Coated Latex Beads as Artificial Cells for Quantitative Investigations of Receptor/Ligand Interactions
Doris Urlaub, Doris Urlaub, Carsten Watzl, Carsten Watzl
Abstract
Cellular interactions are often essential to regulate immune cell activities during an immune response. To understand the details of this process, it is necessary to study individual receptor/ligand interactions in a quantitative fashion. However, this is often very difficult or even impossible when using real cells for stimulation. Here, we present a method to use cell-sized latex beads for such studies. These beads can be coated with agonistic antibodies or specific ligands in a defined and quantifiable fashion. This creates the possibility of titrating the strength of the stimulation for a specific receptor in a three-dimensional system. Using natural killer (NK) cells as an example, we demonstrate how these beads can be used to stimulate NK cell responses. © 2020 The Authors.
Basic Protocol 1 : Covalent coating of latex beads with antibodies
Basic Protocol 2 : Quantification of the amount of antibodies on the beads with the QIFIKIT®
Alternate Protocol 1 : Covalent coating of latex beads with streptavidin to bind biotinylated proteins
Alternate Protocol 2 : Quantification of the amount of protein on the beads with the QIFIKIT®
Support Protocol : Functional testing of the beads in a natural killer cell degranulation assay
INTRODUCTION
In vitro stimulation of immune cells can be carried out using a large variety of methods. Stimulation by cell/cell interaction, e.g., T cells with antigen-presenting cells or natural killer (NK) cells with tumor target cells, mimics the physiological condition most closely. However, using cells to stimulate other cells also entails a number of disadvantages. Even if the cells are properly characterized, there is always the chance that unknown interactions will influence the observed result or that the “stimulating” cell will cause results that will then be attributed to the “stimulated” cell. In addition, it is difficult to study quantitative aspects of the stimulation as it is not easy to vary and to quantify the amounts of stimulating ligands on cells.
Agonistic antibodies have a clear advantage in that only the intended receptor is triggered and that the stimulation strength can be modified by titration of the antibody (Mesecke, Urlaub, Busch, Eils, & Watzl, 2011). In most cases, crosslinking, e.g., with a secondary antibody, is necessary to induce clustering of the receptor and generate activation signals. This is considered to be artificial, and the effects may not be comparable to the outcome of stimulation under physiological conditions. In addition, the affinity of the interaction and the binding site can be different from those of the true ligand, and the stimulation can happen all over the cell surface and is not localized like in an immunological synapse.
Another method with some of the advantages offered by the use of antibodies is to prepare surfaces: either antibody-coated slides or lipid bilayers containing the proteins of interest. Using these surfaces to stimulate immune cells is an option if the readout is going to be by microscopy or if the supernatant will be analyzed for soluble molecules, but it complicates the assay and can distort the results if the analysis is intended to be done by flow cytometry.
Therefore, we developed a method using latex beads that have approximately the same size as immune cells for the stimulation of NK cells (Dorsch et al., 2020). The interaction of these beads with the cells matches the proportions of an immunological synapse, and they easily can be included in flow cytometry measurements. Here, we describe how to coat these beads (Basic Protocol 1) and quantify the exact amounts of antibodies that are bound on the surface (Basic Protocol 2) and how to perform functional testing with NK cells (Support Protocol).
The same method can also be used to coat recombinant proteins, e.g., the extracellular domains of the natural ligands of NK cell receptors, onto the beads. By first coating the beads with streptavidin (Alternate Protocol 1) and using recombinant proteins with a targeted biotinylation, the correct orientation of the proteins on the beads can be ensured (Alternate Protocol 2).
STRATEGIC PLANNING
Each coating needs to be quantified (Basic Protocol 2 and Alternate Protocol 2) because the results may vary between different experiments. Keep in mind that you should prepare enough beads not only for your functional experiment (Support Protocol) but also for the quantification, staining controls, and counting. Given that the beads are very stable, they can be prepared in bigger batches once the method is established. Then, multiple experiments can be performed using the same batch of beads.
