Protein Sequence Analysis Using the MPI Bioinformatics Toolkit
Martin Steinegger, Martin Steinegger, Felix Gabler, Felix Gabler, Seung-Zin Nam, Seung-Zin Nam, Sebastian Till, Sebastian Till, Milot Mirdita, Milot Mirdita, Johannes Söding, Johannes Söding, Andrei N. Lupas, Andrei N. Lupas, Vikram Alva, Vikram Alva
CLANS
cluster analysis
HHpred
HMM
homology
profile hidden Markov models
sequence comparison
sequence similarity searches
structure prediction
Abstract
The MPI Bioinformatics Toolkit (https://toolkit.tuebingen.mpg.de) provides interactive access to a wide range of the best-performing bioinformatics tools and databases, including the state-of-the-art protein sequence comparison methods HHblits and HHpred. The Toolkit currently includes 35 external and in-house tools, covering functionalities such as sequence similarity searching, prediction of sequence features, and sequence classification. Due to this breadth of functionality, the tight interconnection of its constituent tools, and its ease of use, the Toolkit has become an important resource for biomedical research and for teaching protein sequence analysis to students in the life sciences. In this article, we provide detailed information on utilizing the three most widely accessed tools within the Toolkit: HHpred for the detection of homologs, HHpred in conjunction with MODELLER for structure prediction and homology modeling, and CLANS for the visualization of relationships in large sequence datasets. © 2020 The Authors.
Basic Protocol 1 : Sequence similarity searching using HHpred
Alternate Protocol : Pairwise sequence comparison using HHpred
Support Protocol : Building a custom multiple sequence alignment using PSI-BLAST and forwarding it as input to HHpred
Basic Protocol 2 : Calculation of homology models using HHpred and MODELLER
Basic Protocol 3 : Cluster analysis using CLANS
INTRODUCTION
The structure, function, and evolution of new or uncharacterized proteins are routinely inferred based on their homology to proteins with experimentally characterized properties. Sequence searches are a common first step in this process, as sequence similarity is widely accepted as the best marker for substantiating homologous relationships. Over the years, many high-quality sequence search methods [e.g., BLAST (Altschul et al., 1997; Ladunga, 2017), HMMER (Potter et al., 2018; Prakash, Jeffryes, Bateman, & Finn, 2017), HHblits (Remmert, Biegert, Hauser, & Soding, 2011), HHpred (Soding, 2005; Steinegger et al., 2019)]; protein sequence and domain databases [SCOPe (Fox, Brenner, & Chandonia, 2014), ECOD (Cheng et al., 2014; Schaeffer, Liao, & Grishin, 2018), Pfam (Coggill, Finn, & Bateman, 2008; El-Gebali et al., 2019), RefSeq (O'Leary et al., 2016), UniProt (Pundir, Martin, O'Donovan, & The UniProt Consortium, 2016; The UniProt Consortium, 2019)]; and integrative Web resources [the EMBL-EBI Bioinformatics Web Services (Madeira et al., 2019; Madeira, Madhusoodanan, Lee, Tivey, & Lopez, 2019), the SIB Bioinformatics Resource Portal (Swiss Institute of Bioinformatics Members, 2016), National Center for Biotechnology Information Web Resources (NCBI Resource Coordinators, 2018; Gibney & Baxevanis, 2011; Yang, Derbyshire, Yamashita, & Marchler-Bauer, 2020)] have been developed to help researchers make meaningful inferences based on homology. Driven by our work at the interface of computational and experimental biology, we launched the MPI Bioinformatics Toolkit in 2005 to provide researchers in the life sciences with easy, web-based access to the best-performing bioinformatics tools and databases (Biegert, Mayer, Remmert, Soding, & Lupas, 2006). The Toolkit has been in continuous operation ever since, and we replaced the first version with an entirely new one built using more scalable and robust web technologies in 2017 (Alva, Nam, Soding, & Lupas, 2016; Zimmermann et al., 2018). The Toolkit currently includes 35 in-house and external tools for sequence similarity searching [e.g., PSI-BLAST (Altschul et al., 1997), HHblits, HHpred]; calculation of multiple sequence alignments [ClustalΩ (Sievers et al., 2011), Τ-Coffee (Notredame, Higgins, & Heringa, 2000)]; prediction of secondary structure and sequence features [Quick2D, PCOILS (Gruber, Soding, & Lupas, 2006), TPRpred (Karpenahalli, Lupas, & Soding, 2007)]; and sequence classification [CLANS (Frickey & Lupas, 2004), MMseqs2 (Mirdita, Steinegger, & Soding, 2019)].
Over the years, the Toolkit has established itself as an important resource for molecular biology research, mainly due to the sensitive sequence-comparison tools HHblits and HHpred, which, in many instances, can detect homologous relationships that are not readily recognized by other tools. A further strength of the Toolkit lies in the tight interconnection of the tools, allowing the results of one tool to be forwarded as input to others; for instance, the output of a PSI-BLAST search could be forwarded to ClustalΩ to obtain a multiple sequence alignment (MSA) of the identified matches or to MMseqs2 to obtain a reduced set filtered by pairwise sequence identity. Finally, our implementations of some external tools offer enhanced features, such as versions of the NCBI nonredundant (nr) database for PSI-BLAST that are clustered down to 30% (nr30), 50% (nr30), 70% (nr30), or 90% (nr90) sequence identity.
In this article, we describe detailed protocols for the application of the three most frequently used tools. Basic Protocol 1 describes how to use HHpred to search for remote homologs of a protein and make inferences about its domain composition, structure, function, and evolution. The Alternate Protocol describes the pairwise comparison mode of HHpred, which allows two protein sequences or MSAs to be compared with each other. The Support Protocol describes how to build a custom, high-quality MSA starting with a protein sequence and use it as input for HHpred. Basic Protocol 2 describes how to use HHpred in conjunction with MODELLER (Sali & Blundell, 1993) to build a three-dimensional (3D) structural model for a protein sequence of interest. Basic Protocol 3 describes the use of PSI-BLAST in conjunction with CLANS to detect distant homologs of a protein of interest and then visualize the relationships between the detected homologs. To demonstrate these protocols, we use as an example the experimentally uncharacterized FtsZ protein of the Asgard group archaeon Prometheoarchaeum syntrophicum strain MK-D1, which currently represents the closest cultured prokaryotic relative of eukaryotes (Imachi et al., 2020). In most bacteria, many archaea, all chloroplasts, and some mitochondria, with the latter two representing endosymbiosis-derived eukaryotic organelles, FtsZ forms filaments that assemble into a ring (Z-ring) at the future site of cell division (Lowe & Amos, 1998; Margolin, 2005; Szwedziak, Wang, Bharat, Tsim, & Lowe, 2014). Notably, eukaryotic tubulins, which polymerize to form microtubules, a major component of the cytoskeleton, are remotely homologous to FtsZ (Nogales, Downing, Amos, & Lowe, 1998). FtsZ and tubulins are GTPases that comprise an N-terminal GTP-binding domain with a highly conserved GGGTG(T/S)G motif associated with GTP binding and a C-terminal regulatory domain (Erickson, 1998). Strikingly, the pairwise sequence identity between FtsZ and tubulins is lower than 15%. Therefore, most sequence search methods fail to substantiate a homologous relationship between them. We note that the structure, function, and evolution of FtsZ and tubulins have been studied extensively, and that their evolutionary relatedness is also widely accepted (Erickson, 1998; Nogales et al., 1998). However, for instructional purposes, we will assume that the homology between them is unclear. In the following, we show how the Toolkit could be used to investigate the relationship between FtsZ and tubulins.
Basic Protocol 1: SEQUENCE SIMILARITY SEARCHING USING HHpred
An almost ubiquitous first step in the characterization of a protein is the identification of functionally and structurally characterized homologs using BLAST (Altschul et al., 1997) or HMMER (Potter et al., 2018). Frequently, however, these search methods fail to detect statistically significant connections to characterized proteins. In many such cases, the more sensitive sequence search method HHpred (Steinegger et al., 2019), which is based on the comparison of profile hidden Markov models (HMMs), is able to establish connections to remotely homologous, characterized proteins. Starting from a single protein sequence, HHpred builds a multiple sequence alignment using HHblits (Steinegger et al., 2019) or PSI-BLAST (Altschul et al., 1997) and annotates the obtained alignment with the predicted secondary structure using PSIPRED (Jones, 1999). Next, this annotated alignment is converted to a profile HMM and compared to each profile HMM in user-selected target databases, which represent proteins of known structure or annotated protein families. Such databases are, for example, the Pfam (El-Gebali et al., 2019), CDD (Lu et al., 2020), and SMART (Letunic & Bork, 2018) domain databases; the SCOPe (Fox et al., 2014) and ECOD (Cheng et al., 2014) structural classification databases; the Protein Data Bank (Berman et al., 2000); and proteomes of several model organisms. Database HMMs are built using three iterations of HHblits over UniRef30 (Mirdita et al., 2017), which is a version of the UniRef sequence database (Suzek et al., 2015) clustered into groups of similar sequences at a length coverage of at least 80% and a maximum pairwise sequence identity of 30%. Like query HMMs, database HMMs include secondary structure information, either predicted by PSIPRED or assigned based on 3D structure by DSSP (Joosten et al., 2011; Kabsch & Sander, 1983). The inclusion of secondary structure information significantly increases the sensitivity of HHpred. The output of HHpred is a list of the closest homologs, with pairwise alignments.
Necessary Resources
Hardware
- A desktop computer, a laptop, or a tablet with Internet access
Software
- An up-to-date, JavaScript-enabled Web browser (preferably Google Chrome, Mozilla Firefox, or Apple Safari)
Input files
- A protein sequence (in FASTA format or as plain text) or a multiple protein sequence (MSA) alignment (in FASTA, STOCKHOLM, or CLUSTAL format)
Submission page of HHpred
1.Navigate your Web browser to the submission page of HHpred at https://toolkit.tuebingen.mpg.de/tools/hhpred.

2.Paste the amino acid sequence of your protein of interest (in FASTA format or as plain text) or an MSA (in FASTA, CLUSTAL, or STOCKHOLM format) into the large textbox (Fig. 1A). Alternatively, the input sequence or MSA can be uploaded using the ‘Upload File’ option. Follow the Support Protocol to build a custom MSA, starting with a protein sequence of interest.
3.Select target profile HMM database(s) against which you wish to compare the query protein (Fig. 1A).
4.Customize input parameters in the ‘Parameters’ tab (Fig. 1B). The default values for the various parameters are set to yield the best results for most standard cases, and we recommend using them, at least in the initial steps of the analysis.
5.Optionally, assign your job a custom identifier by entering one in the ‘Custom Job ID’ text field (Fig. 1). The identifier should contain at least two characters. If this text field is left empty, an identifier is assigned automatically.
6.Start your search by pressing the ‘Submit’ button.
HHpred search results
7.Typical HHpred searches take about 5 min to run through. However, searches involving long input sequences (>600 residues), large input MSAs, higher MSA generation iterations (4 or more), or multiple target databases could take hours to complete.
