Chloroplast Transformation in Arabidopsis
Stephanie Ruf, Stephanie Ruf, Xenia Kroop, Xenia Kroop, Ralph Bock, Ralph Bock
Arabidopsis
biolistic transformation
chloroplast transformation
plastid transformation
transplastomic plant
Abstract
Plastids (chloroplasts) are the defining organelles of plants and eukaryotic algae. In addition to performing photosynthesis, plastids harbor numerous other metabolic pathways and therefore are often referred to as the biosynthetic center of the plant cell. The chloroplasts of seed plants possess dozens of copies of a circular genome of ∼150 kb that contains a conserved set of 120 to 130 genes. The engineering of this genome by genetic transformation is technically challenging and currently only possible in a small number of species. In this article, we describe the methods involved in generating stable chloroplast-transformed (transplastomic) plants in the model species Arabidopsis (Arabidopsis thaliana). The protocols presented here can be applied to (1) target genes in the Arabidopsis chloroplast genome by reverse genetics and (2) express reporter genes or other foreign genes of interest in plastids of Arabidopsis plants. © 2021 The Authors.
Basic Protocol 1 : Generation of root-derived microcallus material for biolistic transformation
Basic Protocol 2 : Chloroplast transformation by biolistic bombardment of root-derived microcalli
Basic Protocol 3 : Regeneration of transplastomic lines and seed production
INTRODUCTION
The species Arabidopsis thaliana was first described by the German botanist and physician Johannes Thal (1542-1583) in his seminal book Sylva Hercynia (published after his death in 1588; Fig. 1). Sylva Hercynia described the plants occurring around the Harz mountains in Germany and was the first ever published to detail flora (i.e., the first attempt to cover all plant species in a geographic region). In the second half of the twentieth century, Arabidopsis became the model species of plant biology. Several favorable attributes contributed to the quick rise of Arabidopsis as a model plant in genetics and molecular biology, including its small genome size (<150 Mbp; The 1001 Genomes Consortium, 2016), short generation time (<2 months), and small plant size (Fig. 1B, C). Later, it turned out that Arabidopsis was also a lucky choice for transgenic research. Transformation of the nuclear genome is unusually simple (Clough & Bent, 1998) in that it does not even require selection of transgenic cells and their regeneration into transgenic plants in tissue culture. Instead, by a method called “floral dip,” flowers of greenhouse-grown plants are exposed to a suspension of Agrobacterium tumefaciens , followed by selection of transgenic plants by seed germination in the presence of the appropriate selection agent. By contrast, transformation of the chloroplast genome proved to be much more challenging. It required the development of efficient procedures for selection of transplastomic cell lines and workable protocols for in vitro regeneration of plants (Ruf et al., 2019). Although the feasibility of introducing foreign DNA into the Arabidopsis plastid genome was demonstrated more than 20 years ago (Sikdar, Serino, Chaudhuri, & Maliga, 1998), the generation of fertile Arabidopsis plants with stably transformed plastid genomes has become possible only recently (Ruf et al., 2019).

Plastid transformation technology has many applications in both basic and applied research (Bock, 2015; Fuentes, Armarego-Marriott, & Bock, 2018; Maliga, 2004; Maliga & Bock, 2011) and also stirred considerable excitement in synthetic biology (Boehm & Bock, 2019; Scharff & Bock, 2014). Since integration of the transforming DNA into the plastid genome occurs exclusively by homologous recombination, plastid transformation provides an efficient method for functional genomics by reverse genetics. The knockout of open reading frames and the introduction of specific point mutations into chloroplast genes are straightforward. The transplastomic technology also facilitates the study of all aspects of plastid gene expression in vivo, including transcription and transcriptional regulation; RNA processing and turnover; and translation and translational regulation. Finally, the technology can be used to introduce foreign genes into the chloroplast genome, including reporter genes and biotechnologically relevant transgenes.
The generation of transplastomic Arabidopsis plants involves three steps that are detailed below. In Basic Protocol 1, we describe the methods involved in the growth of aseptic Arabidopsis plants and the production of suitable tissue for particle gun-mediated transformation (also referred to as biolistic transformation). In Basic Protocol 2, we provide a step-by-step protocol for biolistic chloroplast transformation. In Basic Protocol 3, we provide a detailed protocol for the selection of transplastomic calli and shoots, rooting, and seed production in sterile conditions.
NOTE : Tissue culture chemicals for preparation of the sterile plant culture media should be certified as “Plant tissue culture grade” (or equivalent; purchased, e. g., from Duchefa Biochemie).
Detailed information about plant tissue culture techniques, preparation, stability, and storage of media and stock solutions can also be found on the websites of several providers of chemical, reagents, and supplies for plant cell and tissue culture (e.g., https://phytotechlab.com/technical-info).
The pH of all media used in this protocol should be adjusted with 0.2 M or 2.0 M KOH.
All solutions that are in contact with aseptic plant tissue must be sterilized by autoclaving or sterile filtration.
Basic Protocol 1: GENERATION OF ROOT-DERIVED MICROCALLUS MATERIAL FOR BIOLISTIC TRANSFORMATION
The most suitable target tissue for biolistic transformation of Arabidopsis is roots of young seedlings. This is because the root contains a meristematic cell layer, the pericycle, that remains diploid and does not undergo somatic endopolyploidization. By contrast, leaf cells of Arabidopsis exhibit a strong tendency to become polyploid (with the average ploidy level in fully expanded rosette leaves being 13C; Preuten et al., 2010). While polyploidy does not pose an obstacle to successful transformation, it leads to problems with plant regeneration, and even if plants can be regenerated, they are usually infertile (Hibberd, 2017; Sikdar et al., 1998; Yu, Lutz, & Maliga, 2017).
For efficient plastid transformation in Arabidopsis , it is highly recommended that (1) an ecotype that displays good regeneration properties in tissue culture and (2) an ACC2 -deficient genotype are used. ACC2 is a nuclear gene encoding an alternative acetyl-CoA carboxylase (ACCase) that can partially replace the plastid ACCase under conditions of inhibited plastid gene expression (Parker, Wang, & Meinke, 2014, 2016). Expression of ACC2 has been shown to be largely responsible for the low sensitivity of cultured Arabidopsis cells to the plastid translational inhibitor spectinomycin (Parker et al., 2014, 2016). Because the spectinomycin resistance gene aadA is the most efficient selectable marker gene for plastid transformation (Bock, 2015; Svab & Maliga, 1993), the poor inhibition of cell division in the presence of spectinomycin greatly impairs the isolation of transplastomic Arabidopsis cells. This problem was solved by generating an ACC2 knockout line (Δ acc2) by CRISPR/Cas9-based genome editing in the easily regenerated Arabidopsis ecotype C24 (Ruf et al., 2019). It is recommended that the Δ acc2 C24 line be used as the recipient for plastid transformation. The protocol below describes the generation of tissue for biolistic plastid transformation by (1) raising Arabidopsis seedlings under aseptic conditions, (2) harvesting roots, and (3) inducing microcallus tissue from roots for biolistic transformation. If executed with the recommended amount of seeds, the protocol will provide material for up to 70 bombardments (i.e., shots with the biolistic gun), which are sufficient for one chloroplast transformation experiment and two control bombardments with a nuclear transformation vector.
NOTE : All materials for plant tissue culture (TC) must be sterilized by autoclaving, heat sterilization, or surface sterilization.
Materials
- Arabidopsis thaliana (At) seeds: cultivar C24, knockout mutant Δ acc2
- Ultrapure water, autoclaved
- EtOH + detergent solution: 25 ml ethanol (70%, v/v, tech. grade) plus 0.75 ml Tween 20 (10%, w/v)
- Hypochlorite + detergent solution: 40 ml NaOCl (13% active chlorine, store at 4°C) plus 1.2 ml Tween 20 (10%, w/v)
- 6 plates (145 × 20 mm) with A. thaliana germination medium (AtGM; see recipe in Table 1)
- 30 plates (100 × 20 mm) with A. thaliana callus induction medium (AtCIM; see recipe in Table 1)
| Medium | Composition | Spec concentration | Tissue culture vessel type and size (mm) | Volume for given vessel | Protocol section |
|---|---|---|---|---|---|
| AtGM |
2.2 g/L MS salt and vitamin mix 10 g/L sucrose 0.5 g/L MES monohydrate, pH adjusted to 5.7 6.8 g/L micro agar |
0 mg/L 100 mg/L |
Petri dish diameter: 1450/H 20 Petri dish diameter: 100/H 20, Square Petri dish 120 × 120 × H 17 |
125 ml 50 ml 100 ml |
Sowing and stratification of seeds Seed assay |
| AtCIM |
3.1 g/L Gamborg B5 salts 0.104 g/L McCown woody plant vitamin mixture (see recipe) 22 g/L glucose monohydrate 0.5 g/L MES monohydrate 0.05 mg/L kinetin 0.5 mg/L 2,4-D 0.025 mg/L PSK-alpha, pH adjusted to 5.7 (see recipe) 5.4 g/L Daishin agar |
0 mg/L |
Petri dish diameter: 100/H 20 |
50 ml | Microcallus induction |
| AtSIM |
3.1 g/L Gamborg B5 salts 0.104 g/L McCown woody plant vitamin mixture (see recipe) 20.0 g/L sucrose 0.5 g/L MES monohydrate 0.15 mg/L IAA 2.0 mg/L 2-iP, pH adjusted to 5.7 5.4 g/L Daishin agar |
50 mg/L |
Petri dish diameter: 100/H 20 |
50 ml | Regeneration of transplastomic lines and seed production |
| AtRIM |
4.4 g/L MS salt and vitamin mix 10 g/L sucrose 0.5 g/L MES monohydrate 1.0 mg/L IBA, pH adjusted to 5.7 6.5 g/L Daishin agar |
10 mg/L |
Tissue culture flat tube Diameter: 30 × H 105 |
10 ml | Root induction |
| AtPM |
4.4 g/L MS salt and vitamin mix 10 g/L sucrose 0.5 g/L MES monohydrate, pH adjusted to 5.7 6.5 g/L Daishin agar |
10 mg/L |
Magenta GA-7 vessel 77 × 77 × 97 Cylindrical jar Diameter: 100 × H 200 |
50 ml 300 ml |
Plant growth and seed production |
-
aAtGM, A. thaliana germination medium; AtCIM, A. thaliana callus induction medium; AtSIM, A. thaliana shoot induction medium; AtRIM, A. thaliana root induction medium; AtPM, A. thaliana propagation medium; IAA, indole-3-acetic acid; 2-iP, N6-(2-isopentenyl)adenine; IBA, indole-3-butyric acid.