Basic Protocol 1: COVALENT COATING OF LATEX BEADS WITH ANTIBODIES
The reaction partners first need to be activated with a chemical crosslinker to allow covalent binding of the antibodies to the beads. The reaction using EDAC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide-hydrochloride] should mainly result in amide bonds between any carboxyl groups of the antibodies and the amine groups of the beads, but EDAC has no specificity for this reaction, so crosslinking of different reactive groups of the antibodies is also possible. The concentration of EDAC, the sequence in which the different reagents are added, and the timing of the incubation, therefore, are critical.
Materials
-
Aliphatic Amine Latex Beads [2% (w/v); ∼3 × 107/ml; Thermo Fisher Scientific, SKU no. A37374; currently only available as custom-order product]
-
1× MES buffer (pH 4.8) with or without 0.1% (w/v) SDS (see recipes)
-
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide-hydrochloride (EDAC; Roth, cat. no. 2156)
-
Anhydrous dimethylsulfoxide (DMSO; Invitrogen, cat. no. D12345)
-
Unconjugated monoclonal mouse IgG antibody
-
Bovine serum album (BSA), fraction V (PAN Biotech, cat. no. P06-1391500)
-
Dulbecco's phosphate-buffered saline (DPBS; no calcium, no magnesium; Gibco/Thermo Fisher Scientific, cat. no. 14190169)
-
Storage buffer: PBS with 0.1% (w/v) glycine (AppliChem, cat. no. 131340) and 0.1% (w/v) sodium azide (Roth, cat. no. K305)
-
Conical centrifuge tube (e.g., 1.5- or 15-ml)
-
Microcentrifuge or centrifuge with swinging bucket
-
Cell counter (e.g., CASY) or hemocytometer and microscope
-
End-over-end rotator
1.Use approximate concentration of the Aliphatic Amine Latex Beads as delivered (3 × 107/ml) to estimate the volume needed for the entire experiment. Mix beads well and place this volume plus ∼10% more (to account for variations and loss during sample processing) in an appropriate conical centrifuge tube (e.g., a 1.5-ml tube for volumes <400 µl or a 15-ml tube for larger volumes).
2.Wash beads twice with an excess (at least twice the volume of the bead suspension) of 1× MES buffer with 0.1% SDS. Mix well and spin down 5 min at 5000 × g in a microcentrifuge or 1000 × g in a centrifuge with a swinging bucket. Aspirate supernatant carefully so as not to disrupt the pellet.
3.Dilute beads in 1× MES buffer with 0.1% SDS, count beads using a cell counter (e.g., CASY) or a hemocytometer and microscope, and wash for a third time (see step 2). Calculate exact amount of beads in the tube and resuspend them in 30 µl of 1× MES buffer with 0.1% SDS per 1 million beads (3.3 × 107/ml).
4.Add EDAC in anhydrous DMSO to beads to a final concentration of 10 mg/ml.
5.Incubate for 30 min at room temperature on an end-over-end rotator.
6.Wash beads once with an excess of 1× MES buffer with 0.1% SDS (see step 2) and resuspend in 30 µl of 1× MES buffer (without SDS) per 1 million beads (3.3 × 107/ml).
7.Add desired amount of unconjugated monoclonal mouse IgG antibody to the beads and incubate for 1 hr at room temperature on an end-over-end rotator.
8.Add 20 µg BSA per million beads to bead/antibody mix and continue incubation on rotator for an additional 30 min.
9.Wash beads twice with an excess of DPBS (see step 2).
10.Use beads directly or resuspend beads in storage buffer, e.g., at a concentration of 1 × 107 beads/ml, and store at 4°C until use (see Basic Protocol 2).
Basic Protocol 2: QUANTIFICATION OF THE AMOUNT OF ANTIBODIES ON THE BEADS WITH THE QIFIKIT®
The manual for the QIFIKIT® provides plenty of information on how different samples can be prepared and analyzed. Nevertheless, we describe here how we adapted this protocol to fit the requirements of our assay.
Materials
-
QIFIKIT®, including tubes containing setup and calibration beads and labeled secondary antibody (Dako/Agilent, cat. no. K0078)
-
Antibody-coated latex beads (see Basic Protocol 1)
-
FACS buffer [PBS with 2% (v/v) fetal bovine serum (FBS)] with or without 2% (w/v) formaldehyde
-
96-V-well plate
-
Centrifuge with swinging bucket
-
Flow cytometer and analysis software
-
Spreadsheet program
1.Vortex tubes containing the setup beads and the calibration beads of the QIFIKIT® and transfer 50 µl of each to a 96-V-well plate.