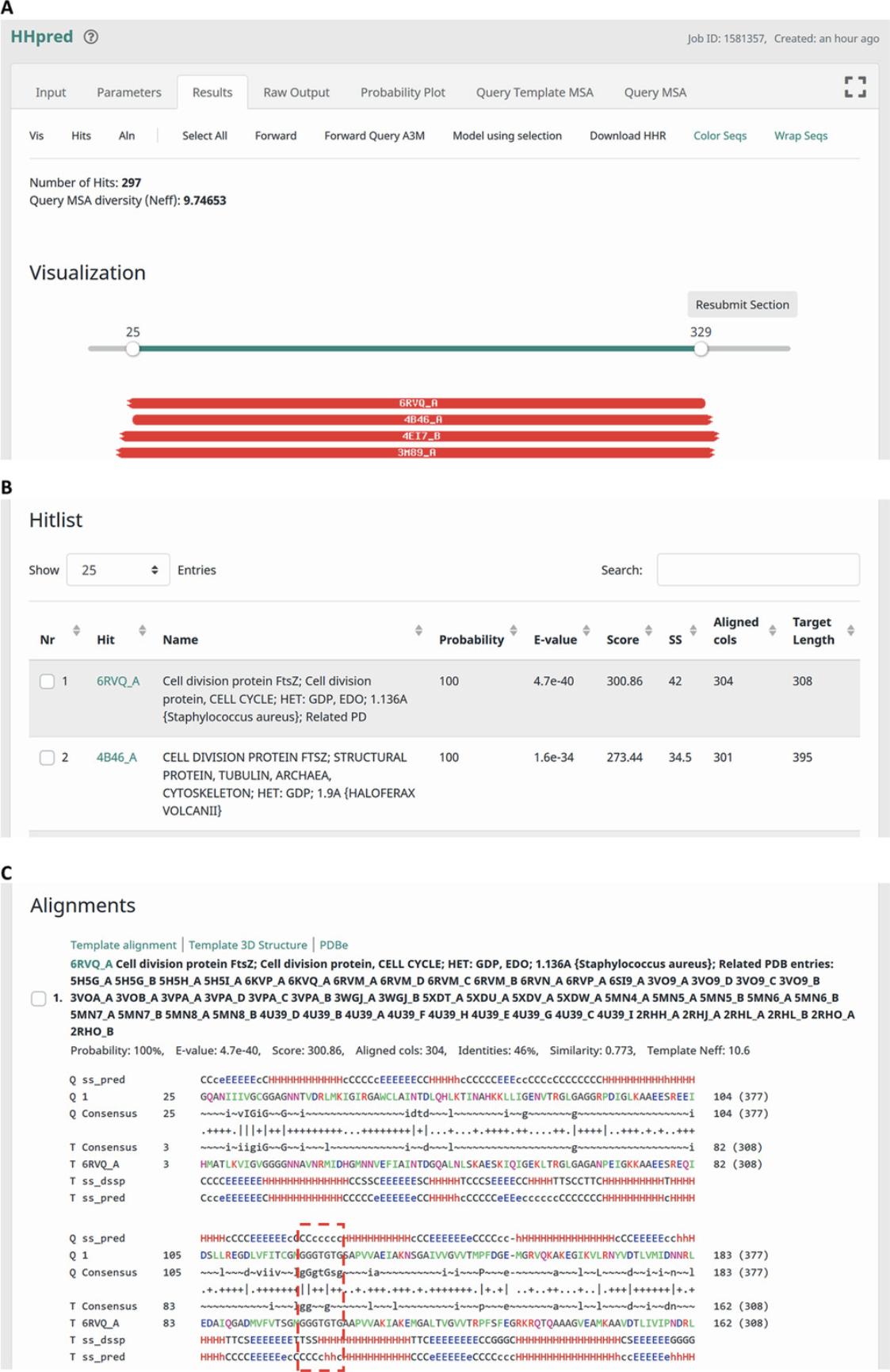
8.The ‘Results’ tab presents information on the detected matches in a user-friendly and interactive manner (Fig. 2).

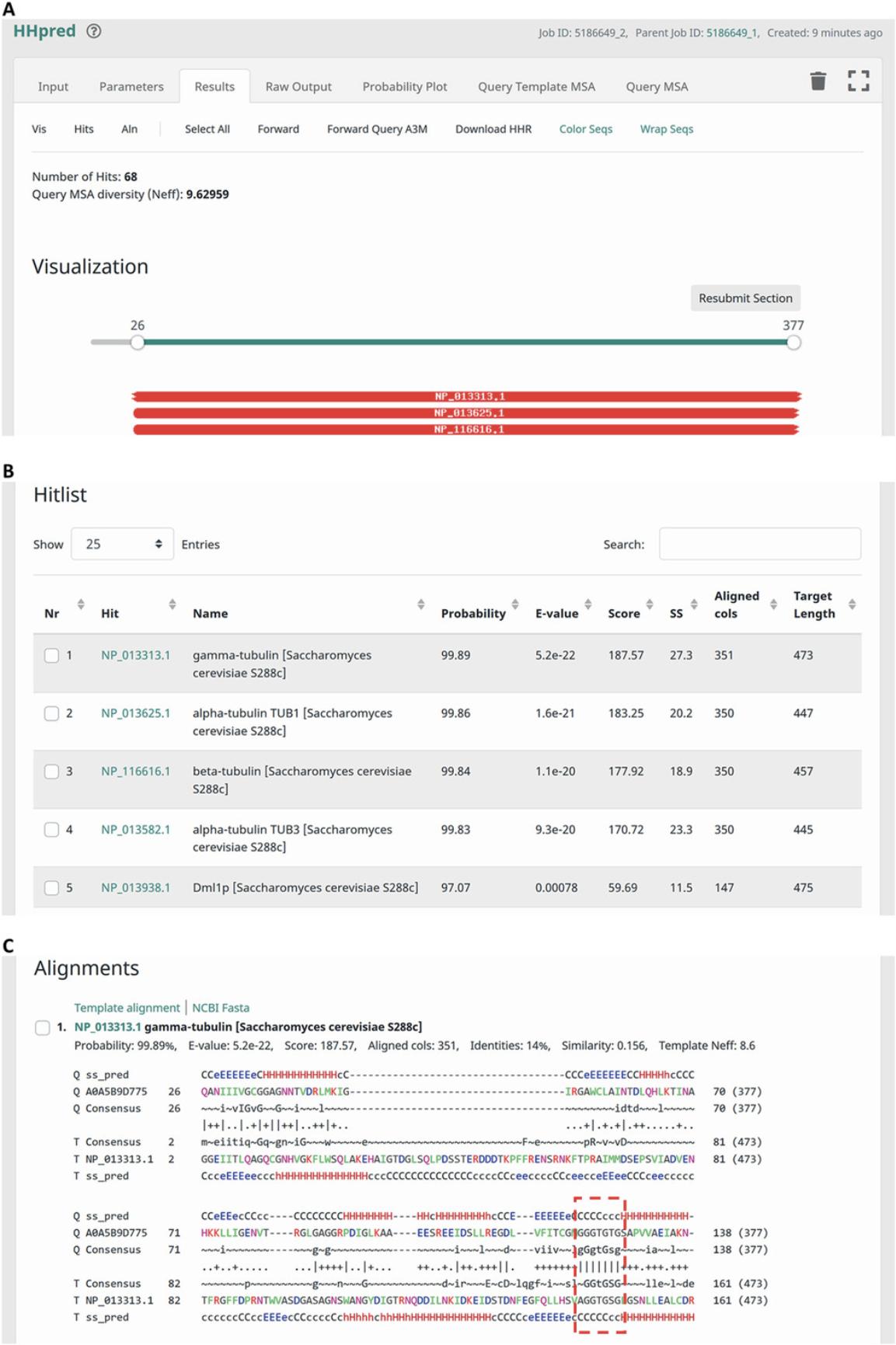
9.The ‘Raw Output’ tab allows visualizing and downloading the raw output file yielded by an HHpred search (Fig. 5A). It is advisable to download and save this file for future reference.

10.The ‘Probability Plot’ tab displays a cumulative histogram of the hits and can be used to obtain a count of matches with probability values higher or lower than a given value.
11.The ‘Query Template MSA’ tab provides access to an MSA comprising the query sequence and sequences of all the obtained hits. It provides options to download the complete alignment (‘Download MSA’) or to forward the alignment to other tools (‘Forward Selected’), either completely (‘Select All’) or only for individually selected sequences.
12.The ‘Query MSA’ tab provides access to the MSA built by the HHpred server for the query (Fig. 5B). The tab displays the 200 most divergent sequences and allows an MSA of selected or all sequences to be forwarded to other tools (‘Forward Selected’). This tab also includes options for downloading this reduced query alignment or the full alignment in A3M format, a space-efficient format that we use internally to store alignments. Alignments in A3M format can be converted to FASTA using the FormatSeq tool offered within our Toolkit (https://toolkit.tuebingen.mpg.de/tools/formatseq).
Alternate Protocol: PAIRWISE SEQUENCE COMPARISON USING HHpred
The pairwise mode of HHpred allows the comparison of two sequences or MSAs. This is particularly useful when you wish to substantiate a homologous relationship between two proteins that you suspect to be homologous, compare proteins that do not exist in our profile HMM databases, or obtain an HMM-HMM based alignment of two distantly related proteins. HHpred builds MSAs for the two input sequences using HHblits or PSI-BLAST, assigns secondary structure using PSIPRED, and converts the annotated MSAs to profile HMMs. In the next step, it compares the computed HMMs and reports an alignment if a match is found that satisfies the cutoffs set in ‘Parameters’. For proteins that contain multiple homologous repeats or domains, it typically reports two or more alignments. For detailed information on using HHpred, please refer to Basic Protocol 1.
Necessary Resources
- Same as for Basic Protocol 1
Submission page of HHpred
1.Navigate your Web browser to the submission page of HHpred at https://toolkit.tuebingen.mpg.de/tools/hhpred. If desired, click on the ‘Reset’ button to reload default values for input parameters.
2.Click on the switch labeled ‘Align two sequences/MSAs’, located below the query textbox, to activate the pairwise comparison mode of HHpred. A second sequence input textbox will be shown (Fig. 6).

3.Paste the amino acid sequences of your proteins of interest (in FASTA format or as plain text) or two MSAs (FASTA, CLUSTAL, or STOCKHOLM format) into the two textboxes. Alternatively, upload them using the ‘Upload File’ option.
4.Customize the input parameters in the ‘Parameters’ tab. Refer to step 4 of Basic Protocol 1 for more information on this.
5.Optionally, assign your job a custom identifier by entering one in the ‘Custom Job ID’ text field.
6.Start your search by pressing the ‘Submit’ button.
HHpred search results
7.Typical pairwise comparisons take about 5 to 10 min. However, searches involving long input sequences (>600 residues) could take several hours to complete.

Support Protocol: BUILDING A CUSTOM MULTIPLE SEQUENCE ALIGNMENT USING PSI-BLAST AND FORWARDING IT AS INPUT TO HHpred
The sensitivity of HHpred searches depends significantly on the quality of MSAs built for the query sequence. For building these MSAs, by default the HHpred server uses three iterations of HHblits over UniRef30 or allows using PSI-BLAST over nr70. Occasionally, however, the query MSAs may not be diverse enough or may be corrupted due to the inclusion of non-homologous sequences, resulting in no statistically significant matches or false positives, respectively. In such cases, using custom-built MSAs as input may significantly increase the sensitivity and reliability of an HHpred search. In the following, we show how to build a custom MSA using PSI-BLAST over a user-selected sequence database, such as the nonredundant protein sequence database (nr), UniProtKB/TrEMBL (uniprot_trembl), and UniProtKB/Swiss-Prot (uniprot_sprot), and forward the obtained alignment as input to HHpred.
Necessary Resources
- Same as for Basic Protocol 1
Submission page of PSI-BLAST
1.Navigate your Web browser to the submission page of PSI-BLAST at https://toolkit.tuebingen.mpg.de/tools/psiblast.
2.Paste the amino acid sequence of your protein of interest (in FASTA format or as plain text) or an MSA (in FASTA, CLUSTAL, or STOCKHOLM format) into the large textbox (Fig. 8A). Alternatively, the input sequence or MSA can be uploaded using the ‘Upload File’ option.

3.Select a target protein sequence database in the drop-down list over which you wish to build the alignment (Fig. 8A).
4.Customize input parameters in the ‘Parameters’ tab (Fig. 8B).
5.Optionally, assign your job a custom identifier.
6.Start your search by pressing the ‘Submit’ button.