- Autoclave, water cooled (i.e., Systec, V150)
- Precision and analytical balances
- Filter unit for syringe, 0.2 µm (e.g., GE Healthcare Life Sciences, Whatman, FP30/0.2 CA-S)
- Syringe, 50 ml, one-way, sterile
- 50-ml screw-cap tubes, self-standing centrifuge tube, polypropylene, 115 × 28 mm (e.g., Sarsteadt, 62.559.001)
- Rack for reaction tubes
- 5-ml pipet and suitable tips (e.g., Eppendorf)
- 1,000, 200, 100, and 20-µl pipets and suitable tips
- Cold room and/or fridge (4°C)
- Sterile laminar flow hood (clean bench)
- Timer
- Disc rotary mixer (e.g., Thermo Fisher Scientific)
- Petri dishes, 145 × 20 mm, with vents (e.g., Greiner Bio-One, 639102)
- Cling film (e.g., Saran wrap), 0.3 × 300 m, cut in 2.5-cm slices
- Tweezers, pointed, length: 15 cm (e.g., Hammacher, HWC 130-15)
- Polyester net, 210 µm, cut in disks of 130 mm diameter (e.g., Hartenstein, PES1)
- Tissue culture Petri dishes, 100 × 20 mm, with vents, nonpyrogenic, polystyrene (e.g., Corning, 430591 or 430167)
- Polyester net, 210 µm, cut in disks of 80 mm diameter (e.g., Hartenstein, PES1)
- Medical tape, narrow, 1.25 cm × 9.2 m (e.g., BSN medical 02453-00)
- Disposable scalpel, sterile, blade shape 21 (e.g., Swann-Morton)
- Tissue culture incubators with temperature control and air circulation optimized for plant tissue culture (e.g., Percival Scientific)
- Light-tight cardboard box for dark incubation
Seed sterilization
1.Fill a 50-ml screw-cap tube up to 5-7 mm (∼700 µl) with At seeds (Fig. 2A).
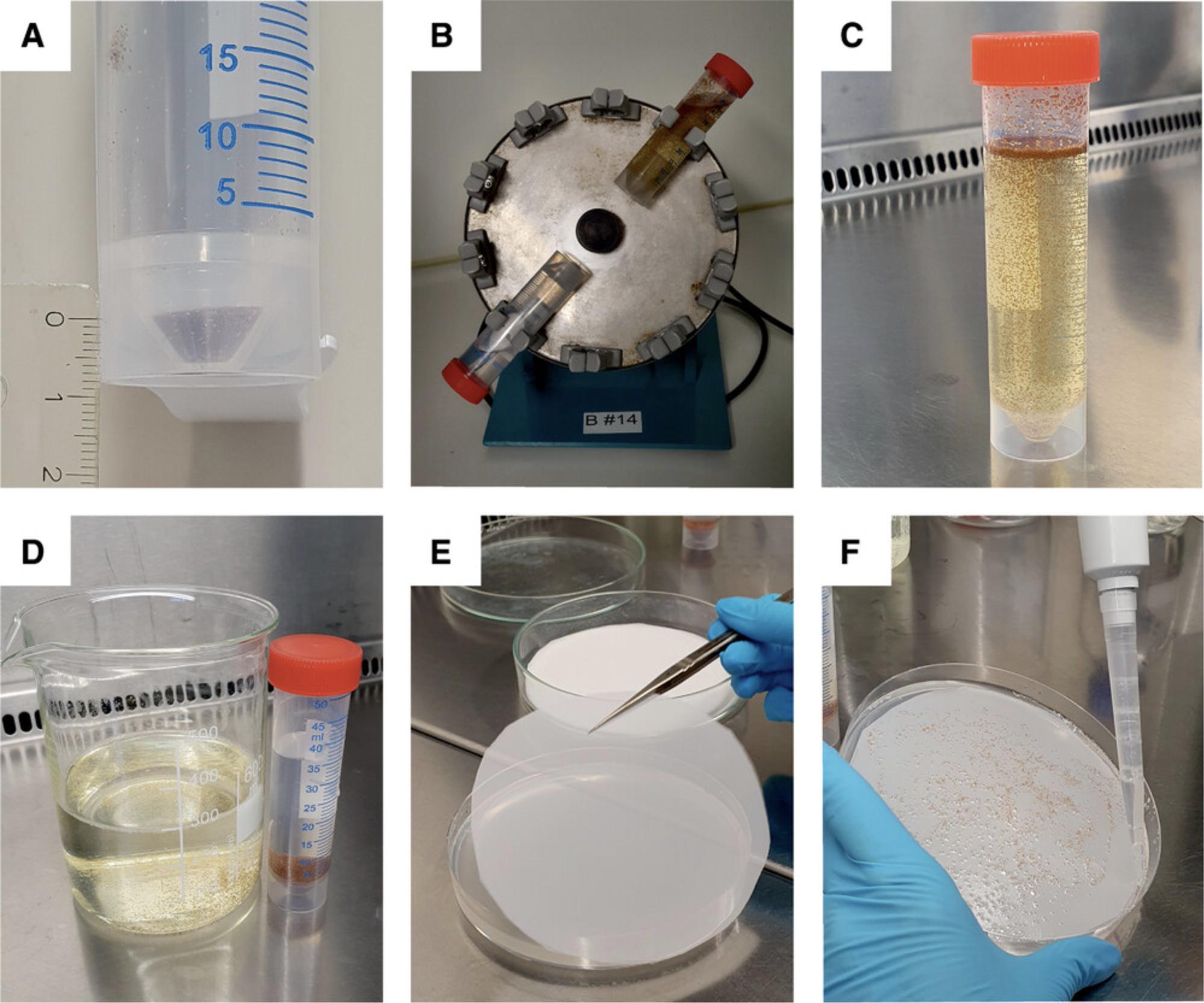
2.Wash seeds with EtOH + detergent solution by adding 25 ml EtOH tech. and 0.75 ml Tween 20 (10%, w/v) to the seeds.
3.Incubate 20 min with shaking (disc rotary mixer) at room temperature (Fig. 2B).
4.Prepare hypochlorite + detergent solution by combining 40 ml NaOCl (cold) and 1.2 ml Tween 20 (10%, w/v).
5.At this point, all work should be done in the sterile laminar flow hood.
6.Remove EtOH + detergent solution from seeds with a 5-ml pipet.
7.Add cold hypochlorite + detergent solution to the seeds (Fig. 2C).
8.Incubate 17 min with shaking (disc rotary mixer in the cold room at 4°C).
9.Remove hypochlorite + detergent solution with a 5-ml pipet. If you are not done after 3 min (i.e., 20 min after start of incubation), quickly add 2 volumes of sterile water to dilute the NaOCl (and prevent killing the seeds).
10.Wash seeds with water by adding 30 ml sterile water. Mix by inverting. Wait a short time until the seeds sink to the bottom. Discard supernatant (Fig. 2D).
11.Repeat this washing step (step 10) at least six times, using 30 ml fresh sterile water each time. After the final step, washing water should be completely clear and odorless.
Sowing and stratification of seeds
12.Prepare six germination plates (145 × 20 mm) by placing an autoclaved polyester net (diameter: 130 mm) on top of the medium AtGM (Fig. 2E). Avoid air bubbles between the netting and the medium.
13.Mix seeds with 30 ml water by pipetting up and down before you plate the seeds (repeat for each plate).