2.Take same amount of antibody-coated latex beads to be quantified from each condition (100,000 to 200,000 beads per sample) and transfer these samples to 96-V-well plate. Include one sample as an unstained control.
3.Wash QIFIKIT® beads and latex beads once by adding 150 µl FACS buffer to each well of the 96-V-well plate, centrifuging 5 min at 500 × g in a centrifuge with a swinging bucket, and aspirating the supernatant.
4.Prepare a master mix of labeled secondary antibody in FACS buffer. Resuspend all beads except unstained control (just QIFIKIT® beads and latex beads) in 50 µl of this master mix. Incubate for 20 min at room temperature in the dark.
5.Wash once with 150 µl FACS buffer (see step 3) and resuspend beads in 100 to 150 µl FACS buffer with or without 2% (w/v) formaldehyde per sample.
6.Analyze samples using a flow cytometer.
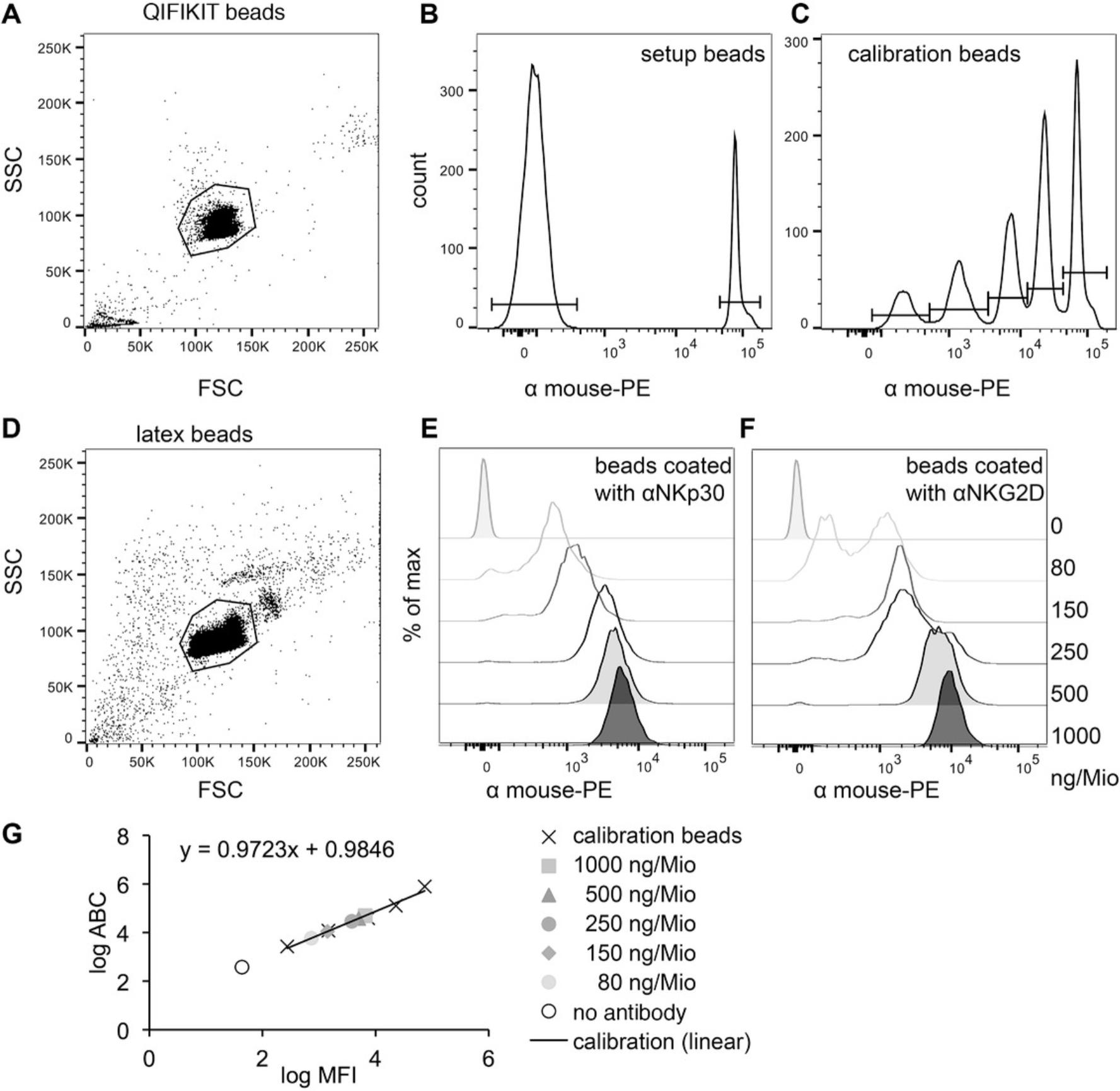
7.Perform analysis using analysis software and a spreadsheet program.