PSI-BLAST search results
7.Typical PSI-BLAST searches take about 3 min. However, searches involving long input sequences (>600 residues) over large databases such as nr could take much longer to complete.

Forwarding an alignment to HHpred
8.Inspect the obtained hits and unselect spurious ones.
9.Click on ‘Forward’ in the floating toolbar to forward an alignment of hits to HHpred. These can be the ones selected in step 8 (‘Selected’), or all hits satisfying an E-value cutoff (‘E-value better than’). In the ‘Forward’ modal, select ‘HHpred’ in the selection list at the bottom left corner and click on the ‘Forward’ button to send the alignment to HHpred (Fig. 9A).
Basic Protocol 2: CALCULATION OF HOMOLOGY MODELS USING HHpred AND MODELLER
The availability of 3D structures is extremely useful for the functional characterization of proteins. However, for many proteins, no experimental structures are available. Since proteins with recognizable sequence similarity generally also have quite similar 3D structures, a structure for a protein of interest can be modeled computationally from its sequence, based on homology to proteins of known structure. This approach is referred to as comparative modeling or homology modeling. In the following, we show how to use HHpred to select homologous templates of known structure for a query protein and how to extract and subsequently forward their alignment to MODELLER (Sali & Blundell, 1993), a popular program for homology modeling. Please refer to Basic Protocol 1 for detailed instructions on using HHpred.
Necessary Resources
- Same as for Basic Protocol 1
Submission page of HHpred
1.Navigate your Web browser to the submission page of HHpred at https://toolkit.tuebingen.mpg.de/tools/hhpred.
2.Paste the amino acid sequence of your protein of interest (in FASTA format or as plain text) or an MSA (in FASTA, CLUSTAL, or STOCKHOLM format) into the large textbox. Alternatively, the input sequence or MSA can be uploaded using the ‘Upload File’ option. Follow Support Protocol to build a custom MSA, starting with a protein sequence of interest.
3.Select either PDB_mmCIF70 or PDB_mmCIF30 as the target database. If other target databases are included, the option for modeling is not offered.
4.Customize input parameters in the ‘Parameters’ tab. The default values for the various parameters are set to yield the best results for most standard cases, and we recommend using them, at least in the initial steps of the analysis.
5.Optionally, assign your job a custom identifier.
6.Start your search by pressing the ‘Submit’ button.
Selecting templates for modeling
7.On the ‘Results’ page of HHpred, analyze the obtained templates and select one or more as wished (Fig. 10A). Next, click on ‘Model using selection’ in the floating toolbar offered at the top of the tab to generate an alignment of the query and the selected templates. If a user clicks on ‘Model using selection’ without having selected any template, the first hit is used for modeling.

8.After clicking on ‘Model using selection’, the query-template alignment is displayed in PIR format in a new job view after a few minutes (Fig. 10B). Click on the ‘Forward to MODELLER’ button to send this alignment as input to MODELLER.
Running MODELLER
9.On the submission page of MODELLER, enter your MODELLER key (Fig. 11A).

10.Optionally, enter a custom job identifier and click on the ‘Submit’ button to start the job.
11.Typical MODELLER jobs take less than 5 min to run through. In the resulting view, the modeled structure is displayed in the NGL Viewer application (Rose et al., 2018) for molecular visualization, and an option is also provided to download the atomic coordinates (Fig. 11B).
Basic Protocol 3: CLUSTER ANALYSIS USING CLANS
Although HHpred is extremely powerful in detecting remote homologs of a protein of interest, it may occasionally not find any meaningful connections because the available target databases contain only a significantly filtered subset of known sequences. Since building profile HMMs is computationally expensive, we currently do not include profile HMMs of large sequence databases for HHpred. In the following, we show how the speed of PSI-BLAST and the power of all-against-all pairwise comparisons can be used to detect remote homologs. For this, we will exploit the observation that the non-significant part of PSI-BLAST searches frequently contains many biologically meaningful connections (indeed, for most proteins, most homologs have non-significant scores). These non-significant pairwise connections nevertheless collectively allow sequences to cluster within a larger sequence space, revealing family relationships. Here, we combine PSI-BLAST with our cluster analysis tool, CLANS (Frickey & Lupas, 2004), to illustrate this approach. CLANS is an implementation of the Fruchterman-Reingold graph-drawing algorithm. It represents protein sequences as points in a virtual 2D or 3D space and allows them to attract or repel each other in proportion to the statistical significance of their all-against-all pairwise comparison. CLANS then searches for the global energy minimum of this landscape of forces, yielding a map in which related sequences group together in connected clusters and unrelated ones drift to the periphery. Here, we will collect input sequences by exploiting the Toolkit implementation of PSI-BLAST, which allows changing the target database between iterations. We will start by building a high-quality PSI-BLAST profile on a small, focused database (nr_arc70) using a strict E-value cutoff (1e-10), then search for all potential homologs up to an E-value of 100 in a more comprehensive database (uniport_prot). We will then forward the obtained hits to the CLANS tool within the Toolkit to carry out an all-against-all sequence comparison. Finally, we will load the resulting CLANS file into the CLANS desktop application to visualize the relationships between the hits and identify groups of related sequences.
Necessary Resources
- Same as for Basic Protocol 1, but additionally, the Java Runtime Environment or Java Development Kit (https://www.oracle.com/java/technologies/javase-jre8-downloads.html or https://openjdk.java.net/install) needs to be installed on the hardware, and at least 4 GB RAM is recommended
PSI-BLAST–Iteration 1
1.Navigate your Web browser to the submission page of PSI-BLAST at https://toolkit.tuebingen.mpg.de/tools/psiblast.
2.Paste the amino acid sequence of your protein of interest (in FASTA format or as plain text; Fig. 12A). Alternatively, the input sequence can be uploaded using the ‘Upload File’ option.

3.Select a target database. For more information on this step, please refer to Support Protocol.
4.Customize input parameters in the ‘Parameters’ tab. Detailed information on each parameter is available in the help pages.
5.Optionally, assign your job a custom identifier and click on the ‘Submit’ button to start the first iteration of PSI-BLAST.
PSI-BLAST–Iteration 2
6.To initiate the second iteration using an MSA of the obtained hits, forward these to PSI-BLAST by clicking on ‘Forward’ in the floating toolbar on the ‘Results’ page of PSI-BLAST (Fig. 12B). ‘PSI-BLAST’ is selected by default. Press the ‘Forward’ button to forward the MSA.
7.On the submission page of PSI-BLAST, select a target protein sequence database (Fig. 13A) and customize input parameters for the second iteration of PSI-BLAST.

8.Optionally, assign your job a custom identifier and click on the ‘Submit’ button to start the second iteration of PSI-BLAST.
Forwarding sequences to CLANS
9.On the ‘Results’ page of PSI-BLAST, click on ‘Select All’ to select all obtained matches (Fig. 13B). By default, only matches with E-values lower than the ‘E-value cutoff for inclusion’ are selected.
10.Click on ‘Forward’ to send sequences of the obtained matches to CLANS. In the ‘Forward’ modal, select ‘CLANS’ in the drop-down list at the bottom left corner, select ‘Full-length sequences’, and click on the ‘Forward’ button (Fig. 13B).
11.On the submission page of CLANS, click on the ‘Submit’ button to start the job (Fig. 14A).
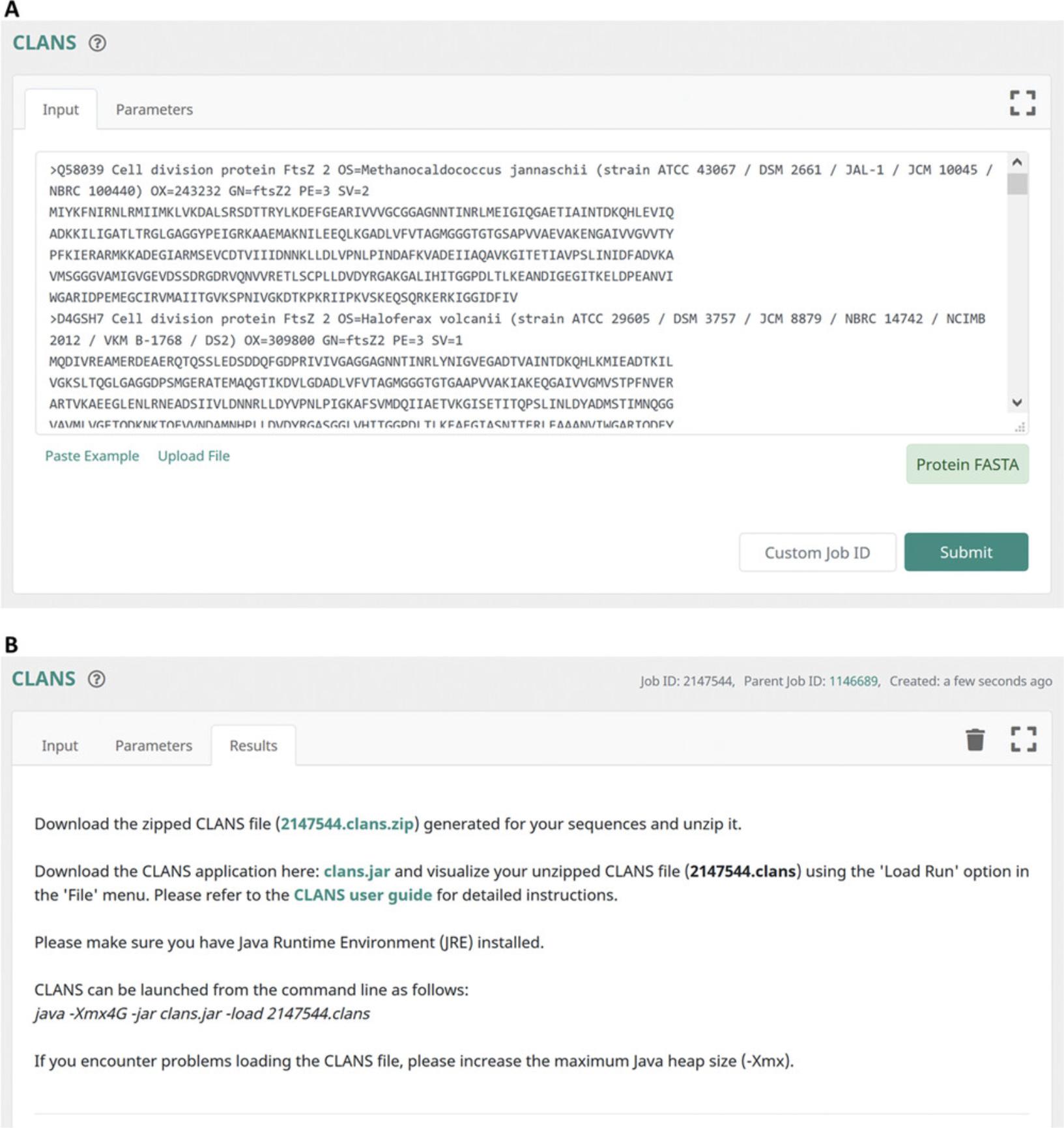
Visualizing sequence relationships in CLANS
12.Depending on the number and length of the input sequences, the CLANS server needs up to several hours to perform all-against-all pairwise PSI-BLAST comparisons. The output page of CLANS offers hyperlinks to the computed raw file in ZIP format, which contains the all-against-all pairwise similarity scores as measured by PSI-BLAST P -values, the CLANS application (clans.jar), and a user guide (Fig. 14B).
13.Download the zipped raw file and extract it.
14.Download the CLANS application and launch it. To run CLANS, you will need to have a Java Runtime Environment (JRE) or a Java Development Kit (JDK) installed.