14.Pipet 5 ml (homogeneous) seed suspension onto the polyester net and spread seeds evenly with the pipet tip. Remove excess water (Fig. 2F).
15.Close plates with cling film and number them (e.g., “Germination plate 1”)
16.Incubate plates 2 days at 4°C in the dark.
17.Transfer plates to a tissue culture incubator with 12 hr light at 20°C/12 hr dark at 18°C, 70-80 µmol of photons m-2 s-1.
18.Incubate plates for 6 days.
Root harvest
Root harvest is done on day 8 (including the 2 days of stratification). The root length should be 6 to 8 mm (Fig. 3A). You will need 20 to 30 plates (100 × 20 mm) with AtCIM for microcallus induction (i.e., for each germination plate up to five plates with AtCIM).
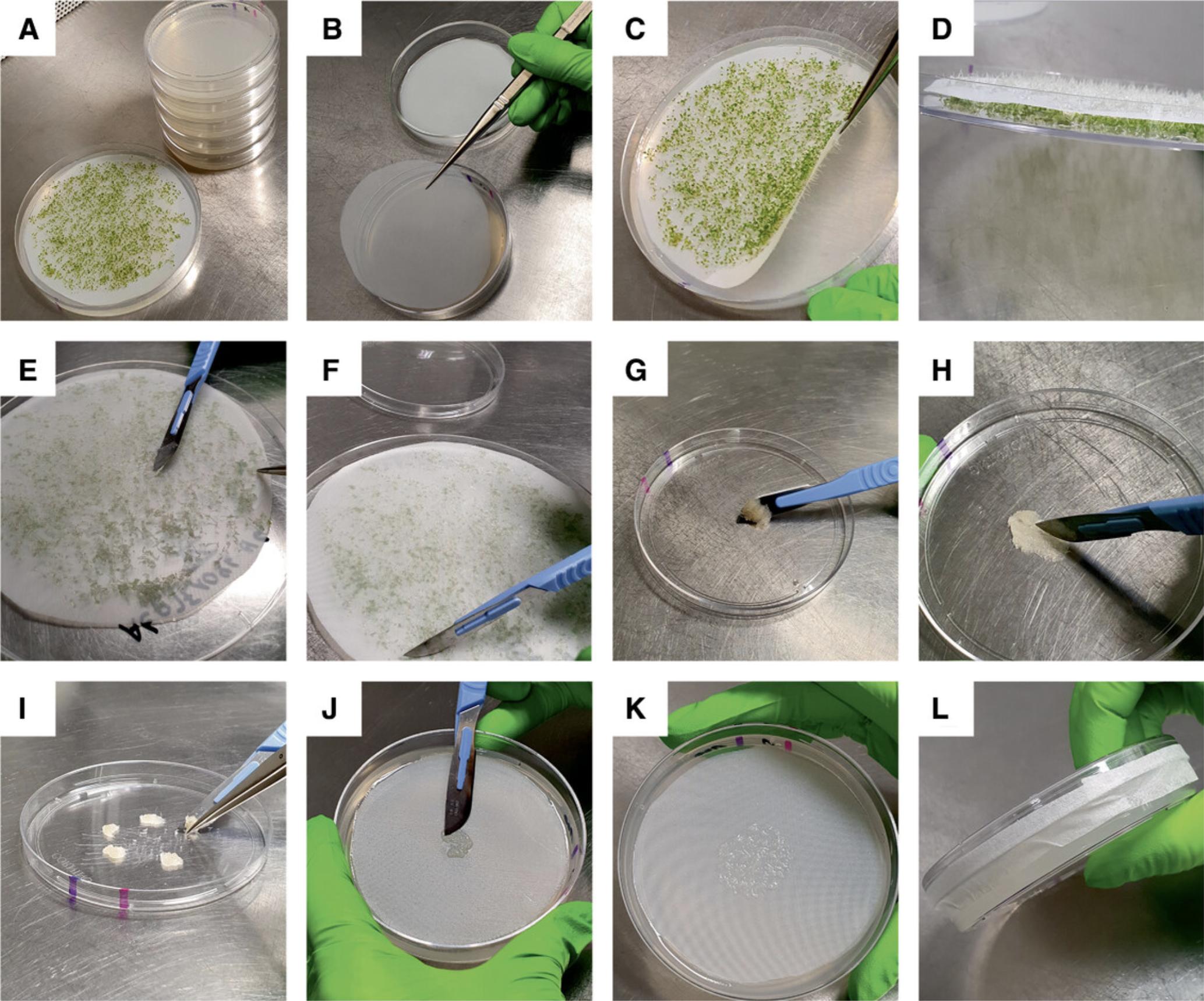
19.Prepare five Petri plates with callus-inducing medium AtCIM per germination plate, by placing an autoclaved polyester net (diameter: 80 mm) onto the medium (Fig. 3B). Avoid air bubbles between the medium and the net.
20.Slowly lift (using tweezers) polyester net with the seedlings to remove roots from the medium (Fig. 3C). Place net upside down into the lid of the Petri dish (Fig. 3D).
21.Check medium remaining in the Petri dish carefully for microbial contamination. If you identify any, discard this plate and seedlings.
22.Carefully remove residual AtGM sticking to the roots with the blunt edge of a scalpel blade (Fig. 3E).
23.Scrape off roots from the net with the scalpel blade. Hold the net with tweezers. Scrape from different directions across the net (Fig. 3F). Use a fresh set of tools for each germination plate to avoid carryover of (potentially overlooked) microbial contaminations.
24.Collect roots in the lid of a sterile Petri dish (Fig. 3G). Use scalpel and tweezers for this step.
25.Hash roots with a scalpel into ∼3 mm long pieces (Fig. 3H).
26.Divide into four to five aliquots (Fig. 3I).
27.Spread (with the scalpel) each aliquot of root material onto the net in the center of an AtCIM plate.
28.Close plate with the lid and seal it with medical tape (Fig. 3L).
29.Number individual plates, so that you know which root plate originates from which germination plate (e.g., “Root plate 1-1”)
Microcallus induction
30.Put AtCIM plates with the root material into a black cardboard box for dark incubation.
31.Incubate plates for 5 days in darkness: 16 hr dark at 25°C/8 hr dark at 20°C before proceeding to Basic Protocol 2.
Basic Protocol 2: CHLOROPLAST TRANSFORMATION BY BIOLISTIC BOMBARDMENT OF ROOT-DERIVED MICROCALLI
Particle gun-mediated transformation (also known as biolistic transformation) represents a nearly universally applicable method for the introduction of foreign DNA into cells and subcellular compartments (Altpeter et al., 2005). It is also the method of choice for the transformation of the plastid genome (Bock, 2001, 2015; Maliga, 2004). Although a few alternative methods for plastid transformation have been proposed, most of them have not been shown to produce stable transplastomic plants. The only exception is PEG-mediated transformation, which, however, is inefficient and available only in those (few) species in which plants can be efficiently regenerated from protoplasts. For plastid transformation of Arabidopsis , biolistic transformation is currently the only option.
NOTE : General information: All materials that will be in contact with the plant tissue must be sterilized by autoclaving, heat sterilization, or surface sterilization.
Materials
-
Gold particles, 0.6 µm diameter (Bio-Rad Laboratories, 1652262)
-
Ethanol p. a., 100%, water free
-
Ultrapure water, autoclaved
-
Chloroplast transformation vector: plasmid DNA, concentration 1.8-2.2 µg/µl purified by NucleoBond Xtra Midi kit for transfection-grade plasmid DNA (Macherey-Nagel, 740410.50)
-
Nuclear transformation vector for kanamycin selection (e.g., pGreen0029GFP) as control for transformation efficiency
-
CaCl2, 2.5 M, cooled to −20°C
-
Spermidine (free base), 100 mM stock solution (Duchefa, S 1369)
-
Ethanol p. a., for surface sterilization of the gun supplies
-
High-quality 1.5-ml reaction tubes, to minimize gold particle adherence to the wall (e.g., Eppendorf, DNA LoBind, 0030 108.051)
-
High-quality pipet tips, to minimize gold particle adherence
-
Table-top centrifuge, e.g., Eppendorf MiniSpin Plus, with rotor F-45-12-11 (maximum centrifuging radius: 6 cm)
-
Test tube shaker (e.g., vortex mixer)
-
Box with crushed ice
-
Waste containers (e.g., beakers) for discarded supernatants
-
Helium-driven particle gun PDS-1000/He (Bio-Rad Laboratories) and vacuum pump
-
Macrocarriers (Bio-Rad Laboratories, 1652335)
-
Rupture disks 1,800 psi (Bio-Rad Laboratories, 1652332)
-
Stopping screens (Bio-Rad Laboratories, 1652336)
-
Glass Petri dishes, 140-150 mm, sterile
-
Mono adapter setup (standard part of PDS-1000/He)
NOTE : Biolistic transformation is performed 5 days after microcallus induction. The typical chloroplast transformation experiment involves the bombardment of 20 to 30 root plates by 49 to 70 mono shots with a chloroplast transformation vector. It is recommended that two controls are always included: One root plate that remains unbombarded and will serve as a tissue culture and regeneration control (in the absence of antibiotic selection) and two plates that are transformed with a nuclear transformation vector. The root plates should be bombarded two to three times with the plasmid DNA-loaded gold particles.
