Modeling the Effects of Maternal Diabetes on the Developing Human Heart Using Pluripotent Stem Cell–Derived Heart Organoids
Yonatan R. Lewis-Israeli, Yonatan R. Lewis-Israeli, Mishref Abdelhamid, Mishref Abdelhamid, Isoken Olomu, Isoken Olomu, Aitor Aguirre, Aitor Aguirre
Abstract
Congenital heart defects (CHD) constitute the most common type of birth defect in humans. Maternal diabetes during the first trimester of pregnancy (pregestational diabetes, or PGD) is one of the most prominent factors contributing to CHD, and is present in a significant population of female patients with diabetes in reproductive age. PGD is challenging to manage clinically due to the extreme sensitivity of the developing embryo to glucose oscillations, and constitutes a critical health problem for the mother and the fetus. The prevalence of PGD-induced CHD is increasing due to the ongoing diabetes epidemic. While studies using animal models and cells in culture have demonstrated that PGD alters critical cellular and developmental processes, the mechanisms remain obscure, and it is unclear to what extent these models recapitulate PGD-induced CHD in humans. Clinical practice precludes direct studies in developing human embryos, further highlighting the need for physiologically relevant models. To bypass many of these technical and ethical limitations, we describe here a human pluripotent stem cell (hPSC)–based method to generate developmentally relevant self-organizing human heart organoids. By using glucose and insulin to mimic the diabetic environment that the embryo faces in PGD, this system allows modeling critical features of PGD in a human system with relevant physiology, structure, and cell types. The protocol starts with the generation of hPSC-derived embryoid bodies in a 96-well plate, followed by a small molecule–based three-step Wnt activation/inhibition/activation strategy. Organoids are then differentiated under healthy (normal insulin and glucose) and diabetic conditions (high insulin and glucose) over time, allowing for the study of the effects of pregestational diabetes on the developing human heart. We also provide an immunofluorescence protocol for comparing, characterizing, and analyzing the differences between the healthy and diabetic organoids, and comment on additional steps for preparing the organoids for analysis by other techniques after differentiation. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation of hPSC-derived embryoid bodies
Basic Protocol 2 : Differentiation of EBs into heart organoids under healthy and diabetes-like conditions
Basic Protocol 3 : Immunofluorescence and organoid preparation for other assays
INTRODUCTION
Congenital heart defects (CHDs) constitute the most common type of congenital defect in humans (Zaidi & Brueckner, 2017). Pregestational diabetes (PGD), defined here as diabetes of the mother before and during the first trimester of pregnancy (regardless of whether it is T1D or T2D), is one of the most prominent non-genetic factors contributing to CHD (Øyen et al., 2016), and is present in a significant and growing fraction of the population of female patients in reproductive age (Centers for Disease Control and Prevention, 2020). Newborns from mothers with PGD have increased risk of CHD (PGD-CHD, 4%-12% depending on the sources) versus ∼1% in the general population (i.e., up to a 12-fold increase; Basu & Garg, 2018; Ornoy, Reece, Pavlinkova, Kappen, & Miller, 2015; Øyen et al., 2016; Tabib et al., 2013). PGD is challenging to manage clinically due to the extreme sensitivity of the developing embryo to glucose oscillations, and constitutes a critical health problem for the mother and the fetus. The prevalence of PGD-CHD is increasing significantly due to the ongoing diabetes epidemic, and although preventative and therapeutic interventions are critically needed, no significant advances have yet been made to treat this condition.
Significant efforts have been devoted to understanding the molecular pathology of PGD-CHD using animal models (Buchanan & Kitzmiller, 1994; Correa, 2016), leading to the identification of increased ROS production, abnormal lipid metabolism, and mitochondrial stress, among other contributing factors (Engineer et al., 2018; Lehtoranta et al., 2013, 2017; Ornoy et al., 2015; Piddington, Joyce, Dhanasekaran, & Baker, 1996). The molecular links and the overall contribution of these abnormalities to the different phenotypes observed in PGD-CHD, however, remain elusive. It also remains unclear to what extent rodent models recapitulate abnormalities present in human PGD-CHD, particularly given critical species differences in heart size as well as cardiac physiology and bioenergetics (Gupta, Chacko, Schär, Akki, & Weiss, 2011; Hamlin & Altschuld, 2011; Loiselle & Gibbs, 1979). For example, there are major species differences between mice and humans in how the heart utilizes glucose, lactate, ketone bodies, and fatty acids. Furthermore, rodent models—and most in vitro cell models—commonly rely on very aggressive diabetic conditions (∼25-50 mM blood glucose), which might lead to cytotoxic phenotypes not relevant to the condition under study. In addition, clinical practice largely precludes studies of PGD-CHD in humans—or severely limits them—and, thus, direct studies of human embryos are not viable. This is understandable given the care that pregnant mothers need, particularly if they suffer from potential increased risk to their fetus. The result, however, is significantly limited access to human tissues for research of early-stage disease and mechanisms of PGD-CHD, resulting in an over-reliance on animal models.