Alternate Protocol 1: COVALENT COATING OF LATEX BEADS WITH STREPTAVIDIN TO BIND BIOTINYLATED PROTEINS
Following the steps of Basic Protocol 1, virtually any protein can be covalently bound to the beads; we use this protocol to coat the beads with streptavidin at one constant concentration. These beads can then bind any biotinylated protein. The amount of protein on the surface can be modified by titrating the recombinant biotinylated protein. Additionally, when the biotinylation of the protein is targeted, correct orientation on the beads can be ensured.
Additional Materials (also see Basic Protocol 1)
- Streptavidin (BioLegend, cat. no. 405150)
- FACS buffer: PBS with 2% (v/v) FBS
- Biotinylated protein (biotin: Sigma, cat. no. B4501)
1.Follow steps 1 to 6 of Basic Protocol 1.
2.Add 0.5 µg streptavidin per million beads and incubate for 1 hr at room temperature on an end-over-end rotator.
3.Continue with steps 8 and 9 of Basic Protocol 1.
4.Resuspend beads in storage buffer, e.g., at a concentration of 1 × 107 beads/ml, and store them at 4°C. To bind biotinylated protein to the beads, incubate beads for 20 min at room temperature in FACS buffer containing the biotinylated protein.
5.Wash beads twice with an excess of FACS buffer (see step 2 of Basic Protocol 1).
6.Use beads directly or keep them at 4°C in storage buffer.
Alternate Protocol 2: QUANTIFICATION OF THE AMOUNT OF PROTEIN ON THE BEADS WITH THE QIFIKIT®
To quantify the protein loaded onto the beads with Alternate Protocol 1, one additional step is needed relative to Basic Protocol 2, but it can be done very easily with the QIFIKIT®.
Additional Materials (also see Basic Protocol 2)
- Streptavidin-coated latex beads with or without biotinylated protein (see Alternate Protocol 1)
- Unconjugated monoclonal mouse IgG antibodies against biotinylated proteins used (see Alternate Protocol 1)
1.Take same amount of streptavidin-coated latex beads with biotinylated protein to be quantified from each condition (100,000 to 200,000 beads per sample) and transfer them to a 96-V-well plate. Include one sample of streptavidin-coated latex beads without biotinylated protein for each antibody that is going to be used. If the beads are in storage buffer, wash them once by adding 150 µl FACS buffer to each well of beads in the 96-V-well plate, centrifuging 5 min at 500 × g , and aspirating supernatant.
2.Resuspend beads in 50 µl FACS buffer containing 5 µg/ml unconjugated monoclonal mouse IgG antibody against the respective biotinylated protein used. Treat sample of streptavidin-only beads in the same way. Incubate for 20 min at room temperature.
3.Wash beads once with 150 µl FACS buffer. Prepare setup and calibration beads from the QIFIKIT® as in Basic Protocol 2, steps 1 and 3.
4.Prepare a master mix of labeled secondary antibody in FACS buffer and resuspend all samples in 50 µl of this master mix. Incubate for 20 min at room temperature in the dark.
5.Follow steps 5 to 7 of Basic Protocol 2.