The smallest possible unit of preparation of plasmid-coated gold particles is for 7 shots (prepared in one 1.5-ml tube). The values given below are calculated for 10 × 7 shots = 70 shots.
Set up particle gun and prepare materials for particle coating
NOTE : Surface sterilization of the disks (rupture disks and macrocarriers; step 3) should be done on a clean bench hosting the sterile particle gun.
1.Fill ice boxes with crushed ice.
2.Put bottles with 100% EtOH, water, and ten sterile 1.5-ml tubes on ice.
3.Work on a clean bench that harbors the particle gun. Pour ethanol into glass Petri dishes for surface sterilization of the stopping screens, macrocarriers, rupture disks (70 each), and holders (Fig. 4A-C). Soak stopping screens 5 min and disks 2 min in ethanol. Remove disks and stopping screens from the ethanol with tweezers and place them into empty Petri dishes for drying (lean them against the wall of the plate).
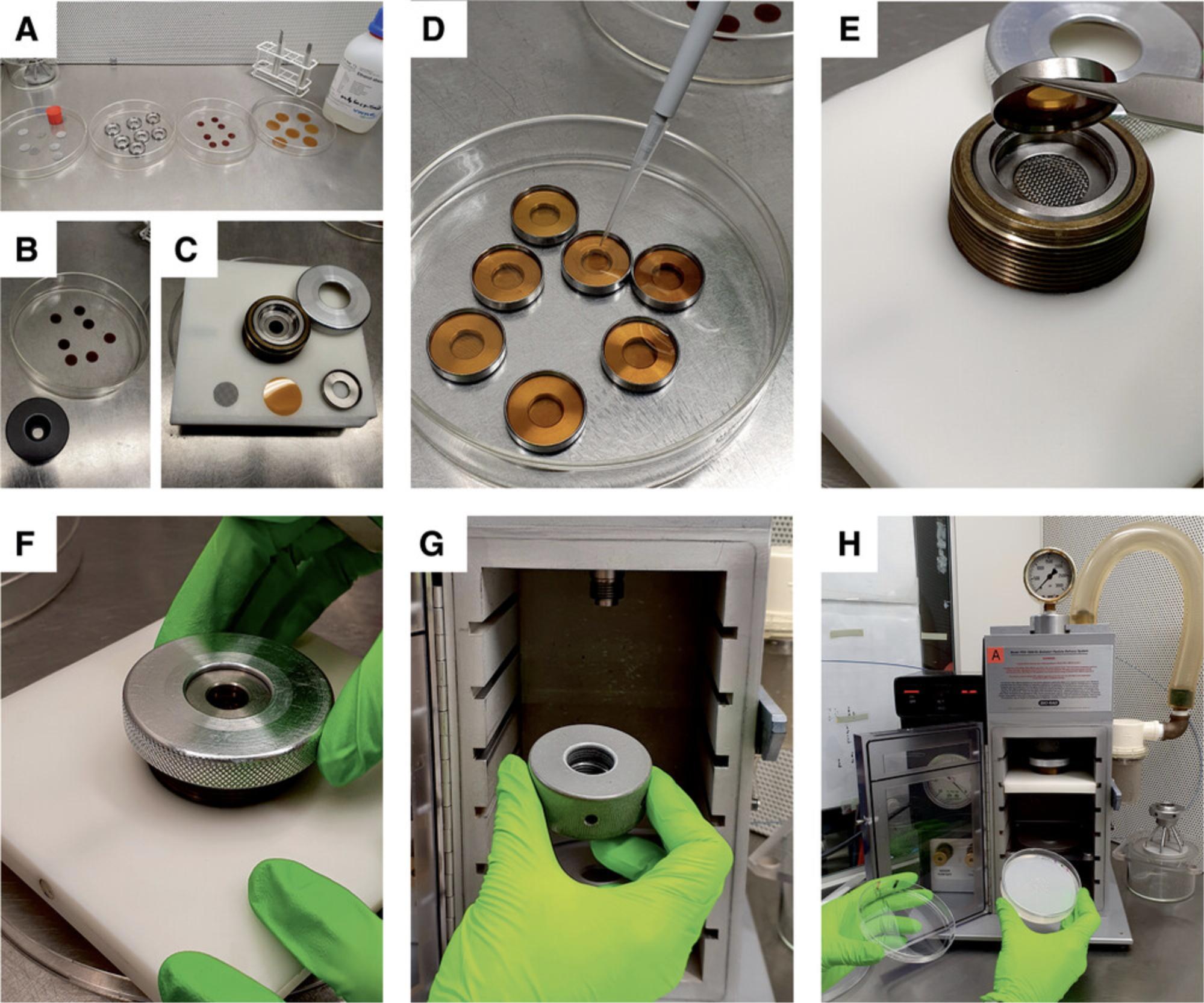
4.Switch on the vacuum pump of the particle gun half an hour before you start with biolistic transformation.
DNA coating of gold particles
5.Weigh gold particles: 8.2-9 mg per tube, two tubes for 70 shots with the mono adapter setup of the gun.
6.Add 950 µl 100% EtOH to each tube.
7.Vortex at least 1 min at maximum power.
8.Pellet particles by centrifugation in a microcentrifuge by allowing the centrifuge to accelerate to 5,000 rpm (1,677 × g). Stop the run after 1-2 s.
9.Remove EtOH with a 200-µl pipet tip.
10.Resuspend particles in 1,000 µl water. Pipet up and down while simultaneously scraping off particles that stick to the wall of the tube.
11.Centrifuge in a microcentrifuge 1-2 s at 5,000 rpm (1,677 × g), remove, and discard supernatant.
12.Resuspend particles very carefully in 875 µl water added to each tube.
13.Aliquot 172 µl particle suspension into each of ten tubes.
14.Add to each aliquot:
1.10 µl plasmid DNA (plastid transformation vector, concentration 2 µg/µl); 2.175 µl 2.5 M CaCl2 (−20°C); 3.35 µl 100 mM spermidine (free base).
Vortex briefly immediately after the addition of each compound.
15.Incubate on ice 10 min with brief vortexing every minute.
16.Centrifuge 1 s at 3,500 rpm (822 × g) in a microcentrifuge and remove supernatant completely.
17.Add 600 µl of 100% EtOH and carefully resuspend particles by pipetting up and down.
18.Centrifuge 1 s at 5,000 rpm (1,677 × g) in a microcentrifuge and remove supernatant completely.
19.Repeat this washing step.
20.Resuspend particles very carefully in 50 µl 100% EtOH by pipetting up and down.
21.Work on a clean bench with the particle gun: Carefully resuspend particles immediately before use. Drop 6.5 µl per shot onto the macrocarrier (Fig. 4D).
Biolistic bombardment of microcalli
22.Follow the instructions in the user manual of the PDS100/He (also see Fig. 4E-H) and use the following settings for shooting:
- Helium pressure at the tank regulator: 2,000-2,050 psi;
- Rupture disks: 1,800 psi;
- Macrocarrier (flying disk) assembly: level one from the top;
- Petri dish holder: level two from the bottom (equivalent to a distance from the macrocarrier assembly of ∼9 cm);
- Vacuum (at the time of the shot): 27-28 in. Hg.
Basic Protocol 3: REGENERATION OF TRANSPLASTOMIC LINES AND SEED PRODUCTION
This protocol describes the procedures involved in the selection of primary transplastomic lines, their regeneration into shoots, and the growth of transplastomic plants to maturity for seed production. Rooting of in vitro regenerated Arabidopsis plants is inefficient (Weigel & Glazebrook, 2006) and it is therefore recommended that the T0 generation be kept under sterile culture until seeds have been obtained. All subsequent generations can be raised from seeds in soil and grown under standard greenhouse conditions.
Materials
-
30 plates (100 × 20 mm) with A. thaliana shoot induction medium (AtSIM) spec50 (see recipe in Table 1; fresh plates are needed every 14 days and the number of plates required will increase with time, as detailed below)
-
Spectinomycin dihydrochloride pentahydrate, 100 mg/ml (stock solution)
-
TC flat tubes (30 × 105 mm) with A. thaliana root induction medium (AtRIM) spec10 (see recipe in Table 1), as needed
-
Magenta vessels GA-7, 77 × 77 × 97 mm with A. thaliana propagation medium (AtPM) spec10 (see recipe in Table 1), as needed
-
Cylindrical jars (100 × 200 mm) with AtPM spec10, as needed
-
Tissue culture (TC) flat tube 10, 30 × 105 mm (e.g., TPP Techno Plastic Products, 91243)
-
Rack for TC flat tubes
-
Magenta vessel GA-7, 77 × 77 × 97 mm (Magenta)
-
Cylindrical jar, 1,590 ml, 100 × 200 mm (e.g., WECK, 1138)
-
Hole puncher (for filter paper), diameter 10 mm
-
Filter paper: diameter 110 mm (e.g., Macherey-Nagel, MN 618, REF 436011); punch a hole (diameter 10 mm) halfway between the center and the edge of the filter paper
-
Medical tape, wide, 2.5 cm × 9.2 m (e.g., BSN medical, 02454-00)
-
Tweezers, extended, 30 cm (e.g., Duchefa Biochemie)
-
Scissors
-
Square Petri dishes, 120 × 120 × 17 mm, with vents (e.g., Greiner Bio-One, 688102)
-
Wooden toothpicks, sterile
Selection of resistant callus, shoot induction, and plant growth
It is highly recommended that you carefully read Critical Parameters and Troubleshooting (especially the sections Media preparation and Handling of tissue cultures) and meticulously follow all instructions therein.