In recent years, novel stem cell–based technologies have enabled the creation of engineered, highly complex human organ–like 3D tissues in vitro , with properties that recapitulate the physiological setting to a significant extent (Kretzschmar & Clevers, 2016; Paşca, 2019; Rossi et al., 2020; Sasai, 2013; Weeber, Ooft, Dijkstra, & Voest, 2017). These organoids (i.e., “resembling an organ”) are particularly useful to study unapproachable disease states in humans (e.g., early disease progression when symptoms are not still present), or states for which animal models are not well-suited (Bredenoord, Clevers, & Knoblich, 2017; Lewis-Israeli et al., 2021; Tuveson & Clevers, 2019). Organoids can also be powerful tools to verify, in human models, what we have learned in animal studies, thus complementing them. While organoids have been used to model a wide range of human tissues and conditions, their applications to cardiovascular studies have been sorely lacking until only very recently (Drakhlis et al., 2021; Hofbauer et al., 2021; Lewis-Israeli et al., 2021; Rossi et al., 2020).
We recently took a crucial technical step forward by engineering one of the first synthetic developmental models of human heart development (Lewis-Israeli et al., 2021). Previous studies into cardiac cell differentiation in 2D have described the use of strategic Wnt pathway activation and subsequent inhibition using small molecules (Lian et al., 2012). More recently, a study into epicardial cell differentiation demonstrated the ability to generate epicardial cell populations in 2D by the use of an additional Wnt pathway activation step 7-9 days after the first activation (Bao et al., 2017). Our approach made use of these findings, and we combined optimization of both the small molecule concentration and time exposure with the addition of strategic growth factors to create a highly reproducible 3D human heart organoid model. Our heart organoids recapitulate critical steps of cardiac development in humans, while displaying high complexity in cardiac cell types and structural organization. The system is highly reproducible and amenable to screening approaches (Lewis-Israeli et al., 2021; Lewis-Israeli, Wasserman, & Aguirre, 2021).
In this article, we describe, in detail, how to utilize our heart organoid technology for the study of PGD-CHD in humans, bridging the gap between rodent studies and the human fetus. First, we describe how to generate embryoid bodies (EBs) from human induced pluripotent stem cells (hiPSCs) in individual wells, allowing for independent samples (Basic Protocol 1). We then describe the process of inducing cardiac mesoderm and subsequent epicardial specification under healthy and diabetic conditions, facilitating a platform to model the effects of pregestational diabetes in the developing human heart (Basic Protocol 2). Finally, in Basic Protocol 3, we describe an immunofluorescence protocol, and also provide the steps to prepare the fetal-like cardiac organoids for other assays, which, together, can be used to study various features of congenital heart defects caused by the hyperglycemic and hyperinsulinemic environment. An overview of the entire workflow is shown in Figure 1.
STRATEGIC PLANNING
hESCs or iPSCs cultured under normal conditions in culture media based on the E8 formulation developed by the Thomson Lab (Chen et al., 2011) are required for this protocol. PSCs can be purchased through commercial vendors such as WiCell (https://www.wicell.org/) or reprogrammed in-house using an iPSC reprogramming protocol (Malik & Rao, 2013; Panopoulos et al., 2017; Takahashi & Yamanaka, 2006). We have validated this protocol with both hESCs and hiPSCs. hPSCs should be cultured and passaged at least twice after thawing, to allow them to grow under the required culture conditions and to monitor for unwanted differentiation. While cells are cultured in 6-well plates in this protocol, other types of cell culture plates can be used, and the number of wells needed would then have to be extrapolated from the details provided in this protocol.
All the work with cells, EBs, and organoids should be conducted in a laminar flow hood under sterile conditions.
This protocol has been optimized for use with E8 medium and related formulations for hPSC growth and expansion (Chen et al., 2011). Less defined cell culture media may result in higher variability and failure to achieve the desired results of this protocol. Media should be allowed to warm to room temperature before use (E8), or warmed up to 37°C in a water bath (RPMI) before coming in contact with cells.