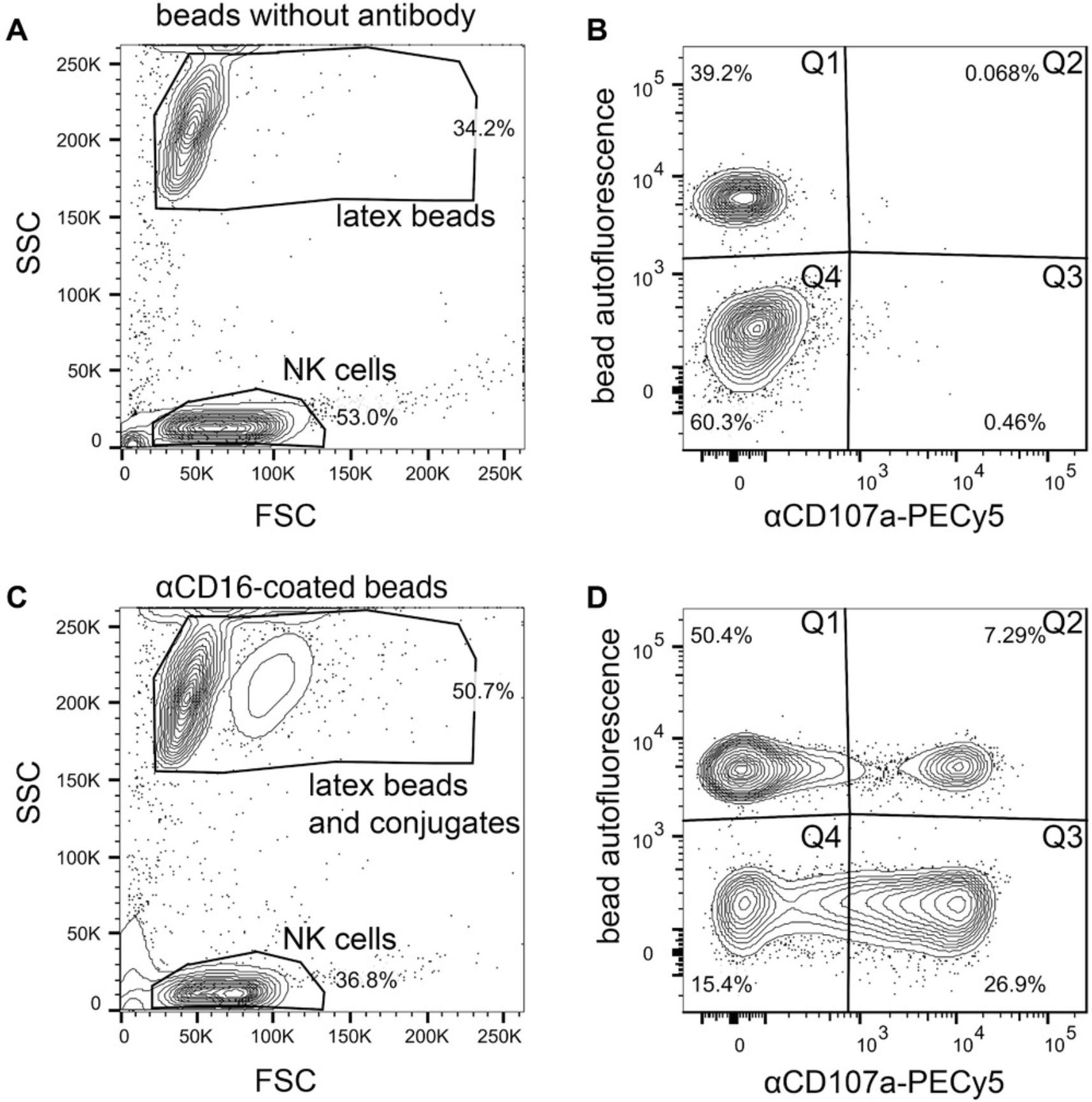
Support Protocol: FUNCTIONAL TESTING OF THE BEADS IN A NATURAL KILLER CELL DEGRANULATION ASSAY
One of the main functional assays that we use and an excellent way to test beads that are coated with antibodies (Basic Protocol 1) that are directed against activating NK cell receptors is a degranulation assay that detects surface CD107a. We use antibodies to coat the beads that have been successfully used in our laboratory to stimulate NK cells via other methods.