On the Linux command line, CLANS can be launched using the following command:
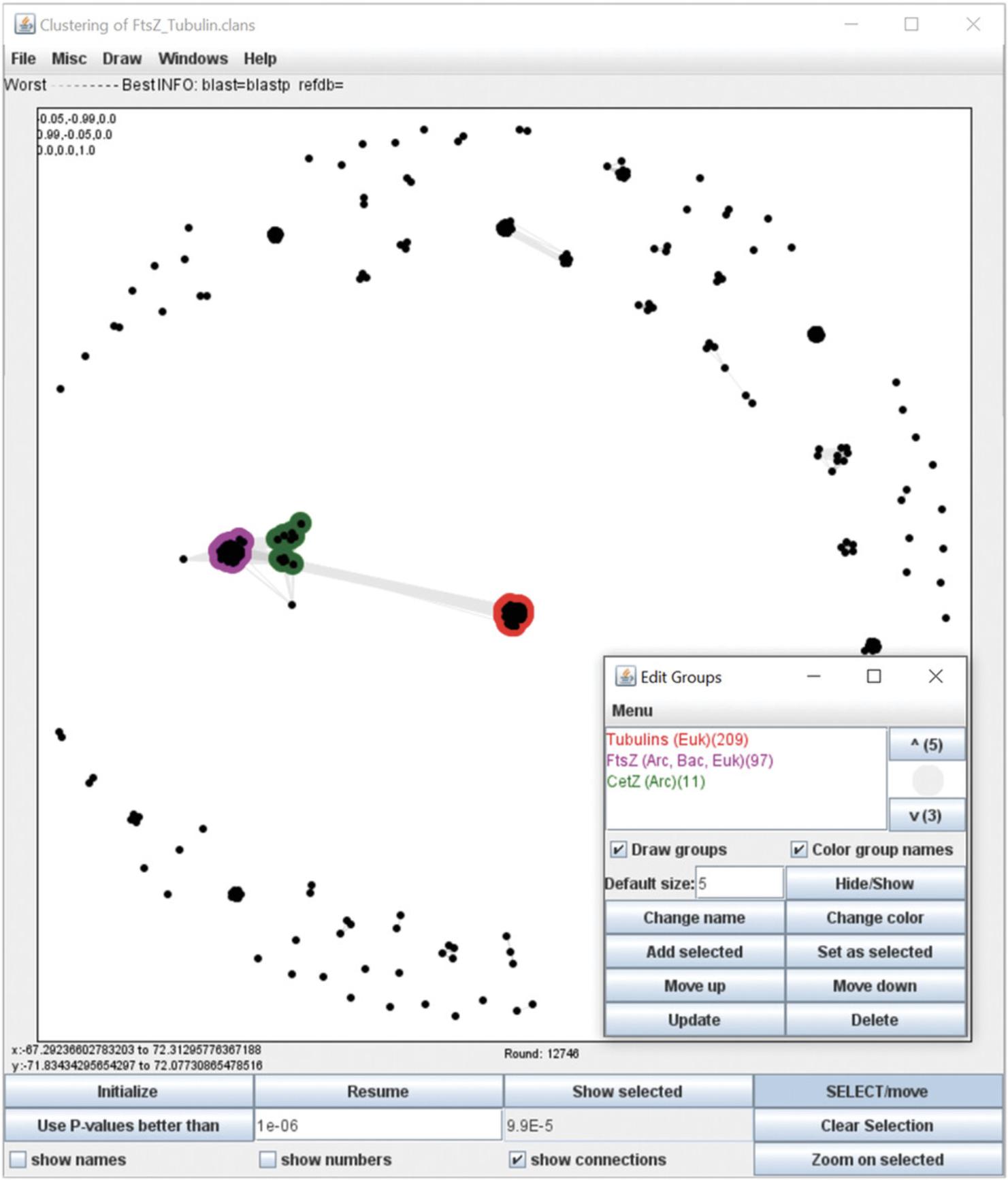
15.In the cluster map loaded within the CLANS application, protein sequences are shown as dots (nodes). Initially, they are placed randomly in a 3D space. Lines (edges) connecting the dots can be shown by selecting ‘show connections’ in the bottom panel. They reflect the significance of the sequence similarity between the sequences; the darker a line, the higher the sequence similarity.
To generate a publication-quality image, we recommend using a 2D space instead of the default 3D space (Misc > Cluster in 2D).
16.Click on ‘Start run’ in the bottom panel to start a clustering run.
17.Analyze the substructure of the map by varying the P -value cutoff used for clustering. To choose a new P -value cutoff, click on ‘Stop’ in the bottom panel to pause the clustering run, enter a value in the ‘Use P -values better than’ text field (e.g., 1e-06 or 0.000001), click on the ‘Use P -values better than’ button, and click on ‘Resume’.
18.Analyze the obtained clusters, annotate them, and produce a publication-quality image of the cluster map.
COMMENTARY
Background Information
Two proteins are said to be homologous if they descended from a common ancestor. Generally, homologous proteins have a similar structure and, depending on the degree of divergence, similar functions, cellular localization, or ligands. Since homology of proteins offers a rich source of functional and structural information, the inference of homology has become an essential tool in molecular biology research and underpins the use of model organisms to study biological processes. The divergent evolution of proteins from hypothetical ancestral forms is generally inferred from the similarity of modern representatives. These comparisons are usually made with sequence data because sequence space is essentially infinite and convergence by chance is, therefore, improbable. In contrast, the number of folded conformations available to the polypeptide chain is limited. Hence, unrelated proteins tend to converge on similar structural solutions, especially at the subdomain level. Also, sequence data are easier to obtain than structural data and, thus, more plentiful by orders of magnitude. Over the years, many different sequence comparison methods have been developed. They achieve different levels of sensitivity, depending on the amount of information they incorporate. Methods that compare individual protein sequences, such as BLAST, are the least sensitive, as they use only the information from the pairwise comparison of two sequences, scored by a global substitution matrix. An additional level of sensitivity is achieved by methods that compare sequence profiles to sequences, such as the iterated version of BLAST, PSI-BLAST. Profiles record the frequencies of the 20 amino acids for each column of an MSA and therefore include family-specific information for the query sequence. Profile-to-profile comparison methods, such as COMPASS (Sadreyev, Tang, Kim, & Grishin, 2009), provide an additional improvement by using family-specific information for both sequences being compared. Incorporation of position-specific insertion and deletion frequencies into profiles yields profile hidden Markov models (HMMs). Methods based on HMM-to-HMM comparison, such as HHpred, are currently our most sensitive tools in the detection of sequence similarity.
Understanding Results
The protocols presented here are meant to allow non-expert users to identify distant homologs to their protein of interest and evaluate potential structural similarities. Since the inference of homology becomes progressively more difficult with decreasing pairwise sequence identity, we focus here on such difficult, divergent protein pairs. The region between 20% and 35% pairwise sequence identity has been named the ‘twilight zone’, and the region below it the ‘midnight zone’ (Rost, 1999); for almost all proteins, most homologs are in these zones. Hence, progress in sequence search tools has been mainly measured by their ability to substantiate homology far into the midnight zone. HHpred, the main tool discussed here, is generally acknowledged as the currently best-performing tool for sequence comparisons. Nevertheless, there are instances where it gives scores indicative of homology to proteins not actually related to the query. It is therefore important to evaluate the plausibility of its results based on a few simple guidelines.
- Check probability and E-value : The probability value reported by HHpred for a match to be a true positive is the most important criterion to infer if a match is homologous to the query or is just a high-scoring chance hit. When it is greater than 95%, evolutionary relatedness is highly likely. Typically, one should give a match serious consideration if it has a probability value >50%, or it has a probability value >30% and is among the top three hits. The E-value is an alternative measure of statistical significance. It is the number of chance hits with a score better than the one for the given match that is expected to be found in the target database. The lower the E-value, the more significant the match is. Unlike the true-positive probability, the HHpred E-value does not take secondary structure similarity into account. Thus, it is a less sensitive measure than the probability. Consequently, even when the E-value is ∼1, matches can be significant by the probability criterion.
- Check secondary structure similarity : If the secondary structure of the query and match is substantially different, the match is probably a false positive.
- Check relationships among top hits : If several of the top matches are homologous to each other, for instance, when they are members of the same SCOPe superfamily or ECOD homology level, then their likelihood of being homologous to the query is very high.
- Check if homology is biologically suggestive : Does the database hit have a function you would also expect for your query? Does it come from an organism that is likely to contain a homolog of your query protein?
- Check for possible conserved motifs : Most homologous pairs of proteins will have at least one (semi-)conserved motif in common. You can identify such putative (semi-)conserved motifs by inspecting HHpred alignments for clusters of three or more well-matching columns (marked with a ‘|’ sign in the row between the query and template consensus sequences) and also by matching consensus sequences. Some false positive matches may have high scores due to possessing an amino acid composition similar to that of the query. In such cases, the alignments tend to be long and lack conserved motifs. You could also scan the alignments for motifs known to be involved in enzymatic function or binding of ligands, such as the GTP-binding motif discussed in this report.
- Check query and template alignments : A corrupted query or template alignment is the main source of high-scoring false positives. The two most common sources of corruption in an alignment are (1) non-homologous sequences, especially repetitive or low-complexity sequences in the alignment, and (2) non-homologous fragments at the ends of the aligned database sequences. Inspect the query and template MSAs for the presence of spurious sequences. In fact, the HHpred server displays an alert message when coiled-coil, transmembrane, or low-complexity segments are detected in the query.
- Check if you can reproduce the results with other parameters : For instance, if you expect the query to be globally homologous to the putative homolog, you could re-run the search using the global alignment mode instead of the local one. You could turn off secondary structure scoring if you suspect that the match between the query and template was scored highly because of a chance similarity of their PSIPRED-predicted or DSSP-determined secondary structures. You can also run the query over other databases to check if similar matches are returned.
Acknowledgments
We would like to thank Andre Noll and Johannes Wörner for their contributions to the development of the Toolkit. This work was supported by institutional funds of the Max Planck Society.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Felix Gabler : Software; writing-review & editing. Seung-Zin Nam : Software. Sebastian Till : Software. Milot Mirdita : Software. Martin Steinegger : Software. Johannes Söeding : Software. Andrei Lupas : Funding acquisition; writing-review & editing. Vikram Alva : Conceptualization; project administration; software; supervision; visualization; writing-original draft; writing-review & editing.
Literature Cited
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research , 25(17), 3389–3402. doi: 10.1093/nar/25.17.3389.
- Alva, V., Nam, S. Z., Soding, J., & Lupas, A. N. (2016). The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Research , 44(W1), W410–415. doi: 10.1093/nar/gkw348.
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., … Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research , 28(1), 235–242. doi: 10.1093/nar/28.1.235.
- Biegert, A., Mayer, C., Remmert, M., Soding, J., & Lupas, A. N. (2006). The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Research , 34(Web Server issue), W335–339. doi: 10.1093/nar/gkl217.
- Cheng, H., Schaeffer, R. D., Liao, Y., Kinch, L. N., Pei, J., Shi, S., … Grishin, N. V. (2014). ECOD: An evolutionary classification of protein domains. PloS Computational Biology , 10(12), e1003926. doi: 10.1371/journal.pcbi.1003926.
- Coggill, P., Finn, R. D., & Bateman, A. (2008). Identifying protein domains with the Pfam database. Current Protocols in Bioinformatics , 23, 2.5.1–2.5.17. doi: 10.1002/0471250953.bi0205s23.
- El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., … Finn, R. D. (2019). The Pfam protein families database in 2019. Nucleic Acids Research , 47(D1), D427–D432. doi: 10.1093/nar/gky995.
- Erickson, H. P. (1998). Atomic structures of tubulin and FtsZ. Trends in Cell Biology , 8(4), 133–137. doi: 10.1016/s0962-8924(98)01237-9.
- Fox, N. K., Brenner, S. E., & Chandonia, J. M. (2014). SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Research , 42(Database issue), D304–D309. doi: 10.1093/nar/gkt1240.