The user should always keep in mind throughout the protocols that the EBs and subsequent organoids are not attached to the plate and are sitting at the very bottom of the well. Therefore, extra care should be employed when changing medium, to minimize any organoid movements that may cause damage or stress. Once organoid differentiation begins (Basic Protocol 2), medium changes should take place in a time-specific manner (i.e., at the same time of day within a 20 min window).
Healthy and diabetes media can be prepared in advance according to the provided recipes, and stored at 4°C for up to 1 month.
NOTE : Users should obtain the necessary permissions and approvals from the relevant institutional and/or national research ethics committee, and research should be conducted according to international guidelines.
Basic Protocol 1: GENERATION OF PSC-DERIVED EMBRYOID BODIES
In this protocol, users will aggregate dissociated PSCs into three-dimensional (3D) spheres termed embryoid bodies (EBs). PSCs are first grown in the appropriate cell culture medium and allowed to reach the appropriate confluency. Then, the PSCs are dissociated into single cells in suspension and are seeded onto a 96-well plate in which the organoids will differentiate and be maintained. The ROCK pathway inhibitor thiazovivin is kept in the medium while cells are being seeded, to improve survival and inhibit apoptosis of the cells in suspension. The 96-well plate is then centrifuged to induce the formation of spherical aggregates from the cells in suspension, resulting in the EBs. The EBs are maintained in cell culture medium until the start of the differentiation protocol, described in Basic Protocol 2.
Materials
-
PSC cells (e.g., iPSCs reprogrammed in the lab (Malik & Rao, 2013; Panopoulos et al., 2017; Takahashi & Yamanaka, 2006) or obtained from commercial sources (see Strategic Planning)
-
Essential 8 Flex medium (Gibco, cat. no. A2858501)
-
Penicillin-Streptomycin (Gibco, cat. no. 15140122)
-
Thiazovivin (Millipore Sigma, cat. no. SML1045)
-
Matrigel GFR (Corning, cat. no. CB40230)
-
PBS (Gibco, cat. no. 10010049)
-
ReLeSRTM (STEMCELL Technologies, cat. no. 05872)
-
Accutase (Innovative Cell Technologies, cat. no. NC9464543)
-
HERAcell VIOS 160i CO2 incubator (Thermo Fisher Scientific, cat. no. 51030285)
-
Bead bath
-
15-ml Falcon tubes (Fisher Scientific, cat. no. 1495970C)
-
Sorvall Legend ×1 centrifuge (Thermo Fisher Scientific, cat. no. 75004261)
-
6-well flat-bottom cell culture plates (Corning, cat. no. 0720083)
-
Light microscope (Olympus, cat. no. CKX53)
-
P-1000 and P-200 micropipettes and corresponding [wide-bore] pipette tips
-
50-ml reagent reservoirs (Fisherbrand, cat. no. 13681502)
-
Multichannel pipettes, 30-300 µl (Fisherbrand, cat. no. FBE1200300)
-
1.5-ml microcentrifuge tubes (Fisher Scientific, cat. no. 02682002)
-
Moxi Cell Counter (Orflo Technologies, cat. no. MXZ001) or hemocytometer
-
Serological pipettes (Fisher Scientific, cat. no. 14-955-233, 14-955-234)
-
96-well clear ultra low attachment microplates (Costar, cat. no. 07201680)
Stem cell dissociation
1.Culture the PSCs.
-
Thaw a cryovial containing the desired cell line by taking it out of liquid nitrogen storage and placing it in a bead bath for 2-4 min until most of the medium is thawed.
-
Transfer medium from the cryovial to a 15-ml Falcon tube containing 8 ml of Essential 8 Flex medium (E8 Flex) with 1% (v/v) penicillin/streptomycin (5 ml penicillin/streptomycin in 500 ml E8 Flex) and 2 µM thiazovivin (TZV).
-
Centrifuge for 5 min at 300 ×g, discard supernatant, and resuspend pellet in 4 ml of E8 Flex containing 2 µM TZV.
-
Seed cells onto two wells of a 6-well plate coated with Matrigel (0.5 mg of Matrigel per 6-well plate, according to manufacturer's instructions) at a volume of 2 ml per well.
-
Replace medium after 24 hr with E8 Flex medium (without TZV) and culture cells until sub-confluency (70%-80%) is reached, changing medium with fresh E8 Flex (without TZV) every 48 hr.
Perform medium changes on the cells every 24-48 hr based on the culture medium manufacturer's recommendation (e.g., E8 Flex can be changed every 48 hr).
Cells are seeded onto two wells to avoid high seeding density, although only one well will be required for the remainder of the protocol.
2.Passage iPSCs.
-
Aspirate medium from sub-confluent iPSCs and wash cells with 1 ml/well of PBS.