Additional Materials (also see Basic Protocol 2)
- Preferred culture medium for NK cells
- NK cells (e.g., primary human NK cells or NK-92 cell line)
- αCD107a (clone H4A3) PE-Cy5 (BD Pharmingen, cat. no. 555802)
NOTE : All solutions and equipment coming into contact with cells must be sterile, and proper sterile technique should be used accordingly.
NOTE : All culture incubations are performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified.
1.Take same amount of antibody-coated latex beads from each sample to test (e.g., 1 × 105) and transfer them to a 96-V-well plate.
2.Wash beads once with 100 µl of the preferred culture medium for NK cells to remove the storage buffer. Centrifuge 5 min at 500 × g in a centrifuge with a swinging bucket and aspirate supernatant.
3.Count NK cells, centrifuge 5 min at 500 × g if necessary, and resuspend the same number of cells as beads (1:1 ratio) at a concentration of 2 × 106 cells/ml in culture medium.
4.Allocate NK cells (e.g., 1 × 105 NK cells in 50 µl) as an unstained control in an empty well of the 96-V-well plate. Add 0.2 µg/ml αCD107a PE-Cy5 to remaining NK cell suspension and mix by carefully inverting tube. Remove NK cells (e.g., 1 × 105 NK cells in 50 µl) as an unstimulated control. Then, resuspend beads in 50 µl NK cell mix per sample.
5.Incubate 96-V-well plate containing control cells and samples mixed with beads for 3 hr in a humidified incubator at 37°C and 5% CO2.
6.Wash all samples once with 150 µl FACS buffer and resuspend them in 150 µl FACS buffer.
7.Analyze samples directly using a flow cytometer or store them for a short time at 4°C protected from light before flow cytometry.
8.Perform analysis using analysis software and a spreadsheet program.
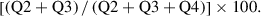
REAGENTS AND SOLUTIONS
MES buffer (pH 4.8), 1×
To prepare a 10× stock solution of MES, dissolve MES [2-(N -morpholino)ethanesulfonic acid sodium salt, Calbiochem, cat. no. 475894] in ultrapure water to 0.25 M, adjust pH to 4.8 with 1 M HCl, and filter through a 0.22-µm filter. Store sterile 10× stock for ≤1 year at room temperature. Dilute to 1× (0.025 M MES) in ultrapure water and check pH again. Store ≤1 year at room temperature.
MES buffer (pH 4.8), 1×, with 0.1% SDS
Prepare a 5% (w/v) SDS stock solution from sodium dodecyl sulfate (SDS; grained pure; AppliChem, cat. no. A7249) in ultrapure water. Warm solution to 37°C if necessary and stir well until SDS is completely dissolved. Store SDS stock for ≤1 year at room temperature. Add this SDS stock 1:50 to 1× MES buffer, pH 4.8 (see recipe). Store ≤2 months at room temperature.
COMMENTARY
Background Information
Various beads have been used to stimulate NK cells or other immune cells. Some commercial kits use beads to expand cells in culture (e.g., Miltenyi B Cell Expansion Kit). Magnetic beads that are manufactured to isolate cells or proteins can be diverted from this use and coated with antibodies or proteins to stimulate cells (Sim et al., 2019, Todros-Dawda, Kveberg, Vaage, & Inngjerdingen, 2014). However, none of these methods has all properties in the combination that we aim for in our experiments: we want beads that have the same size as cells to mimic a real immunological synapse, that have covalent coating for stability, that support the possibility of titrating and quantifying the coating, and that have an autofluorescence comparable to that of cells to facilitate flow cytometric analysis.
Critical Parameters
Detergent use and centrifugation of the latex beads
The latex beads are delivered as a surfactant-free suspension. We tested various detergents to find one that facilitates bead handling without negative effects on the coating efficiency. However, even with the addition of SDS to the MES buffer, the beads are difficult to handle during all steps before they are coated with protein (Basic Protocol 1 and Alternate Protocol 1). The centrifugation steps should be performed in a centrifuge with a swinging bucket to minimize loss of beads during the washing steps or must be performed at higher speed. The settling speed of the beads is not the limiting factor, but rather their tendency to stick to all surfaces and to not form a proper pellet. Once the bead surface is coated with protein, slower centrifugation speeds are sufficient, but we still recommend using a centrifuge with a swinging bucket.