- Frickey, T., & Lupas, A. (2004). CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics , 20(18), 3702–3704. doi: 10.1093/bioinformatics/bth444.
- Gibney, G., & Baxevanis, A. D. (2011). Searching NCBI databases using Entrez. Current Protocols in Bioinformatics , 34, 1.3.1–1.3.25. doi: 10.1002/0471250953.bi0103s34.
- Gruber, M., Soding, J., & Lupas, A. N. (2006). Comparative analysis of coiled-coil prediction methods. Journal of Structural Biology , 155(2), 140–145. doi: 10.1016/j.jsb.2006.03.009.
- Imachi, H., Nobu, M. K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., … Takai, K. (2020). Isolation of an archaeon at the prokaryote-eukaryote interface. Nature , 577(7791), 519–525. doi: 10.1038/s41586-019-1916-6.
- Jones, D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology , 292(2), 195–202. doi: 10.1006/jmbi.1999.3091.
- Joosten, R. P., te Beek, T. A., Krieger, E., Hekkelman, M. L., Hooft, R. W., Schneider, R., … Vriend, G. (2011). A series of PDB related databases for everyday needs. Nucleic Acids Research , 39(Database issue), D411–D419. doi: 10.1093/nar/gkq1105.
- Kabsch, W., & Sander, C. (1983). Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers , 22(12), 2577–2637. doi: 10.1002/bip.360221211.
- Karpenahalli, M. R., Lupas, A. N., & Soding, J. (2007). TPRpred: A tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics , 8, 2. doi: 10.1186/1471-2105-8-2.
- Ladunga, I. (2017). Finding homologs in amino acid sequences using network BLAST searches. Current Protocols in Bioinformatics , 59, 3 4 1–3 4 24. doi: 10.1002/cpbi.34.
- Letunic, I., & Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Research , 46(D1), D493–D496. doi: 10.1093/nar/gkx922.
- Lowe, J., & Amos, L. A. (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature , 391(6663), 203–206. doi: 10.1038/34472.
- Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., … Marchler-Bauer, A. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Research , 48(D1), D265–D268. doi: 10.1093/nar/gkz991.
- Madeira, F., Madhusoodanan, N., Lee, J., Tivey, A. R. N., & Lopez, R. (2019). Using EMBL-EBI Services via Web Interface and Programmatically via Web Services. Current Protocols in Bioinformatics , 66(1), e74. doi: 10.1002/cpbi.74.
- Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., … Lopez, R. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research , 47(W1), W636–W641. doi: 10.1093/nar/gkz268.
- Margolin, W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nature Reviews Molecular Cell Biology , 6(11), 862–871. doi: 10.1038/nrm1745.
- Mirdita, M., Steinegger, M., & Soding, J. (2019). MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics , 35(16), 2856–2858. doi: 10.1093/bioinformatics/bty1057.
- Mirdita, M., von den Driesch, L., Galiez, C., Martin, M. J., Soding, J., & Steinegger, M. (2017). Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Research , 45(D1), D170–D176. doi: 10.1093/nar/gkw1081.
- NCBI Resource Coordinators. (2018). Database resources of the National Center for Biotechnology Information. Nucleic Acids Research , 46(D1), D8–D13. doi: 10.1093/nar/gkx1095.
- Nogales, E., Downing, K. H., Amos, L. A., & Lowe, J. (1998). Tubulin and FtsZ form a distinct family of GTPases. Nature Structural Biology , 5(6), 451–458. doi: 10.1038/nsb0698-451.
- Notredame, C., Higgins, D. G., & Heringa, J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology , 302(1), 205–217. doi: 10.1006/jmbi.2000.4042.
- O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., … Pruitt, K. D. (2016). Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Research , 44(D1), D733–D745. doi: 10.1093/nar/gkv1189.
- Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., & Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Research , 46(W1), W200–W204. doi: 10.1093/nar/gky448.
- Prakash, A., Jeffryes, M., Bateman, A., & Finn, R. D. (2017). The HMMER Web Server for Protein Sequence Similarity Search. Current Protocols in Bioinformatics , 60, 3 15 11–13 15 23. doi: 10.1002/cpbi.40.
- Pundir, S., Martin, M. J., O'Donovan, C., & The UniProt Consortium. (2016). UniProt tools. Current Protocols in Bioinformatics , 53, 1.29.21–21.29.15. doi: 10.1002/0471250953.bi0129s53.
- Remmert, M., Biegert, A., Hauser, A., & Soding, J. (2011). HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature Methods , 9(2), 173–175. doi: 10.1038/nmeth.1818.
- Rose, A. S., Bradley, A. R., Valasatava, Y., Duarte, J. M., Prlic, A., & Rose, P. W. (2018). NGL viewer: Web-based molecular graphics for large complexes. Bioinformatics , 34(21), 3755–3758. doi: 10.1093/bioinformatics/bty419.
- Rost, B. (1999). Twilight zone of protein sequence alignments. Protein Engineering , 12(2), 85–94. doi: 10.1093/protein/12.2.85.
- Sadreyev, R. I., Tang, M., Kim, B. H., & Grishin, N. V. (2009). COMPASS server for homology detection: Improved statistical accuracy, speed and functionality. Nucleic Acids Research , 37(Web Server issue), W90–W94. doi: 10.1093/nar/gkp360.
- Sali, A., & Blundell, T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology , 234(3), 779–815. doi: 10.1006/jmbi.1993.1626.
- Schaeffer, R. D., Liao, Y., & Grishin, N. V. (2018). Searching ECOD for homologous domains by sequence and structure. Current Protocols in Bioinformatics , 61(1), e45. doi: 10.1002/cpbi.45.
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., … Higgins, D. G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology , 7, 539. doi: 10.1038/msb.2011.75.
- Soding, J. (2005). Protein homology detection by HMM-HMM comparison. Bioinformatics , 21(7), 951–960. doi: 10.1093/bioinformatics/bti125.
- Steinegger, M., Meier, M., Mirdita, M., Vohringer, H., Haunsberger, S. J., & Soding, J. (2019). HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics , 20(1), 473. doi: 10.1186/s12859-019-3019-7.
- Suzek, B. E., Wang, Y., Huang, H., McGarvey, P. B., Wu, C. H., & UniProt, C. (2015). UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics , 31(6), 926–932. doi: 10.1093/bioinformatics/btu739.
- Swiss Institute of Bioinformatics Members. (2016). The SIB Swiss Institute of Bioinformatics' resources: Focus on curated databases. Nucleic Acids Research , 44(D1), D27–D37. doi: 10.1093/nar/gkv1310.
- Szwedziak, P., Wang, Q., Bharat, T. A., Tsim, M., & Lowe, J. (2014). Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife , 3, e04601. doi: 10.7554/eLife.04601.
- The UniProt Consortium. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research , 47(D1), D506–D515. doi: 10.1093/nar/gky1049.
- Yang, M., Derbyshire, M. K., Yamashita, R. A., & Marchler-Bauer, A. (2020). NCBI's conserved domain database and tools for protein domain analysis. Current Protocols in Bioinformatics , 69(1), e90. doi: 10.1002/cpbi.90.
- Zimmermann, L., Stephens, A., Nam, S. Z., Rau, D., Kubler, J., Lozajic, M., … Alva, V. (2018). A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. Journal of Molecular Biology , 430(15), 2237–2243. doi: 10.1016/j.jmb.2017.12.007.
Citing Literature
Number of times cited according to CrossRef: 234
- Qian Zhao, Xiaoning Li, Yu Jiao, Ying Chen, Yanfang Yan, Yuzhu Wang, Cyril Hamiaux, Yule Wang, Fengwang Ma, Ross G. Atkinson, Pengmin Li, Identification of two key genes involved in flavonoid catabolism and their different roles in apple resistance to biotic stresses, New Phytologist, 10.1111/nph.19644, 242 , 3, (1238-1256), (2024).
- Corinne Känel, Silke Oeljeklaus, Christoph Wenger, Philip Stettler, Anke Harsman, Bettina Warscheid, André Schneider, Intermembrane space‐localized TbTim15 is an essential subunit of the single mitochondrial inner membrane protein translocase of trypanosomes, Molecular Microbiology, 10.1111/mmi.15262, 121 , 6, (1112-1126), (2024).
- Bianca Oliva, Athanasios Zervas, Peter Stougaard, Peter Westh, Mariane Schmidt Thøgersen, Metagenomic exploration of cold‐active enzymes for detergent applications: Characterization of a novel, cold‐active and alkali‐stable GH8 endoglucanase from ikaite columns in SW Greenland, Microbial Biotechnology, 10.1111/1751-7915.14466, 17 , 6, (2024).
- Jimin Pei, Lisa N. Kinch, Qian Cong, Computational analysis of propeptide‐containing proteins and prediction of their post‐cleavage conformation changes, Proteins: Structure, Function, and Bioinformatics, 10.1002/prot.26702, 92 , 10, (1206-1219), (2024).
- Giuseppe Cortone, Melissa A. Graewert, Manil Kanade, Antonio Longo, Raghurama Hegde, Amaia González‐Magaña, Belén Chaves‐Arquero, Francisco J. Blanco, Luisa M. R. Napolitano, Silvia Onesti, Structural and biochemical characterization of the C‐terminal region of the human RTEL1 helicase, Protein Science, 10.1002/pro.5093, 33 , 9, (2024).
- Gustavo Pierdominici‐Sottile, Juliana Palma, María Leticia Ferrelli, Pablo Sobrado, The dynamics of the flavin, NADPH, and active site loops determine the mechanism of activation of class B flavin‐dependent monooxygenases, Protein Science, 10.1002/pro.4935, 33 , 4, (2024).
- Qiming Yang, Te-Wen Lo, Katjuša Brejc, Caitlin Schartner, Edward J Ralston, Denise M Lapidus, Barbara J Meyer, X-chromosome target specificity diverged between dosage compensation mechanisms of two closely related Caenorhabditis species, eLife, 10.7554/eLife.85413, 12 , (2023).
- Nata Bakuradze, Maia Merabishvili, Ia Kusradze, Pieter-Jan Ceyssens, Jolien Onsea, Willem-Jan Metsemakers, Nino Grdzelishvili, Guliko Natroshvili, Tamar Tatrishvili, Davit Lazvliashvili, Nunu Mitskevich, Jean-Paul Pirnay, Nina Chanishvili, Characterization of a Bacteriophage GEC_vB_Bfr_UZM3 Active against Bacteroides fragilis, Viruses, 10.3390/v15051042, 15 , 5, (1042), (2023).
- Peter Evseev, Irina Tikhonova, Andrei Krasnopeev, Ekaterina Sorokovikova, Anna Gladkikh, Oleg Timoshkin, Konstantin Miroshnikov, Olga Belykh, Tychonema sp. BBK16 Characterisation: Lifestyle, Phylogeny and Related Phages, Viruses, 10.3390/v15020442, 15 , 2, (442), (2023).
- Peter Evseev, Anna Lukianova, Rashit Tarakanov, Anna Tokmakova, Anastasia Popova, Eugene Kulikov, Mikhail Shneider, Alexander Ignatov, Konstantin Miroshnikov, Prophage-Derived Regions in Curtobacterium Genomes: Good Things, Small Packages, International Journal of Molecular Sciences, 10.3390/ijms24021586, 24 , 2, (1586), (2023).
- Suniti Bhaumik, Marzena Łazarczyk, Norwin Kubick, Pavel Klimovich, Agata Gurba, Justyna Paszkiewicz, Patrycja Teodorowicz, Tomasz Kocki, Jarosław Olav Horbańczuk, Gina Manda, Mariusz Sacharczuk, Michel-Edwar Mickael, Investigation of the Molecular Evolution of Treg Suppression Mechanisms Indicates a Convergent Origin, Current Issues in Molecular Biology, 10.3390/cimb45010042, 45 , 1, (628-648), (2023).