-
Aspirate PBS and passage cells using ReLeSRTM, according to the manufacturer's instructions. Passage the cells at a 1:6 well ratio by resuspending one well of detached cells in 12 ml of E8 Flex containing 2 µM of TZV, and then seeding onto six wells of a 6-well plate coated with Matrigel, at a volume of 2 ml per well.
-
Replace medium after 24 hr and culture cells in E8 medium until sub-confluency (70%-80%) is reached. If more cells are desired, repeat for the other well; if not, discard the cells in the second well.
Repeat step 2 to maintain cells in culture until you are ready to create organoids. iPSCs can be passaged for >50 passages without any karyotypic abnormalities.
3.Aspirate medium from sub-confluent iPSCs and wash cells with 1 ml of PBS. Use 2-3 wells of the 6-well plate from step 2 for each desired 96-well plate of organoids.
4.Aspirate PBS from each of the wells you will use and add 1 ml of room temperature Accutase to each of them. Gently tap the side of the plate 3-5 times every minute to induce cell detachment. Continue doing so for ∼5 min or until most cells have detached. Monitor the cells under a light microscope to ensure cells have detached.
5.Prepare 1 ml per well of E8 Flex containing 2 µM TZV. Stop the reaction by adding 1 ml of this mix to each well. Gently pipette the cells up and down 2-3 times using a wide-tip p1000 pipette to dissociate any remaining cell aggregates, and transfer medium from all wells to a single conical 15-ml Falcon tube.
6.Centrifuge the tube for 5 min at 300 × g , room temperature.
7.Aspirate supernatant and resuspend the pellet in 1 ml of E8 Flex + 2 µM TZV, gently pipetting up and down 2-3 times. Make sure to pipette up and down enough so that the pellet is resuspended.
8.Take 10 µl of medium with cells and transfer to a microcentrifuge tube containing 990 µl of PBS, to prepare a 1:100 dilution. Count the number of dissociated cells using a Moxi Cell Counter.
9.Gently mix the medium with cells using a wide-tip p1000 pipette to create a uniform cell suspension, and transfer the cells to fresh E8 Flex + 2 µM TZV in a 15-ml Falcon tube. Adjust to a final cell concentration of 100,000 cells/ml.
Cell aggregation into embryoid bodies
10.For a single 96-well plate, and using a serological pipette, transfer 11 ml of E8 Flex + TZV containing 100,000 PSCs per ml into a reagent reservoir.
11.Using a multichannel pipette, transfer 100 µl of cell-containing medium to each well of a 96-well clear ultra low attachment microplate, for a total of 10,000 cells per well.
12.Centrifuge the plate for 3 min at 100 × g , room temperature.
13.Carefully remove plate from the centrifuge, check aggregates under a light microscope, and place the plate in an incubator at 37°C with 5% CO2 for 24 hr.
14.After 24 hr, check EBs under a light microscope. Carefully remove 50 µl of medium from the wells using a multichannel pipette and add 200 µl of fresh E8 Flex medium. Return to incubator at 37°C with 5% CO2 for an additional 24 hr.
15.After the 24 hr incubation, proceed to Basic Protocol 2.
Basic Protocol 2: DIFFERENTIATION OF EBs INTO HEART ORGANOIDS UNDER HEALTHY AND DIABETES-LIKE CONDITIONS
Here, the user will begin a time-specific (i.e., same time of day for every required medium change) differentiation protocol on the PSC-derived EBs generated in Basic Protocol 1, based on three Wnt pathway modulations. The basal medium of differentiation will differ across organoids, with half of the plate placed in healthy glucose and insulin levels and half in hyperglycemic and hyperinsulinemic levels, the latter to simulate diabetes conditions. Aside from the levels of glucose and insulin in the medium, all other conditions are identical, and the organoids are differentiated in the same manner at the same time, side by side. Care should be taken when performing medium changes on the developing organoids, as they are in suspension at the bottom of the well and very delicate and, therefore, sensitive to agitation. The user is advised to conduct the removal and addition of medium slowly on the side of the well, and to vary the angle of the multichannel pipette in accordance with the level of medium. See Figure 1 for a summary of the protocol steps.