EDAC stability
EDAC (Basic Protocol 1 and Alternate Protocol 1) hydrolyzes and loses its activity in the presence of even small amounts of humidity. Solutions in aqueous media have to be prepared freshly for each experiment. We observed that even when condensation was avoided, EDAC lost its activity after the container had been opened several times. Therefore, we make “aliquots” of ∼10 mg and store them dry at −20°C with desiccant. From these aliquots, you can prepare a fresh solution in MES buffer for each experiment. Because small amounts of EDAC are required frequently, especially while establishing the method, preparing a stock solution in water-free DMSO is the most practical option. EDAC does not dissolve well in DMSO, but a concentration of 100 mg/ml is possible, and this solution can be stored at −20°C and reused several times without a reduction in activity.
Bead stability
In the storage buffer, which contains sodium azide as a biocide, the beads (Basic Protocol 1 and Alternate Protocol 1) can be stored for ≥2 months at 4°C without loss of activity. In our initial experiments, we quantified the beads a second time after 8 or 9 weeks and found no substantial changes: neither the ABC nor the functional activity was reduced. Even after 2 years, the ABC of antibody-coated beads (Basic Protocol 1) was within the same range as directly after the coating reaction.
Troubleshooting
Buffer conditions for bead coating
The buffer used before and during coating (Basic Protocol 1 and Alternate Protocol 1) can influence the efficiency drastically. MES buffer with pH 6 is recommended by the bead manufacturer, but different proteins might require different pH conditions. The results for coating antibodies were most stable when we used MES buffer with pH 4.8; therefore, we use this pH throughout our experiments. If you have to optimize the protocol because the coating is not sufficient, changing the pH of the MES buffer might work.
As mentioned above, a detergent in the buffer facilitates handling of the beads during the initial washing steps. We tested various detergents and found some that did not disturb the coating process and with which the efficiency was comparable to the coating without detergent. SDS even increased antibody binding to the beads when used before coating, but during the incubation of beads with the antibodies, MES without detergent should be used. If the coating does not work properly, reducing the SDS concentration or using other detergents like Triton-X-100 or Igepal might help, but we have not tested beads coated this way as thoroughly as the ones created with the protocols described (Basic Protocol 1 and Alternate Protocol 1).
Concentration of EDAC and timing of EDAC addition
The concentration of EDAC used and when it is applied are also critical (Basic Protocol 1 and Alternate Protocol 1). We thought that co-incubating EDAC together with beads and antibody should lead to a more efficient coating of the beads, but this did not work for various EDAC concentrations tested. The beads generated this way indeed had a high density of antibodies on the surface, but these antibodies were not functional. We suspect that reactions between reactive groups of the antibodies destroyed their structure, including the antigen-binding site. We found that pre-incubation of the beads with a quite high EDAC concentration and washing it out before the antibodies are added leads to the best results. Thus, if you observe high coating of the beads but without the expected functional effects of the antibody, reducing the EDAC concentration or intensifying the washing after the EDAC incubation could be the solution.
Heterogeneous coating
Especially at lower antibody concentrations, we often observe heterogeneous coating of the beads (Basic Protocol 1 and Alternate Protocol 1). To reduce this, mixing of samples throughout the incubations with EDAC and with antibodies is essential. To ensure equal distribution of the antibody, we prepare the dilution in MES buffer. After the incubation with EDAC, the beads are washed and resuspended directly in the antibody dilution.
Some antibodies just do not work
Maybe due to their individual amino acid sequences or due to the positions of the reactive groups in the structure of the antibodies, some antibody clones just do not bind efficiently to the beads irrespectively of all coating conditions (Basic Protocol 1 and Alternate Protocol 1). Among the different antibodies that we use, the maximal coating differs a lot and is specific to the individual antibody. As long as the coating density is sufficient to induce the functional effects in NK cells (Support Protocol), we still can use those beads in our experiments. In contrast, some antibodies have such a low coating density that we cannot use them to stimulate our cells and have to exclude them from our experiments.