- Peter Evseev, Daria Gutnik, Mikhail Shneider, Konstantin Miroshnikov, Use of an Integrated Approach Involving AlphaFold Predictions for the Evolutionary Taxonomy of Duplodnaviria Viruses, Biomolecules, 10.3390/biom13010110, 13 , 1, (110), (2023).
- Xinran Zhang, Yantao Liang, Kaiyang Zheng, Ziyue Wang, Yue Dong, Yundan Liu, Linyi Ren, Hongmin Wang, Ying Han, Andrew McMinn, Yeong Yik Sung, Wen Jye Mok, Li Lian Wong, Jianfeng He, Min Wang, Characterization and genomic analysis of phage vB_ValR_NF, representing a new viral family prevalent in the Ulva prolifera blooms, Frontiers in Microbiology, 10.3389/fmicb.2023.1161265, 14 , (2023).
- Luca A. Robinson, Alice C. Z. Collins, Ronan A. Murphy, Jane C. Davies, Luke P. Allsopp, Diversity and prevalence of type VI secretion system effectors in clinical Pseudomonas aeruginosa isolates, Frontiers in Microbiology, 10.3389/fmicb.2022.1042505, 13 , (2023).
- Sebiha Cevik, Xiaoyu Peng, Tina Beyer, Mustafa S Pir, Ferhan Yenisert, Franziska Woerz, Felix Hoffmann, Betul Altunkaynak, Betul Pir, Karsten Boldt, Asli Karaman, Miray Cakiroglu, S Sadik Oner, Ying Cao, Marius Ueffing, Oktay I Kaplan, WDR31 displays functional redundancy with GTPase-activating proteins (GAPs) ELMOD and RP2 in regulating IFT complex and recruiting the BBSome to cilium, Life Science Alliance, 10.26508/lsa.202201844, 6 , 8, (e202201844), (2023).
- Erwin Tantoso, Birgit Eisenhaber, Swati Sinha, Lars Juhl Jensen, Frank Eisenhaber, About the dark corners in the gene function space of Escherichia coli remaining without illumination by scientific literature, Biology Direct, 10.1186/s13062-023-00362-0, 18 , 1, (2023).
- Jia-Wen Wu, Jin-Town Wang, Tzu-Lung Lin, Ya-Zhu Liu, Lii-Tzu Wu, Yi-Jiun Pan, Identification of three capsule depolymerases in a bacteriophage infecting Klebsiella pneumoniae capsular types K7, K20, and K27 and therapeutic application, Journal of Biomedical Science, 10.1186/s12929-023-00928-0, 30 , 1, (2023).
- Humaira Aziz Sawal, Shagufta Nighat, Tanzeela Safdar, Laiba Anees, Comparative In Silico Analysis and Functional Characterization of TANK-Binding Kinase 1–Binding Protein 1, Bioinformatics and Biology Insights, 10.1177/11779322231164828, 17 , (117793222311648), (2023).
- Atikur Rahman, Md. Takim Sarker, Md Ashiqul Islam, Mohammad Uzzal Hossain, Mahmudul Hasan, Tasmina Ferdous Susmi, Targeting Essential Hypothetical Proteins of Pseudomonas aeruginosa PAO1 for Mining of Novel Therapeutics: An In Silico Approach, BioMed Research International, 10.1155/2023/1787485, 2023 , 1, (2023).
- Shoshana J. Wodak, Sandor Vajda, Marc F. Lensink, Dima Kozakov, Paul A. Bates, Critical Assessment of Methods for Predicting the 3D Structure of Proteins and Protein Complexes, Annual Review of Biophysics, 10.1146/annurev-biophys-102622-084607, 52 , 1, (183-206), (2023).
- Dorothee Serian, Yury Churin, Jens André Hammerl, Manfred Rohde, Arne Jung, Anja Müller, Min Yue, Corinna Kehrenberg, Characterization of Temperate LPS-Binding Bordetella avium Phages That Lack Superinfection Immunity, Microbiology Spectrum, 10.1128/spectrum.03702-22, (2023).
- Yvonne Marvellous Akpudo, Oliver K. Bezuidt, Thulani P. Makhalanyane, Metagenome-Assembled Genomes of Four Southern Ocean Archaea Harbor Multiple Genes Linked to Polyethylene Terephthalate and Polyhydroxybutyrate Plastic Degradation, Microbiology Resource Announcements, 10.1128/mra.01098-22, 12 , 3, (2023).
- Skylar M. Weiss, Kezia K. Happy, Faith W. Baliraine, Abigail K. Beach, Sean M. Brobston, Claire P. Martinez, Kaitlyn J. Menard, Savannah M. Orton, Angela L. Salazar, Gregory D. Frederick, Frederick N. Baliraine, Complete Genome Sequences and Characteristics of Seven Novel Mycobacteriophages Isolated in East Texas, Microbiology Resource Announcements, 10.1128/mra.00335-23, (2023).
- Huang Huang, Xiangmin Hua, Xidan Pang, Zhongmei Zhang, Jingyi Ren, Jiasen Cheng, Yanping Fu, Xueqiong Xiao, Yang Lin, Tao Chen, Bo Li, Huiquan Liu, Daohong Jiang, Jiatao Xie, Discovery and Characterization of Putative Glycoprotein-Encoding Mycoviruses in the Bunyavirales , Journal of Virology, 10.1128/jvi.01381-22, 97 , 1, (2023).
- Oksana Koshla, Lea-Marie Vogt, Oleksandr Rydkin, Yuliia Sehin, Iryna Ostash, Mark Helm, Bohdan Ostash, Landscape of Post-Transcriptional tRNA Modifications in Streptomyces albidoflavus J1074 as Portrayed by Mass Spectrometry and Genomic Data Mining, Journal of Bacteriology, 10.1128/jb.00294-22, 205 , 1, (2023).
- Shaohui Wang, Xianghong Ju, Joshua Heuler, Keshan Zhang, Zhibian Duan, Hiran Malinda Lamabadu Warnakulasuriya Patabendige, Song Zhao, Xingmin Sun, Recombinant Fusion Protein Vaccine Containing Clostridioides difficile FliC and FliD Protects Mice against C. difficile Infection, Infection and Immunity, 10.1128/iai.00169-22, 91 , 4, (2023).
- Vipin S. Rana, Chrysoula Kitsou, Shraboni Dutta, Michael H. Ronzetti, Min Zhang, Quentin Bernard, Alexis A. Smith, Julen Tomás-Cortázar, Xiuli Yang, Ming-Jie Wu, Oleksandra Kepple, Weizhong Li, Jennifer E. Dwyer, Jaqueline Matias, Bolormaa Baljinnyam, Jonathan D. Oliver, Nallakkandi Rajeevan, Joao H F Pedra, Sukanya Narasimhan, Yan Wang, Ulrike Munderloh, Erol Fikrig, Anton Simeonov, Juan Anguita, Utpal Pal, Dome1–JAK–STAT signaling between parasite and host integrates vector immunity and development, Science, 10.1126/science.abl3837, 379 , 6628, (2023).
- Nina Urbelienė, Matas Tiškus, Giedrė Tamulaitienė, Renata Gasparavičiūtė, Ringailė Lapinskaitė, Vykintas Jauniškis, Jurgis Sūdžius, Rita Meškienė, Daiva Tauraitė, Emilija Skrodenytė, Gintaras Urbelis, Justas Vaitekūnas, Rolandas Meškys, Cytidine deaminases catalyze the conversion of N ( S , O ) 4 -substituted pyrimidine nucleosides , Science Advances, 10.1126/sciadv.ade4361, 9 , 5, (2023).
- Ana Laura Salinas, Aurora Osorio, Tonatiuh Legorreta‐Hissner, Reyna Lara‐Martinez, Luis Felipe Jimenez‐Garcia, Laura Camarena, Sebastian Poggio, A new type of phasin characterized by the presence of a helix‐hairpin‐helix domain is required for normal polyhydroxybutyrate accumulation and granule organization in Caulobacter crescentus, Molecular Microbiology, 10.1111/mmi.15124, 120 , 3, (307-323), (2023).
- Dariusz Czernecki, Antonin Nourisson, Pierre Legrand, Marc Delarue, Reclassification of family A DNA polymerases reveals novel functional subfamilies and distinctive structural features, Nucleic Acids Research, 10.1093/nar/gkad242, 51 , 9, (4488-4507), (2023).
- Tanja Zahn, Zihao Zhu, Niklas Ritoff, Jonathan Krapf, Astrid Junker, Thomas Altmann, Thomas Schmutzer, Christian Tüting, Panagiotis L Kastritis, Steve Babben, Marcel Quint, Klaus Pillen, Andreas Maurer, Novel exotic alleles of EARLY FLOWERING 3 determine plant development in barley , Journal of Experimental Botany, 10.1093/jxb/erad127, (2023).
- Yonah A Radousky, Michael T J Hague, Sommer Fowler, Eliza Paneru, Adan Codina, Cecilia Rugamas, Grant Hartzog, Brandon S Cooper, William Sullivan, Distinct Wolbachia localization patterns in oocytes of diverse host species reveal multiple strategies of maternal transmission , GENETICS, 10.1093/genetics/iyad038, 224 , 1, (2023).
- Lucía Blasco, Manuel González de Aledo, Concha Ortiz-Cartagena, Inés Blériot, Olga Pacios, María López, Laura Fernández-García, Antonio Barrio-Pujante, Marta Hernández-Garcia, Rafael Cantón, María Tomás, Study of 32 new phage tail-like bacteriocins (pyocins) from a clinical collection of Pseudomonas aeruginosa and of their potential use as typing markers and antimicrobial agents, Scientific Reports, 10.1038/s41598-022-27341-1, 13 , 1, (2023).
- Alex Reed, Timothy Ware, Haoxin Li, J. Fernando Bazan, Benjamin F. Cravatt, TMEM164 is an acyltransferase that forms ferroptotic C20:4 ether phospholipids, Nature Chemical Biology, 10.1038/s41589-022-01253-7, 19 , 3, (378-388), (2023).
- Morgan Gaïa, Lingjie Meng, Eric Pelletier, Patrick Forterre, Chiara Vanni, Antonio Fernandez-Guerra, Olivier Jaillon, Patrick Wincker, Hiroyuki Ogata, Mart Krupovic, Tom O. Delmont, Mirusviruses link herpesviruses to giant viruses, Nature, 10.1038/s41586-023-05962-4, 616 , 7958, (783-789), (2023).
- Rafael Laso-Pérez, Fabai Wu, Antoine Crémière, Daan R. Speth, John S. Magyar, Kehan Zhao, Mart Krupovic, Victoria J. Orphan, Evolutionary diversification of methanotrophic ANME-1 archaea and their expansive virome, Nature Microbiology, 10.1038/s41564-022-01297-4, 8 , 2, (231-245), (2023).
- Robert E. Jefferson, Aurélien Oggier, Andreas Füglistaler, Nicolas Camviel, Mahdi Hijazi, Ana Rico Villarreal, Caroline Arber, Patrick Barth, Computational design of dynamic receptor—peptide signaling complexes applied to chemotaxis, Nature Communications, 10.1038/s41467-023-38491-9, 14 , 1, (2023).
- Marietta S. Kaspers, Vivian Pogenberg, Christian Pett, Stefan Ernst, Felix Ecker, Philipp Ochtrop, Michael Groll, Christian Hedberg, Aymelt Itzen, Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b, Nature Communications, 10.1038/s41467-023-37621-7, 14 , 1, (2023).
- Li Mi, Ming Shi, Yu-Xuan Li, Gang Xie, Xichen Rao, Damu Wu, Aimin Cheng, Mengxiao Niu, Fengli Xu, Ying Yu, Ning Gao, Wensheng Wei, Xianhua Wang, Yangming Wang, DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing, Nature Communications, 10.1038/s41467-023-36600-2, 14 , 1, (2023).