1.Transfer polyester net with the bombarded microcalli to AtSIM with 50 mg/L spectinomycin.
2.Seal plates with medical tape.
3.Incubate under a diurnal cycle of 16 hr light at 25°C/8 hr dark at 20°C with a light intensity of 4-5 µmol of photons m-2 s-1.
4.Change medium every 14 days by placing the polyester net with the calli onto fresh medium (Fig. 5A). Seal plates with medical tape.
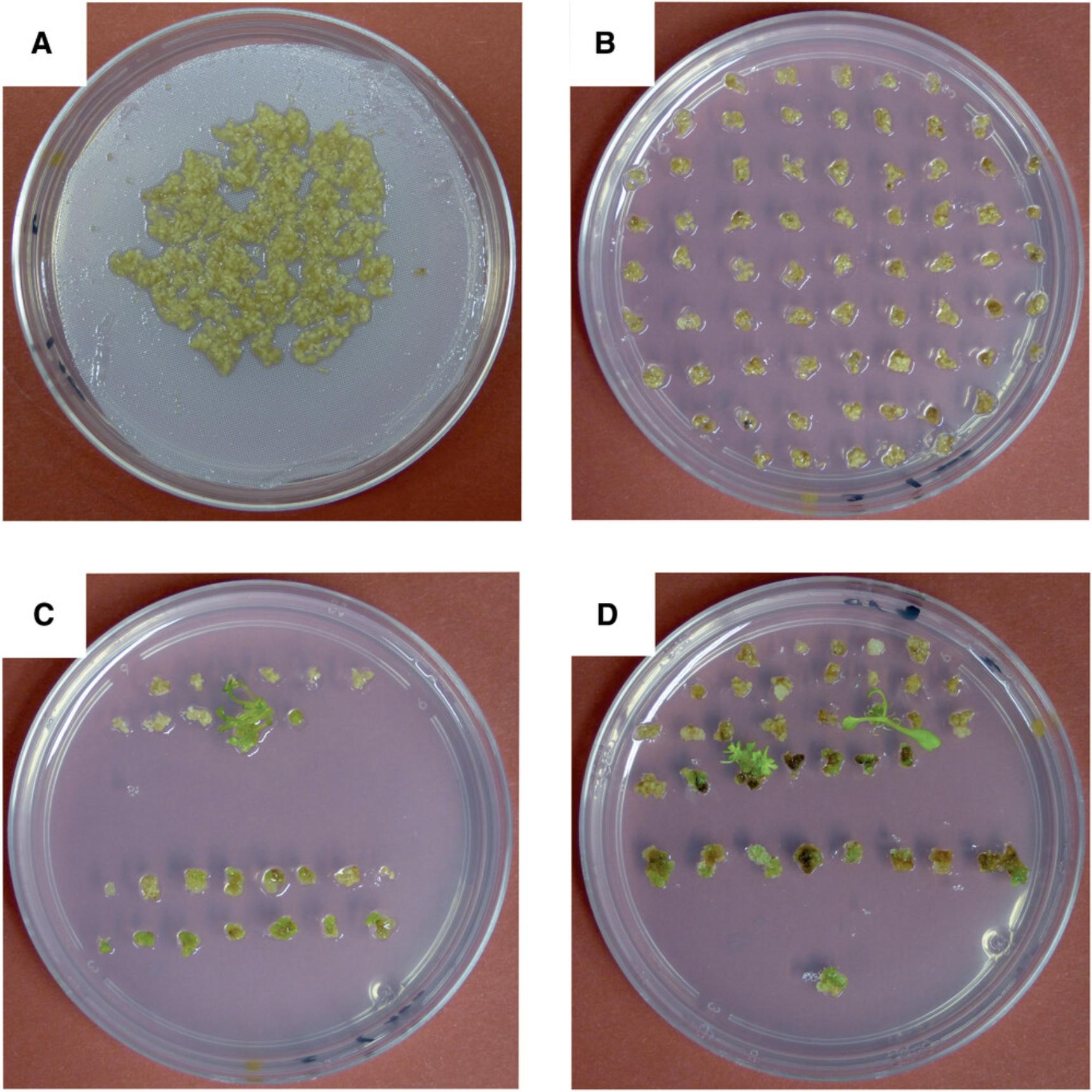
5.When calli start to overgrow each other (Fig. 5A), separate them from each other and place callus pieces of ∼5-8 mm diameter directly (without polyester netting) onto fresh AtSIM spec50 plates (Fig. 5B).
6.When green resistant callus appears, wait until it is ∼2 mm in size. Then separate green callus from the yellow-beige callus material and transfer green callus to a fresh AtSIM spec50 plate. When green calli exceed a diameter of ∼5 mm, break them into smaller pieces with two pairs of fine-tipped tweezers.
7.Remove shoots from callus by breaking them off with two fine-tipped pairs of tweezers. Place them onto a fresh plate with AtSIM spec50. Culture multiple shoots in a single plate to stimulate growth (see comment under step 6).
8.Incubate under a diurnal cycle of 16 hr light at 25°C/8 hr dark at 20°C, with a light intensity of 4-5 µmol of photons m-2 s-1.
9.Change medium every 14 days by transferring green calli and shoots to plates with fresh medium. Seal plates with medical tape.
10.Repeat steps 8 and 9 until seeds have been obtained from the transplastomic line and the seeds germinate on medium with spectinomycin (see below).
11.Start taking shoots for root induction when they have a height of ∼8 mm (or when you have multiple smaller shoots from the same transplastomic line and can sacrifice a few shoots without risk of losing the line).
Root induction
12.Remove shoots regenerating from the calli by gently breaking apart the callus tissue with two fine-tipped pairs of tweezers. The extracted shoots must not have any residual callus material at the base of the shoot.
13.Transfer shoots to TC flat tubes with AtRIM spec10.Stick two to four shoots per tube into the agar slant (Fig. 6A, B). Place tubes upright in a tube rack.

14.Incubate for 2 weeks under a diurnal cycle of 12 hr light at 20°C and 12 hr dark at 18°C with a light intensity of 50-70 µmol m-2 s-1.
15.Transfer shoots, independent of them having formed roots or not, to Magenta boxes with AtPM spec10. Stick three to four shoots per box into the medium (Fig. 6C).
16.Close Magenta box with a punched filter paper (with a hole of 10 mm diameter). Cover hole with medical tape and fix filter paper with medical tape to the wall of the Magenta box (Fig. 6C).
Plant growth and seed production
17.Incubate plants in Magenta boxes (Fig. 6C) for 1-2 weeks under a diurnal cycle of 16 hr light at 22°C and 8 hr dark at 22°C with a light intensity of 35-65 µmol m-2 s-1. Water plants every day or every other day (depending on the humidity in your growth facility) with 3-5 ml sterile water.
18.Transfer shoots to jars with AtPM spec10 (Fig. 6D). Grow three to four plants in each jar.
19.Close jar with a layer of filter paper with a punched hole. Cover hole and fix filter paper to the jar with medical tape.
20.Water plants twice a week with 10-20 ml sterile water.
21.When the plants flower, support self-pollination by daily tapping with your fingers on the wall of the jar until you see the flowers shaking.
22.When the siliques are ripe, harvest seeds and collect them in a sterile tube.
Seed assay as test for homoplasmy and maternal inheritance
Successful transformation of the chloroplast genome and integration of the transgenes by homologous recombination should be confirmed by Southern blot analysis (e.g., Ruf et al., 2019; Ruf, Hermann, Berger, Carrer, & Bock, 2001; Tabatabaei, Ruf, & Bock, 2017) using labeled probes flanking the transgene insertion site in the plastid genome. For several reasons (including the possibility of vector integration into the nuclear genome and the presence of promiscuous plastid DNA in the nuclear genome; Hager, Biehler, Illerhaus, Ruf, & Bock, 1999; Koop et al., 1996; Ruf, Biehler, & Bock, 2000), PCR is not suitable for this purpose. Southern blot analysis is also essential to assess whether or not all wild-type copies of the highly polyploidy plastid genome (Golczyk et al., 2014; Greiner et al., 2020) have been successfully eliminated by spectinomycin selection, a status referred to as “homoplasmic transplastomic.” Homoplasmy is essential for the genetic stability of the transplastomic lines, as heteroplasmic lines can lose the transgene through (random) segregation of plastid genomes in the absence of selection (Drescher, Ruf, Calsa, Carrer, & Bock, 2000). Independent confirmation of plastid transformation and homoplasmy should be provided by seed assays in which the uniparentally maternal inheritance of the (chloroplast encoded) spectinomycin resistance and the lack of segregation (i.e., absence of antibiotic-sensitive seedlings from the progeny) are demonstrated (Bock, 2001; Ruf, Karcher, & Bock, 2007).