Understanding Results
The quantification with the QIFIKIT® (Basic Protocol 2 and Alternate Protocol 2) gives you the total number of antibodies or proteins on one bead. However, depending on the cell type that you use and the cells’ activation state, cells may only make contact with a part of the bead surface. You could estimate the average contact area for your cells by microscopic analysis. Therefore, the number of antibodies that stimulate a certain effect will be lower than the total number of antibodies on a bead.
Time Considerations
The coating of the beads (Basic Protocol 1 and Alternate Protocol 1) typically requires <2.5 hr of total incubation time. Depending on how many different conditions you plan to prepare, it can be done within 3 hr. The staining for the quantification of antibody-coated beads and beads coated with biotinylated protein (Basic Protocol 2 and Alternate Protocol 2) can be done in 1 to 1.5 hr. The time that you need for measuring the samples and analyzing the result (Basic Protocol 2 and Alternate Protocol 2) then depends mainly on the number of samples that you prepared. Because the beads can be stored after coating, you do not need to perform these experiments the same day. The functional assays (Support Protocol) that you want to perform with those beads also can be planned independently once you have prepared the beads. A small degranulation assay with fewer than 10 samples requires <30 min preparation time and can be completed within 5 hr, including the incubation time, measurement, and analysis.
Acknowledgments
We thank Madeleine Dorsch and Vivian Bönnemann for their help in establishing these methods.
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Doris Urlaub Investigation; methodology; writing-original draft. Carsten Watzl Conceptualization; funding acquisition; project administration; supervision;writing-review & editing.
Literature Cited
- Dorsch, M., Urlaub, D., Bonnemann, V., Brode, P., Sandusky, M., & Watzl, C. (2020). Quantitative analysis of human NK cell reactivity using latex beads coated with defined amounts of antibodies. European Journal of Immunology , 50(5), 656–665. doi: 10.1002/eji.201948344.
- Mesecke, S., Urlaub, D., Busch, H., Eils, R., & Watzl, C. (2011). Integration of activating and inhibitory receptor signaling by regulated phosphorylation of Vav1 in immune cells. Science Signaling , 4(175), ra36. doi: 10.1126/scisignal.2001325.
- Sim, M. J. W., Rajagopalan, S., Altmann, D. M., Boyton, R. J., Sun, P. D., & Long, E. O. (2019). Human NK cell receptor KIR2DS4 detects a conserved bacterial epitope presented by HLA-C. Proceedings of the National Academy of Sciences of the United States of America , 116(26), 12964–12973. doi: 10.1073/pnas.1903781116.
- Todros-Dawda, I., Kveberg, L., Vaage, J. T., & Inngjerdingen, M. (2014). The tetraspanin CD53 modulates responses from activating NK cell receptors, promoting LFA-1 activation and dampening NK cell effector functions. PLOS One , 9(5), e97844. doi: 10.1371/journal.pone.0097844.
Citing Literature
Number of times cited according to CrossRef: 3
- Anna Luise Grab, Peter S. Kim, Lukas John, Kamlesh Bisht, Hongfang Wang, Anja Baumann, Helgi Van de Velde, Irene Sarkar, Debarati Shome, Philipp Reichert, Calin Manta, Stefanie Gryzik, Rogier M. Reijmers, Niels Weinhold, Marc S. Raab, Pre-Clinical Assessment of SAR442257, a CD38/CD3xCD28 Trispecific T Cell Engager in Treatment of Relapsed/Refractory Multiple Myeloma, Cells, 10.3390/cells13100879, 13 , 10, (879), (2024).
- Maria Dede, Annemieke van Dam, Conjugation of visual enhancers in lateral flow immunoassay for rapid forensic analysis: A critical review, Analytical and Bioanalytical Chemistry, 10.1007/s00216-024-05565-6, (2024).
- Erick Sánchez-Salguero, Karina Corona-Cervantes, Hector Armando Guzmán-Aquino, María Fernanda de la Borbolla-Cruz, Víctor Contreras-Vargas, Alberto Piña-Escobedo, Jaime García-Mena, Leopoldo Santos-Argumedo, Maternal IgA2 Recognizes Similar Fractions of Colostrum and Fecal Neonatal Microbiota, Frontiers in Immunology, 10.3389/fimmu.2021.712130, 12 , (2021).