- Leticia C. Beltran, Virginija Cvirkaite-Krupovic, Jessalyn Miller, Fengbin Wang, Mark A. B. Kreutzberger, Jonasz B. Patkowski, Tiago R. D. Costa, Stefan Schouten, Ilya Levental, Vincent P. Conticello, Edward H. Egelman, Mart Krupovic, Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery, Nature Communications, 10.1038/s41467-023-36349-8, 14 , 1, (2023).
- Ao Sun, Cheng-Ping Li, Zhihang Chen, Shouyue Zhang, Dan-Yuan Li, Yun Yang, Long-Qi Li, Yuqian Zhao, Kaichen Wang, Zhaofu Li, Jinxia Liu, Sitong Liu, Jia Wang, Jun-Jie Gogo Liu, The compact Casπ (Cas12l) ‘bracelet’ provides a unique structural platform for DNA manipulation, Cell Research, 10.1038/s41422-022-00771-2, 33 , 3, (229-244), (2023).
- Zhan Liu, Yi Li, Daijing Wei, Jing Wang, Chong Qiao, Guo-you Li, Guolin Zhang, Yinggang Luo, Biosensor-Enabled Discovery of CaERG6 Inhibitors and Their Antifungal Mode of Action against Candida albicans , ACS Infectious Diseases, 10.1021/acsinfecdis.2c00490, 9 , 4, (785-800), (2023).
- Xiaomin Hu, Yuanyuan Shi, Bingya Jiang, Jie Fu, Xingxing Li, Shufen Li, Guizhi Sun, Weicong Ren, Xinxin Hu, Xuefu You, Zhiyong Liu, Xingli Han, Tianyu Zhang, Bin Hong, Linzhuan Wu, Iterative Methylation Leads to 3-Methylchuangxinmycin Production in Actinoplanes tsinanensis CPCC 200056 , Journal of Natural Products, 10.1021/acs.jnatprod.2c00360, 86 , 1, (1-7), (2023).
- Xanthe Vafopoulou, Logan W. Donaldson, Colin G.H. Steel, The prothoracicotropic hormone (PTTH) of Rhodnius prolixus (Hemiptera) is noggin-like: Molecular characterisation, functional analysis and evolutionary implications, General and Comparative Endocrinology, 10.1016/j.ygcen.2022.114184, 332 , (114184), (2023).
- Sonia Fieulaine, Thibault Tubiana, Stéphane Bressanelli, De novo modelling of HEV replication polyprotein: Five-domain breakdown and involvement of flexibility in functional regulation, Virology, 10.1016/j.virol.2022.12.002, 578 , (128-140), (2023).
- K.A. Yashica, S. Samanta, R. Balaji, V. Jawalagatti, M. Silamparasan, S. Anandu, A. Rialch, S.C. Gupta, Anup Kumar Tewari, Molecular characterization and serodiagnostic evaluation of the Echinococcus ortleppi recombinant glutaredoxin 1 protein for cystic echinococcosis in buffalo (Bubalus bubalis), Veterinary Parasitology, 10.1016/j.vetpar.2023.109941, 319 , (109941), (2023).
- Benjamin T. Donovan, Hengye Chen, Priit Eek, Zhiyuan Meng, Caroline Jipa, Song Tan, Lu Bai, Michael G. Poirier, Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions, Molecular Cell, 10.1016/j.molcel.2023.03.006, 83 , 8, (1251-1263.e6), (2023).
- Derek N. Woolfson, Understanding a protein fold: The physics, chemistry, and biology of α-helical coiled coils, Journal of Biological Chemistry, 10.1016/j.jbc.2023.104579, 299 , 4, (104579), (2023).
- Jethro L. Hemmann, Philipp Keller, Lucas Hemmerle, Thomas Vonderach, Andrea M. Ochsner, Miriam Bortfeld-Miller, Detlef Günther, Julia A. Vorholt, Lanpepsy is a novel lanthanide-binding protein involved in the lanthanide response of the obligate methylotroph Methylobacillus flagellatus, Journal of Biological Chemistry, 10.1016/j.jbc.2023.102940, 299 , 3, (102940), (2023).
- Hiroshi Arai, Hisashi Anbutsu, Yohei Nishikawa, Masato Kogawa, Kazuo Ishii, Masahito Hosokawa, Shiou-Ruei Lin, Masatoshi Ueda, Madoka Nakai, Yasuhisa Kunimi, Toshiyuki Harumoto, Daisuke Kageyama, Haruko Takeyama, Maki N. Inoue, Combined actions of bacteriophage-encoded genes in Wolbachia-induced male lethality, iScience, 10.1016/j.isci.2023.106842, 26 , 6, (106842), (2023).
- Suzanne M McDermott, Vy Pham, Isaac Lewis, Maxwell Tracy, Kenneth Stuart, mt-LAF3 is a pseudouridine synthase ortholog required for mitochondrial rRNA and mRNA gene expression in Trypanosoma brucei, International Journal for Parasitology, 10.1016/j.ijpara.2023.04.002, (2023).
- Haishan Qi, Yihang Gao, Lin Zhang, Zhongxin Cui, Xiaojie Sui, Jianfan Ma, Jing Yang, Zhiquan Shu, Lei Zhang, Rational Design of and Mechanism Insight into an Efficient Antifreeze Peptide for Cryopreservation, Engineering, 10.1016/j.eng.2023.01.015, (2023).
- Emma E. George, Dovilė Barcytė, Gordon Lax, Sam Livingston, Daria Tashyreva, Filip Husnik, Julius Lukeš, Marek Eliáš, Patrick J. Keeling, A single cryptomonad cell harbors a complex community of organelles, bacteria, a phage, and selfish elements, Current Biology, 10.1016/j.cub.2023.04.010, 33 , 10, (1982-1996.e4), (2023).
- Jessica Kimmel, Marius Schmitt, Alexej Sinner, Pascal Wilhelmus Theodorus Christianus Jansen, Sheila Mainye, Gala Ramón-Zamorano, Christa Geeke Toenhake, Jan Stephan Wichers-Misterek, Jakob Cronshagen, Ricarda Sabitzki, Paolo Mesén-Ramírez, Hannah Michaela Behrens, Richárd Bártfai, Tobias Spielmann, Gene-by-gene screen of the unknown proteins encoded on Plasmodium falciparum chromosome 3, Cell Systems, 10.1016/j.cels.2022.12.001, 14 , 1, (9-23.e7), (2023).
- Stéphanie Renaud, Audrey Dussutour, Fayza Daboussi, Denis Pompon, Characterization of chitinases from the GH18 gene family in the myxomycete Physarum polycephalum, Biochimica et Biophysica Acta (BBA) - General Subjects, 10.1016/j.bbagen.2023.130343, 1867 , 6, (130343), (2023).
- Yadveer Kaur, Niranjan Das, Gibberellin 2-Oxidases in Potato (Solanum tuberosum L.): Cloning, Characterization, In Silico Analysis and Molecular Docking, Molecular Biotechnology, 10.1007/s12033-023-00745-8, (2023).
- Trishna Jarambasa, Preetom Regon, Sabnoor Yeasrin Jyoti, Divya Gupta, Sanjib Kumar Panda, Bhaben Tanti, Genome-wide identification and expression analysis of the Pisum sativum (L.) APETALA2/ethylene-responsive factor (AP2/ERF) gene family reveals functions in drought and cold stresses, Genetica, 10.1007/s10709-023-00190-0, (2023).
- Behzad Hajieghrari, Ali Niazi, Phylogenetic and Evolutionary Analysis of Plant Small RNA 2′-O-Methyltransferase (HEN1) Protein Family, Journal of Molecular Evolution, 10.1007/s00239-023-10109-0, (2023).
- Dea Gogishvili, Eva Illes‐Toth, Matthew J. Harris, Christopher Hopley, Charlotte E. Teunissen, Sanne Abeln, Structural flexibility and heterogeneity of recombinant human glial fibrillary acidic protein (GFAP), Proteins: Structure, Function, and Bioinformatics, 10.1002/prot.26656, 92 , 5, (649-664), (2023).
- Burcu Ozden, Andriy Kryshtafovych, Ezgi Karaca, The impact of AI‐based modeling on the accuracy of protein assembly prediction: Insights from CASP15, Proteins: Structure, Function, and Bioinformatics, 10.1002/prot.26598, 91 , 12, (1636-1657), (2023).
- Annika Topitsch, Torsten Schwede, Joana Pereira, Outer membrane β‐barrel structure prediction through the lens of AlphaFold2, Proteins: Structure, Function, and Bioinformatics, 10.1002/prot.26552, 92 , 1, (3-14), (2023).
- Florian Michel, Sergio Romero‐Romero, Birte Höcker, Retracing the evolution of a modern periplasmic binding protein, Protein Science, 10.1002/pro.4793, 32 , 11, (2023).
- Norma E. Padilla‐Mejia, Mark C. Field, Evolutionary, structural and functional insights in nuclear organisation and nucleocytoplasmic transport in trypanosomes, FEBS Letters, 10.1002/1873-3468.14747, 597 , 20, (2501-2518), (2023).
- Desislava P Staneva, Stefan Bresson, Tatsiana Auchynnikava, Christos Spanos, Juri Rappsilber, A Arockia Jeyaprakash, David Tollervey, Keith R Matthews, Robin C Allshire, The SPARC complex defines RNAPII promoters in Trypanosoma brucei, eLife, 10.7554/eLife.83135, 11 , (2022).
- Inês Gomes Castro, Shawn P Shortill, Samantha Katarzyna Dziurdzik, Angela Cadou, Suriakarthiga Ganesan, Rosario Valenti, Yotam David, Michael Davey, Carsten Mattes, Ffion B Thomas, Reut Ester Avraham, Hadar Meyer, Amir Fadel, Emma J Fenech, Robert Ernst, Vanina Zaremberg, Tim P Levine, Christopher Stefan, Elizabeth Conibear, Maya Schuldiner, Systematic analysis of membrane contact sites in Saccharomyces cerevisiae uncovers modulators of cellular lipid distribution, eLife, 10.7554/eLife.74602, 11 , (2022).
- Lei Song, Jingjing Luo, Hongou Wang, Dan Huang, Yunhao Tan, Yao Liu, Yingwu Wang, Kaiwen Yu, Yong Zhang, Xiaoyun Liu, Dan Li, Zhao-Qing Luo, Legionella pneumophila regulates host cell motility by targeting Phldb2 with a 14-3-3ζ-dependent protease effector, eLife, 10.7554/eLife.73220, 11 , (2022).
- Feng Wang, Xiaochen Feng, Ren Kong, Shan Chang, Generating new protein sequences by using dense network and attention mechanism, Mathematical Biosciences and Engineering, 10.3934/mbe.2023195, 20 , 2, (4178-4197), (2022).
- Susan M. Lehman, Rohit Kongari, Adam M. Glass, Matthew Koert, Melissa D. Ray, Roger D. Plaut, Scott Stibitz, Phage K gp102 Drives Temperature-Sensitive Antibacterial Activity on USA300 MRSA, Viruses, 10.3390/v15010017, 15 , 1, (17), (2022).
- Zi-Lu Wei, Feng Yang, Bo Li, Pu Hou, Wen-Wen Kong, Jie Wang, Yuxing Chen, Yong-Liang Jiang, Cong-Zhao Zhou, Structural Insights into the Chaperone-Assisted Assembly of a Simplified Tail Fiber of the Myocyanophage Pam3, Viruses, 10.3390/v14102260, 14 , 10, (2260), (2022).