23.Incubate seeds harvested from the T0 generation plants (representing the T1 generation) for 3 hr in sterile water.
24.Place seeds on AtGM spec100 in a Petri dish (round Petri dishes 100 × 20 mm or squared Petri dishes 120 × 120 × 17 mm; Fig. 7). To be able to pick individual seeds, pour seed suspension onto a sterile filter paper in a Petri dish. Individual seeds can then be conveniently picked with a sterile toothpick and placed onto germination medium (evenly spaced, to avoid cross-protection from the antibiotic; Fig. 7). Seal plate with cling film.

25.Incubate plates for 2 days in the dark at 4°C.
26.Transfer plates to an incubator with 12 hr light at 20°C and 12 hr dark at 18°C, light intensity 70-80 µmol of photons m-2 s-1.
REAGENTS AND SOLUTIONS
Use ultrapure water for preparation of all media and solutions.
McCown woody plant vitamin mixture, 1000×
- Add 85 ml water to 10.4 g vitamin mixture, dissolve powder, and fill up to 100 ml.
- Sterile filter and aliquot in 2-ml tubes.
- Store at −20°C for up to 1 year.
- If after thawing, a white precipitate is observed, it should be carefully redissolved by vortexing.
Peptide hormone phytosulfokine-alpha (PSK-alpha), 100 µM
- Prepare a stock solution of 84.69 mg/L.
Note that if the peptide is not 100% pure (which is normally the case), the PSK-alpha amount needs to be adjusted.
- Dissolve 0.85 mg active PSK-alpha in 10 ml water.
- Sterile filter and aliquot stock solution.
- Store at −80°C for up to 1 year.
Avoiding repeated freezing and thawing of the stock solution.
- Use 0.3 ml stock solution per liter of medium, adding it after autoclaving the medium.
Tissue culture (TC) medium
- Prepare 1 to 20 L of medium without agar. See Table 1 for recipes for the following media: A. thaliana germination medium (AtGM); A. thaliana callus induction medium (AtCIM); A. thaliana shoot induction medium (AtSIM); A. thaliana root induction medium (AtRIM); A. thaliana propagation medium (AtPM).
- Weigh agar for aliquots of 500 ml and add agar to autoclavable 1-L bottles (i.e., 2 to 40 bottles).
- Pour 500 ml medium into each bottle and shake gently (to avoid clumping of the agar).
- Autoclave (in a water-cooled autoclave).
- Shake autoclaved bottles to distribute melted agar equally in the medium.
- Cool to 55°C in a 55°C water bath.
- From now on, work on a clean bench. Add appropriate amounts of stock solutions of heat-labile chemicals (i.e., antibiotics, hormones).
- Pour appropriate volume of medium into tissue culture vessels (see Table 1).
- Let medium solidify and dry overnight on a clean bench (Fig. 8) with the light switched off.
- If medium is not used immediately, store at 4°C in the dark for up to 2 weeks. Petri dishes with medium should not be stored upside down, due to softness of the medium.
The medium should be of soft consistency (much softer than standard plates with medium for bacterial growth), as plant regeneration will be very poor on hard medium.
- Maximum storage times depend on the stability of the critical (i.e., most labile) components of the medium. In general, storage for >2 weeks is not recommended. When using stored medium, allow for sufficient time to equilibrate to room temperature.
- The following chemicals and mixtures are added as powders: MS salt and vitamin mix (e.g., Murashige & Skoog medium M 0222 from Duchefa; Murashige & Skoog, 1962), sucrose, 2-(N -morpholino)ethanesulfonic acid (MES), agar, and Gamborg B5 salts (e.g., G 0209 from Duchefa; see Table 2).
- The following chemicals are added to medium as stock solutions: 2,4-dichlorophenoxyacetic acid (2,4-D), kinetin, indole-3-acetic acid (IAA), N6-(2-isopentenyl)adenine (2-iP), indole-3-butyric acid (IBA), and McCown woody plant vitamin mixture (e.g., M 0408 from Duchefa; see recipe), PSK-alpha (see recipe).

| Substance, abbreviation | Substance, full name | CAS number | MW | Stock solution (recommended concentration) | Solvent | Filter-sterilize before storage | Storage temperature | Add to medium before or after autoclaving |
|---|---|---|---|---|---|---|---|---|
| 2,4-D | 2,4-Dichlorophenoxyacetic acid | 94-75-7 | 221.0 | 1.0 mg/ml | 0.1 M NaOH | Yes | 4°C | Before |
| 2-iP | N6-(2-isopentenyl)adenine | 2365-40-4 | 203.2 | 10.0 mg/ml | 0.1 M NaOH | Yes | −20°C | Before |
| EtOH, p. a. | ethanol, analytical grade | 64-17-5 | 46.07 | ≥99.8% | NA | No | 20°C | NA |
| EtOH tech. | ethanol, technical grade | 64-17-5 | 46.07 | 70% (w/v) | Water | No | 20°C | NA |
| Hypochlorite | Sodium hypochlorite | 7681-52-9 | 74.44 | 13% active chlorine | NA | No | 4°C | NA |
| IAA | Indole-3-acetic acid | 87-51-4 | 175.2 | 1.0 mg/ml | 0.1 M NaOH | Yes | −20°C | Before |
| IBA | Indole-3-butyric acid | 133-32-4 | 203.2 | 1.0 mg/ml | 0.1 M NaOH | Yes | −20°C | Before |
| kan | Kanamycin A monosulfate monohydrate | 25389-94-0 | 600.6 | 100 mg/ml | Water | Yes | 4°C | After |
| kin | Kinetin | 525-79-1 | 215.2 | 1.0 mg/ml | 0.1 M NaOH | Yes | −20°C | Before |
| MES | 2-(N-morpholino)ethanesulfonic acid monohydrate | 4432-31-9 | 213.2 | NA | NA | NA | Room temperature | Before |
| spec | Spectinomycin dihydrochloride pentahydrate | 22189-32-8 | 495.3 | 100 mg/ml | Water | Yes | 4°C | After |
| suc | Sucrose | 57-50-1 | 342.3 | NA | NA | NA | Room temperature | Before |
| Tween 20 | Polyoxyethylene (20) sorbitan monolaurate | 9005-64-5 | 1227.72 | 10% (w/v) | Water | Yes | Room temperature | NA |
-
aNA, not applicable.
COMMENTARY
Background Information
The general feasibility of introducing foreign DNA into the Arabidopsis plastid genome was demonstrated more than 20 years ago (Sikdar et al., 1998). However, the generation of fertile transplastomic Arabidopsis plants has turned out to be challenging and has become possible only recently (Ruf et al., 2019). As evident from the described protocols, the generation of transplastomic Arabidopsis plants involves laborious and time-consuming procedures. It is highly recommended that the provided protocols are followed exactly, as even small deviations can result in a drastic decline of transformation efficiency or complete failure of the transformation experiments.
For an overview of the applications of the plastid transformation technology in basic and applied research, the interested reader is referred to recent review articles that exhaustively cover this topic (Maliga & Bock, 2011; Bock, 2015; Fuentes et al., 2018; Scharff & Bock, 2014; Boehm & Bock, 2019).
Critical Parameters and Troubleshooting
Seed production of the recipient line in the greenhouse
Batches of high-quality seeds need to be produced (i.e., seed with low load of bacterial and fungal spores and a high germination rate). To achieve high germination rates, seeds should be freshly harvested from well-dried siliques. After harvest, seeds should be dried and stored at low humidity (≤15%).
Arabidopsis cultivar
The use of Arabidopsis cultivar C24 is highly recommended because the tissue culture and regeneration properties of C24 are superior to most other Arabidopsis ecotypes.
Genotype of the recipient strain for transformation
Use of the Δ acc2 genotype is a must for efficient selection of spectinomycin-resistant transplastomic Arabidopsis lines. Most wild-type strains of Arabidopsis display limited sensitivity to spectinomycin, due to expression of the nucleus-encoded and plastid-targeted ACC2 protein (Parker et al., 2016; Ruf et al., 2019; Small, 2019; Yu et al., 2017).
Seed sterilization with sodium hypochlorite
Sodium hypochlorite solutions age over time, resulting in decreased sterilization efficiency. It is recommended that fresh sodium hypochlorite is purchased every 6 months.
Media preparation
- The optimal amount of agar in the medium needs to be carefully determined (and may change with every fresh batch of agar purchased!). The medium should be just solid but still soft.
