda.hhs.gov) , and then click Add Collaborator.