- Cyril J. Versoza, Abigail A. Howell, Tanya Aftab, Madison Blanco, Akarshi Brar, Elaine Chaffee, Nicholas Howell, Willow Leach, Jackelyn Lobatos, Michael Luca, Meghna Maddineni, Ruchira Mirji, Corinne Mitra, Maria Strasser, Saige Munig, Zeel Patel, Minerva So, Makena Sy, Sarah Weiss, Susanne P. Pfeifer, Comparative Genomics of Closely-Related Gordonia Cluster DR Bacteriophages, Viruses, 10.3390/v14081647, 14 , 8, (1647), (2022).
- Jennifer Dahan, Yuri I. Wolf, Gardenia E. Orellana, Erik J. Wenninger, Eugene V. Koonin, Alexander V. Karasev, A Novel Flavi-like Virus in Alfalfa (Medicago sativa L.) Crops along the Snake River Valley, Viruses, 10.3390/v14061320, 14 , 6, (1320), (2022).
- Peter Evseev, Mikhail Shneider, Konstantin Miroshnikov, Evolution of Phage Tail Sheath Protein, Viruses, 10.3390/v14061148, 14 , 6, (1148), (2022).
- Miguel F. Gonzales, Denish K. Piya, Brian Koehler, Kailun Zhang, Zihao Yu, Lanying Zeng, Jason J. Gill, New Insights into the Structure and Assembly of Bacteriophage P1, Viruses, 10.3390/v14040678, 14 , 4, (678), (2022).
- Aleksandra Nakonieczna, Paweł Rutyna, Magdalena Fedorowicz, Magdalena Kwiatek, Lidia Mizak, Małgorzata Łobocka, Three Novel Bacteriophages, J5a, F16Ba, and z1a, Specific for Bacillus anthracis, Define a New Clade of Historical Wbeta Phage Relatives, Viruses, 10.3390/v14020213, 14 , 2, (213), (2022).
- Rashit I. Tarakanov, Anna A. Lukianova, Peter V. Evseev, Stepan V. Toshchakov, Eugene E. Kulikov, Alexander N. Ignatov, Konstantin A. Miroshnikov, Fevzi S.-U. Dzhalilov, Bacteriophage Control of Pseudomonas savastanoi pv. glycinea in Soybean, Plants, 10.3390/plants11070938, 11 , 7, (938), (2022).
- Kianoush Jeiran, Scott M. Gordon, Denis O. Sviridov, Angel M. Aponte, Amanda Haymond, Grzegorz Piszczek, Diego Lucero, Edward B. Neufeld, Iosif I. Vaisman, Lance Liotta, Ancha Baranova, Alan T. Remaley, A New Structural Model of Apolipoprotein B100 Based on Computational Modeling and Cross Linking, International Journal of Molecular Sciences, 10.3390/ijms231911480, 23 , 19, (11480), (2022).
- Romain Launay, Elin Teppa, Carla Martins, Sophie S. Abby, Fabien Pierrel, Isabelle André, Jérémy Esque, Towards Molecular Understanding of the Functional Role of UbiJ-UbiK2 Complex in Ubiquinone Biosynthesis by Multiscale Molecular Modelling Studies, International Journal of Molecular Sciences, 10.3390/ijms231810323, 23 , 18, (10323), (2022).
- Ilona Michalik, Kamil J. Kuder, Katarzyna Kieć-Kononowicz, Jadwiga Handzlik, Structure Prediction, Evaluation, and Validation of GPR18 Lipid Receptor Using Free Programs, International Journal of Molecular Sciences, 10.3390/ijms23147917, 23 , 14, (7917), (2022).
- Agnieszka Bednarek, Agata Cena, Wioleta Izak, Joanna Bigos, Małgorzata Łobocka, Functional Dissection of P1 Bacteriophage Holin-like Proteins Reveals the Biological Sense of P1 Lytic System Complexity, International Journal of Molecular Sciences, 10.3390/ijms23084231, 23 , 8, (4231), (2022).
- Marius Rehanek, David G. Karlin, Martina Bandte, Rim Al Kubrusli, Shaheen Nourinejhad Zarghani, Thierry Candresse, Carmen Büttner, Susanne von Bargen, The Complex World of Emaraviruses—Challenges, Insights, and Prospects, Forests, 10.3390/f13111868, 13 , 11, (1868), (2022).
- Abu Saim Mohammad Saikat, Apurbo Kumar Paul, Dipta Dey, Ranjit Chandra Das, Madhab Chandra Das, In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus , The 26th International Electronic Conference on Synthetic Organic Chemistry, 10.3390/ecsoc-26-13522, (21), (2022).
- Peter Evseev, Anna Lukianova, Rashit Tarakanov, Anna Tokmakova, Mikhail Shneider, Alexander Ignatov, Konstantin Miroshnikov, Curtobacterium spp. and Curtobacterium flaccumfaciens: Phylogeny, Genomics-Based Taxonomy, Pathogenicity, and Diagnostics, Current Issues in Molecular Biology, 10.3390/cimb44020060, 44 , 2, (889-927), (2022).
- Lionel Ballut, Sébastien Violot, Frédéric Galisson, Isabelle R. Gonçalves, Juliette Martin, Santosh Shivakumaraswamy, Loïc Carrique, Hemalatha Balaram, Nushin Aghajari, Tertiary and Quaternary Structure Organization in GMP Synthetases: Implications for Catalysis, Biomolecules, 10.3390/biom12070871, 12 , 7, (871), (2022).
- Thomas Tarenzi, Giovanni Mattiotti, Marta Rigoli, Raffaello Potestio, In Search of a Dynamical Vocabulary: A Pipeline to Construct a Basis of Shared Traits in Large-Scale Motions of Proteins, Applied Sciences, 10.3390/app12147157, 12 , 14, (7157), (2022).
- Jack Fleet, Mujtaba Ansari, Jon K. Pittman, Phylogenetic analysis and structural prediction reveal the potential functional diversity between green algae SWEET transporters, Frontiers in Plant Science, 10.3389/fpls.2022.960133, 13 , (2022).
- Rowan Herridge, Tyler McCourt, Jeanne M. E. Jacobs, Peter Mace, Lynette Brownfield, Richard Macknight, Identification of the genes at S and Z reveals the molecular basis and evolution of grass self-incompatibility, Frontiers in Plant Science, 10.3389/fpls.2022.1011299, 13 , (2022).
- Ana Laura Ramos, Maria Aquino, Gema García, Miriam Gaspar, Cristina de la Cruz, Anaid Saavedra-Flores, Susana Brom, Ramón Cervantes-Rivera, Clara Elizabeth Galindo-Sánchez, Rufina Hernandez, Andrea Puhar, Andrei N. Lupas, Edgardo Sepulveda, RpuS/R Is a Novel Two-Component Signal Transduction System That Regulates the Expression of the Pyruvate Symporter MctP in Sinorhizobium fredii NGR234, Frontiers in Microbiology, 10.3389/fmicb.2022.871077, 13 , (2022).
- Christopher A. Beaudoin, Martin Bartas, Adriana Volná, Petr Pečinka, Tom L. Blundell, Are There Hidden Genes in DNA/RNA Vaccines?, Frontiers in Immunology, 10.3389/fimmu.2022.801915, 13 , (2022).
- Boris Shaskolskiy, Dmitry Kravtsov, Ilya Kandinov, Sofya Gorshkova, Alexey Kubanov, Victoria Solomka, Dmitry Deryabin, Ekaterina Dementieva, Dmitry Gryadunov, Comparative Whole-Genome Analysis of Neisseria gonorrhoeae Isolates Revealed Changes in the Gonococcal Genetic Island and Specific Genes as a Link to Antimicrobial Resistance, Frontiers in Cellular and Infection Microbiology, 10.3389/fcimb.2022.831336, 12 , (2022).
- Martin P. Schwalm, Lena M. Berger, Maximilian N. Meuter, James D. Vasta, Cesear R. Corona, Sandra Röhm, Benedict-Tilman Berger, Frederic Farges, Sebastian M. Beinert, Franziska Preuss, Viktoria Morasch, Vladimir V. Rogov, Sebastian Mathea, Krishna Saxena, Matthew B. Robers, Susanne Müller, Stefan Knapp, A Toolbox for the Generation of Chemical Probes for Baculovirus IAP Repeat Containing Proteins, Frontiers in Cell and Developmental Biology, 10.3389/fcell.2022.886537, 10 , (2022).
- Julia L Daiß, Michael Pilsl, Kristina Straub, Andrea Bleckmann, Mona Höcherl, Florian B Heiss, Guillermo Abascal-Palacios, Ewan P Ramsay, Katarina Tlučková, Jean-Clement Mars, Torben Fürtges, Astrid Bruckmann, Till Rudack, Carrie Bernecky, Valérie Lamour, Konstantin Panov, Alessandro Vannini, Tom Moss, Christoph Engel, The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans, Life Science Alliance, 10.26508/lsa.202201568, 5 , 11, (e202201568), (2022).
- Nupur Sharma, Christof Osman, Yme2, a putative RNA recognition motif and AAA+ domain containing protein, genetically interacts with the mitochondrial protein export machinery, Biological Chemistry, 10.1515/hsz-2021-0398, 403 , 8-9, (807-817), (2022).
- Ran Mo, Wenhui Ma, Weijie Zhou, Beile Gao, Polar localization of CheO under hypoxia promotes Campylobacter jejuni chemotactic behavior within host, PLOS Pathogens, 10.1371/journal.ppat.1010953, 18 , 11, (e1010953), (2022).
- Roland Pfoh, Adithya S. Subramanian, Jingjing Huang, Dustin J. Little, Adam Forman, Benjamin R. DiFrancesco, Negar Balouchestani-Asli, Elena N. Kitova, John S. Klassen, Régis Pomès, Mark Nitz, P. Lynne Howell, The TPR domain of PgaA is a multifunctional scaffold that binds PNAG and modulates PgaB-dependent polymer processing, PLOS Pathogens, 10.1371/journal.ppat.1010750, 18 , 8, (e1010750), (2022).
- Patrick Günther, Dennis Quentin, Shehryar Ahmad, Kartik Sachar, Christos Gatsogiannis, John C. Whitney, Stefan Raunser, Structure of a bacterial Rhs effector exported by the type VI secretion system, PLOS Pathogens, 10.1371/journal.ppat.1010182, 18 , 1, (e1010182), (2022).
- Maryam Rafiqi, Lukas Jelonek, Aliou Moussa Diouf, AbdouLahat Mbaye, Martijn Rep, Alhousseine Diarra, Profile of the in silico secretome of the palm dieback pathogen, Fusarium oxysporum f. sp. albedinis, a fungus that puts natural oases at risk, PLOS ONE, 10.1371/journal.pone.0260830, 17 , 5, (e0260830), (2022).
- Ran Mo, Siqi Zhu, Yuanyuan Chen, Yuqian Li, Yugeng Liu, Beile Gao, The evolutionary path of chemosensory and flagellar macromolecular machines in Campylobacterota, PLOS Genetics, 10.1371/journal.pgen.1010316, 18 , 7, (e1010316), (2022).
- Liu He, Lotte van Beem, Berend Snel, Casper C. Hoogenraad, Martin Harterink, PTRN-1 (CAMSAP) and NOCA-2 (NINEIN) are required for microtubule polarity in Caenorhabditis elegans dendrites, PLOS Biology, 10.1371/journal.pbio.3001855, 20 , 11, (e3001855), (2022).
- Sarah M. Roelle, Nidhi Shukla, Anh T. Pham, Anna M. Bruchez, Kenneth A. Matreyek, Expanded ACE2 dependencies of diverse SARS-like coronavirus receptor binding domains, PLOS Biology, 10.1371/journal.pbio.3001738, 20 , 7, (e3001738), (2022).
- Xue Zhang, Yuxi Yang, Yuxuan Wei, Qingshun Zhao, Xin Lou, blf and the drl cluster synergistically regulate cell fate commitment during zebrafish primitive hematopoiesis , Development, 10.1242/dev.200919, 149 , 24, (2022).
- See more