- Use of a small (∼150 L) autoclave with water cooling is preferred. Rapid heating and cooling is recommended to reduce decomposition of agar and other critical ingredients, and avoid excessive caramelization of the sugar in the medium.
- Shake the medium after autoclaving carefully but gently. Cool to 55°C (in a water bath) before adding the antibiotics. Shake again carefully but gently. Avoid producing air bubbles.
Handling of tissue cultures
- Optimize the agar concentration of the AtCIM: The concentration should be just high enough to solidify the medium. A sufficiently soft medium is important to facilitate efficient provision of nutrients, hormones, and antibiotics to root cells and calli across the polyester net.
- Use freshly prepared medium plates. The medium needs to have sufficient moisture to ensure high humidity in the airspace above the medium. However, make sure that there is no condensed water accumulating in the lid.
- The root-derived calli must be protected from stagnant moisture. Excess water can be removed upon medium exchange, by pulling the net with the roots over the side wall of the Petri dish (thus stripping of any liquid) before transferring it onto a new plate. Make sure that there are no bubbles between the netting and the medium. Do not use plates with (solidified) bubbles at the surface of the medium.
- Always handle the cultures in dim light. High light intensities cause photooxidative stress to the cells and will decrease growth and regeneration efficiency.
- For each type of culture vessel, there is an optimum amount of medium (see Figures 1 to 8). If the amount of medium is too low for the culture vessel used, the medium will dry out, leading to poor growth of the cultures. There is also an optimal density of the tissue explants or calli per plate (and shoots per tube or jar; see Figures 1 to 8 and detailed instructions in the protocols).
- Do not cut the calli with the scalpel. Instead, break them apart by using two pairs of fine-tipped tweezers. Breaking induces less cell damage, leading to better shoot regeneration.
- It can happen that some plates deteriorate in that the roots or calli turn orange or brownish. The reasons are not entirely clear (with suboptimal humidity and/or air exchange likely contributing to the phenomenon) and as long as only a small proportion of the plates are affected, this is not a major concern.
- Keep any spontaneous spectinomycin-resistant mutants you may obtain (Bock, 2001; Svab & Maliga, 1991). They can serve as a perfect internal control for tissue culture efficiency and also provide useful controls for inheritance assays and the molecular characterization of transplastomic clones. Finally, the tissue of such mutants also can be used to adjust the density of calli and shoots to the optimum (see above). It is therefore recommended that the spontaneous mutants are passed through the entire protocol until seeds are obtained and reciprocal crosses have been performed.
Biolistic bombardment
- To adjust the settings of the particle gun, optimize the particle preparation procedure and control for its quality; transient nuclear transformation assays using a β-glucuronidase (GUS) reporter gene construct should be used. Any standard plant transformation vector containing a GUS reporter gene under the control of a strong constitutive promoter can be used for this purpose. The frequency of transient GUS expression as evidenced by histochemical staining provides a suitable proxy for plastid transformation efficiency. Detailed protocols for GUS staining of bombarded leaves have been published elsewhere (e.g., Bock, 2004).
- Detailed instructions for how to optimize the bombardment parameters are provided in the manual of the biolistic gun.
- As a control for tissue culture and selection efficiency and an additional control for transformation efficiency, it is highly recommended that in each Arabidopsis plastid transformation experiment a few shots for stable nuclear transformation of root-derived microcalli are included.
Keeping track of independent transformation events
Reporting of plastid transformation experiments requires generation of at least two to three independent transplastomic lines. It is therefore essential to keep track of the origin of each transplastomic clone by appropriate numbering of the bombarded plates and the resulting callus cultures.
Note that after collecting a resistant callus from the netting, it may be difficult to know with certainty whether any additional green callus appearing later represent an independent event or originated from the same initially transformed cell. In case of doubt, multiple resistant calli harvested from the same bombarded plate should be considered one and the same event.
Achieving homoplasmy
In most transformable species, primary transplastomic lines are heteroplasmic (i.e., contain a mix of transformed plastid genome copies and residual wild-type genomes). Elimination of the undesired wild-type genomes is achieved by repeated plant regeneration under selection pressure (Bock, 2001, 2015). However, in Arabidopsis , there is usually no need to perform additional rounds of regeneration and selection to purify transplastomic clones to homoplasmy. Typically, the regenerating shoots are already homoplasmic (Ruf et al., 2019). This is likely due to the stringency of the antibiotic selection in the callus-based transformation and regeneration system.
Seed production in sterile culture
Arabidopsis flowers require low humidity for successful seed set. By using filter paper to cover the culture vessels, the (typically very high) humidity in tissue culture can be reduced, thus allowing successful seed production. To avoid rapid drying of the medium, it is important to water the plantlets regularly with sterile water.
Confirmation of plastid transformation by molecular analyses
Faithful integration of the transforming DNA into the plastid genome and homoplasmy of the transplastomic material should be confirmed by restriction fragment length polymorphism (RFLP) analysis using Southern blotting. Relying on PCR tests is strongly discouraged for multiple reasons, including the risk of false-positive results (running contamination-free PCRs in a transformation laboratory is often not a trivial exercise), the possibility of transgene integration into the nuclear genome, and various other common PCR-related artifacts (e.g., template switching).
Understanding Results
Under optimal conditions, up to 10 transplastomic lines can be obtained from 27 bombarded plates with root-derived microcalli (Figures 3 and 5). However, it should be noted that the number of transplastomic lines recovered per experiment can vary greatly, for reasons that are not always obvious. Even small deviations from the described protocols can cause dramatic reductions in transformation efficiency (and often result in complete failure of the experiment). Transformation efficiencies can also be low if (1) expression of the transgene(s) to be introduced is deleterious to the cell (Magee et al., 2004; Petersen & Bock, 2011) or (2) knockout of essential plastid genes (Alkatib et al., 2012; Drescher et al., 2000; Rogalski, Ruf, & Bock, 2006) is attempted.
For the reasons explained above, it is very strongly recommended that a nuclear transformation control is included in each chloroplast transformation experiment. The nuclear control should yield, on average, five to fifteen independent transgenic lines per bombarded plate.
Not all spectinomycin-resistant regenerants will represent true transplastomic events. Spontaneous resistance to spectinomycin can arise from point mutations in plastid genes encoding specific components of the plastid ribosome, especially the 16S ribosomal RNA (rRNA; Svab & Maliga, 1991). However, for reasons that are not entirely clear, spontaneous spectinomycin-resistant mutants are relatively rare in Arabidopsis compared to tobacco, the most popular model species for plastid transformation. Nonetheless, spontaneous mutants will occasionally appear (especially if the plastid transformation frequency is low) and need to be distinguished from true transplastomic lines by molecular methods (i.e., RFLP analysis by Southern blotting).
Time Considerations
On the regeneration control (untransformed Δ acc2 roots, regenerated in the absence of antibiotic selection), shoot regeneration is typically observed after 1 to 2 weeks on AtSIM.
The nuclear transformation control (Δ acc2 roots, transformed with a nuclear transformation vector such as pGreen0029-GFP; Hellens, Edwards, Leyland, Bean, & Mullineaux, 2000) is subjected to selection with the appropriate antibiotic (typically kanamycin at a concentration of 50 mg/L). Appearance of regenerating transgenic shoots usually starts after 2 to 3 weeks on AtSIM kan50, with the number of events peaking between week 3 and week 5.
In a successful plastid transformation experiment (using spectinomycin selection), green antibiotic-resistant calli will appear after 7 to 52 weeks. Spontaneous spectinomycin-resistant mutants appear at a similar time and can be distinguished from true transplastomic events with molecular methods (RFLP analysis).
Estimates of the duration of the individual steps in the Arabidopsis plastid transformation protocol are given below:
| Seed sterilization and sowing | 0.5 day |
| Stratification | 2 days |
| Germination | 6 days |
| Root harvest | 0.5 day |
| Microcallus induction | 5 days |
| Biolistic bombardment | 1 day |
| Selection of resistant calli and shoot induction | 7 to 52 weeks |
| Root induction | 2 weeks |
| Plant growth and seed production | 5 to 13 weeks. |
Acknowledgments
We thank Laura Schollbach and Sandra Stegemann (Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm) for excellent photography. Research on plastid genome engineering in the authors’ laboratory is financed by the Max Planck Society and a grant from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (ERC-ADG-2014; grant agreement 669982) to R.B.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Stephanie Ruf: Conceptualization, Writing-original draft, Writing-review & editing, Xenia Kroop: Writing-review & editing, Ralph Bock: Conceptualization, Funding acquisition, Writing-original draft, Writing-review & editing
Conflict of Interest statement
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study
Literature Cited
- Alkatib, S., Scharff, L. B., Rogalski, M., Fleischmann, T. T., Matthes, A., Seeger, S., … Bock, R. (2012). The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genetics , 8, e1003076. doi: 10.1371/journal.pgen.1003076.
- Altpeter, F., Baisakh, N., Beachy, R., Bock, R., Capell, T., Christou, P., … Visser, R. (2005). Particle bombardment and the genetic enhancement of crops: Myths and realities. Molecular Breeding , 15, 305–327. doi: 10.1007/s11032-004-8001-y.
- Bock, R. (2001). Transgenic plastids in basic research and plant biotechnology. Journal of Molecular Biology , 312, 425–438. doi: 10.1006/jmbi.2001.4960.
- Bock, R. (2004). Studying RNA editing in transgenic chloroplasts of higher plants. Methods in Molecular Biology , 265, 345–356. doi: 10.1385/1-59259-775-0:345
- Bock, R. (2015). Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annual Review of Plant Biology , 66, 211–241. doi: 10.1146/annurev-arplant-050213-040212.
- Boehm, C. R., & Bock, R. (2019). Recent advances and current challenges in synthetic biology of the plastid genetic system and metabolism. Plant Physiology , 179, 794–802. doi: 10.1104/pp.18.00767.
- Clough, S. J., & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant Journal , 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x.
- Drescher, A., Ruf, S., Calsa, Jr., T., Carrer, H., & Bock, R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant Journal , 22, 97–104. doi: 10.1046/j.1365-313x.2000.00722.x.
- Fuentes, P., Armarego-Marriott, T., & Bock, R. (2018). Plastid transformation and its application in metabolic engineering. Current Opinion in Biotechnology , 49, 10–15. doi: 10.1016/j.copbio.2017.07.004.
- Golczyk, H., Greiner, S., Wanner, G., Weihe, A., Bock, R., Börner, T., & Herrmann, R. G. (2014). Chloroplast DNA in mature and senescing leaves: A reappraisal. Plant Cell , 26, 847–854. doi: 10.1105/tpc.113.117465.
- Greiner, S., Golczyk, H., Malinova, I., Pellizzer, T., Bock, R., Börner, T., & Herrmann, R. H. (2020). Chloroplast nucleoids are highly dynamic in ploidy, number, and structure during angiosperm leaf development. Plant Journal , 102, 730–746. doi: 10.1111/tpj.14658.
- Hager, M., Biehler, K., Illerhaus, J., Ruf, S., & Bock, R. (1999). Targeted inactivation of the smallest plastid genome-encoded open reading frame reveals a novel and essential subunit of the cytochrome b6f complex. EMBO Journal , 18, 5834–5842. doi: 10.1093/emboj/18.21.5834.
- Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., & Mullineaux, P. M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium -mediated plant transformation. Plant Molecular Biology , 42, 819–832. doi: 10.1023/A:1006496308160.
- Hibberd, J. M. (2017). A major advance in plastid transformation. Plant Physiology , 175, 5. doi: 10.1104/pp.17.00992.
- Koop, H.-U., Steinmüller, K., Wagner, H., Rößler, C., Eibl, C., & Sacher, L. (1996). Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta , 199, 193–201. doi: 10.1007/BF00196559.
- Magee, A. M., Coyne, S., Murphy, D., Horvath, E. M., Medgyesy, P., & Kavanagh, T. A. (2004). T7 RNA polymerase-directed expression of an antibody fragment transgene in plastids causes a semi-lethal pale-green seedling phenotype. Transgenic Research , 13, 325–337. doi: 10.1023/B:TRAG.0000040019.35147.a4.
- Maliga, P. (2004). Plastid transformation in higher plants. Annual Review of Plant Biology , 55, 289–313. doi: 10.1146/annurev.arplant.55.031903.141633.
- Maliga, P., & Bock, R. (2011). Plastid biotechnology: Food, fuel, and medicine for the 21st century. Plant Physiology , 155, 1501–1510. doi: 10.1104/pp.110.170969.
- Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiologia Plantarum , 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x.
- Parker, N., Wang, Y., & Meinke, D. (2014). Natural variation in sensitivity to a loss of chloroplast translation in Arabidopsis. Plant Physiology , 166, 2013–2027. doi: 10.1104/pp.114.249052.
- Parker, N., Wang, Y., & Meinke, D. (2016). Analysis of Arabidopsis accessions hypersensitive to a loss of chloroplast translation. Plant Physiology , 172, 1862–1875. doi: 10.1104/pp.16.01291.
- Petersen, K., & Bock, R. (2011). High-level expression of a suite of thermostable cell wall-degrading enzymes from the chloroplast genome. Plant Molecular Biology , 76, 311–321. doi: 10.1007/s11103-011-9742-8.
- Preuten, T., Cincu, E., Fuchs, J., Zoschke, R., Liere, K., & Börner, T. (2010). Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant Journal , 64, 948–959. doi: 10.1111/j.1365-313X.2010.04389.x.
- Rogalski, M., Ruf, S., & Bock, R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Research , 34, 4537–4545. doi: 10.1093/nar/gkl634.
- Ruf, S., Biehler, K., & Bock, R. (2000). A small chloroplast-encoded protein as a novel architectural component of the light-harvesting antenna. Journal of Cell Biology , 149, 369–377. doi: 10.1083/jcb.149.2.369.
- Ruf, S., Forner, J., Hasse, C., Kroop, X., Seeger, S., Schollbach, L., … Bock, R. (2019). High-efficiency generation of fertile transplastomic Arabidopsis plants. Nature Plants , 5, 282–289. doi: 10.1038/s41477-019-0359-2.
- Ruf, S., Hermann, M., Berger, I. J., Carrer, H., & Bock, R. (2001). Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nature Biotechnology , 19, 870–875. doi: 10.1038/nbt0901-870.
- Ruf, S., Karcher, D., & Bock, R. (2007). Determining the transgene containment level provided by chloroplast transformation. Proceedings of the National Academy of Sciences of the United States of America , 104, 6998–7002. doi: 10.1073/pnas.0700008104.
- Scharff, L. B., & Bock, R. (2014). Synthetic biology in plastids. Plant Journal , 78, 783–798. doi: 10.1111/tpj.12356.
- Sikdar, S. R., Serino, G., Chaudhuri, S., & Maliga, P. (1998). Plastid transformation in Arabidopsis thaliana. Plant Cell Reports , 18, 20–24. doi: 10.1007/s002990050525.
- Small, I. (2019). Transplastomic Arabidopsis plants at last. Nature Plants , 5, 249–250. doi: 10.1038/s41477-019-0388-x.
- Svab, Z., & Maliga, P. (1991). Mutation proximal to the tRNA binding region of the Nicotiana plastid 16S rRNA confers resistance to spectinomycin. Molecular and General Genetics , 228, 316–319. doi: 10.1007/BF00282483.
- Svab, Z., & Maliga, P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proceedings of the National Academy of Sciences of the United States of America , 90, 913–917. doi: 10.1073/pnas.90.3.913.
- Tabatabaei, I., Ruf, S., & Bock, R. (2017). A bifunctional aminoglycoside acetyltransferase/phosphotransferase conferring tobramycin resistance provides an efficient selectable marker for plastid transformation. Plant Molecular Biology , 93, 269–281. doi: 10.1007/s11103-016-0560-x.
- Thal, J. (1588). Sylva hercynia, Frankfurt am Main: Johannes Feyerabend.
- The 1001 Genomes Consortium (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell , 166, 481–491. doi: 10.1016/j.cell.2016.05.063.
- Weigel, D., & Glazebrook, J. (2006). Root transformation of Arabidopsis. CSH Protocols , 7. doi: 10.1101/pdb.prot4671.
- Yu, Q., Lutz, K. A., & Maliga, P. (2017). Efficient plastid transformation in Arabidopsis. Plant Physiology , 175, 186–193. doi: 10.1104/pp.17.00857.
Citing Literature
Number of times cited according to CrossRef: 6
- Erum Shoeb, Srividhya Venkataraman, Uzma Badar, Kathleen Hefferon, Genome Editing for Crop Biofortification, Applications of Genome Engineering in Plants, 10.1002/9781394183913.ch6, (91-121), (2024).
- Deserah D. Strand, Berkley J. Walker, Energetic considerations for engineering novel biochemistries in photosynthetic organisms, Frontiers in Plant Science, 10.3389/fpls.2023.1116812, 14 , (2023).
- Faisal Saeed, Sarbesh Das Dangol, Muneeb Hassan Hashmi, Md Jakir Hossain, Allah Bakhsh, Role of genetic engineering in improving potato production, Potato Production Worldwide, 10.1016/B978-0-12-822925-5.00006-2, (303-315), (2023).
- Yaqi An, Yue Wang, Xinwei Wang, Jianwei Xiao, Development of chloroplast transformation and gene expression regulation technology in land plants, Frontiers in Plant Science, 10.3389/fpls.2022.1037038, 13 , (2022).
- Katherine Domb, Nan Wang, Guillaume Hummel, Chang Liu, Spatial Features and Functional Implications of Plant 3D Genome Organization, Annual Review of Plant Biology, 10.1146/annurev-arplant-102720-022810, 73 , 1, (173-200), (2022).
- Gideon Sadikiel Mmbando, Recent Advances in Antibiotic-Free Markers; Novel Technologies to Enhance Safe Human Food Production in the World, Molecular Biotechnology, 10.1007/s12033-022-00609-7, 65 , 7, (1011-1022), (2022).