Materials
-
Healthy differentiation medium (see recipe)
-
Diabetes differentiation medium (see recipe)
-
10 mM CHIR-99021 (Selleck, cat. no. 442310)
-
29.4 µM ( 1 µg/ml) Recombinant Human BMP-4 (Gibco, cat. no. PHC9534)
-
76.9 µM ( 1 µg/ml) Activin A Recombinant Protein (R&D Systems, cat. no. 338AC010)
-
Embryoid bodies generated in Basic Protocol 1
-
10 mM WNT-C59 (Selleck, cat. no. NC0710557)
-
Serological pipette (Fisher Scientific, cat. no. 14-955-233, 14-955-234)
-
Multichannel pipettes, 30-300 µl (Fisherbrand, cat. no. FBE1200300)
-
50-ml reagent reservoirs (Fisherbrand, cat. no. 13681502)
-
P-200, and P-200 micropipettes and corresponding tips
-
HERAcell VIOS 160i CO2 incubator (Thermo Fisher Scientific, cat. no. 51030285)
Cardiac mesoderm organoid differentiation
1.Prepare an aliquot of healthy differentiation medium and a separate aliquot of diabetes differentiation medium. Prepare 10 ml of each for a single 96-well plate. To each 10-ml aliquot, add 6 µl of 10 mM CHIR99021, 18.8 µl of 29.4 µM BMP4, and 15 µl of 76.9 µM Activin A, for a final medium concentration of 6 µM CHIR99021, 36 nM BMP4 (1.25 ng/ml), and 79.9 nM Activin A (1 ng/ml).
2.Take the 96-well plate containing the EBs (from Basic Protocol 1, step 14, containing 250 µl of medium per well) out of the incubator and remove 166 µl of medium (i.e., 2/3 of total volume) using a multichannel pipette from each well. To half of the plate (48 wells), add 166 µl of the healthy differentiation medium prepared in step 1 per well. To the other half, add 166 µl of the diabetes medium per well.
3.Incubate the plate at 37°C with 5% CO2 for exactly 24 hr.
4.On day 1 of differentiation, take the plate out of the incubator, remove 166 µl of medium from each well (i.e., 2/3 of the total volume) using a multichannel pipette, and add 166 µl of fresh healthy or diabetes differentiation medium to the corresponding wells, making sure to change tips between the healthy and diabetic condition. Return the plate to the incubator at 37°C with 5% CO2 for exactly 24 hr.
5.On day 2, prepare 10 ml of healthy differentiation medium and 10 ml of diabetes differentiation medium per 96-well plate, each with 3 µl of 10 mM WNT-C59, for a final medium concentration of 3 µM WNT-C59.Take the plate out of the incubator and remove 166 µl of medium from each well (i.e., 2/3 of the volume) using a multichannel pipette. Add 166 µl of fresh healthy or diabetes differentiation medium containing 3 µM of to the corresponding wells, resulting in a final concentration of 2 µM WNT-C59 in each well. Return the plate to the incubator at 37°C with 5% CO2 for exactly 48 hr.
6.On day 4, take the plate out of the incubator and remove 166 µl of medium from each well (i.e., 2/3 of the volume) using a multichannel pipette, and add 166 µl of fresh healthy or diabetes differentiation medium to the corresponding wells. Return the plate to the incubator at 37°C with 5% CO2 for exactly 48 hr.
Proepicardial induction and cardiac organoid maintenance
7.On differentiation day 6, remove 166 µl of medium from each well (i.e., 2/3 of the volume) using a multichannel pipette and add 166 µl of healthy or diabetes maintenance medium to the corresponding healthy and diabetic conditioned wells. Return to incubator at 37°C with 5% CO2 for exactly 24 hr.
8.On day 7, prepare 10 ml of healthy differentiation medium and 10 ml of diabetes differentiation medium per 96-well plate, each with 3 µl of 10 mM CHIR99021, for a final medium concentration of 3 µM CHIR99021.Take the plate out of the incubator and remove 166 µl of medium from each well (i.e., 2/3 of the volume) using a multichannel pipette. Add 166 µl of fresh healthy or diabetes maintenance medium containing 3 µM of CHIR99021 to the corresponding wells, resulting in a final concentration of 2 µM CHIR99021 in each well. Return to incubator at 37°C with 5% CO2 for 1 hr.
9.Take the plate out of the incubator and remove 166 µl of medium from each well (i.e., 2/3 of the total volume) using a multichannel pipette. Add 166 µl of fresh healthy or diabetes maintenance medium to the corresponding wells. Return the plate to the incubator at 37°C with 5% CO2.
10.Perform medium changes every 48 hr by removing 166 µl of medium from each well (i.e., 2/3 of the total volume) using a multichannel pipette and adding 166 µl of fresh healthy or diabetes maintenance medium to the corresponding wells. Proceed to downstream analyses (Basic Protocol 3) on differentiation day 15, or continue culturing for longer-term organoid maintenance.
Basic Protocol 3: IMMUNOFLUORESCENCE AND ORGANOID PREPARATION FOR OTHER ASSAYS
This protocol describes the steps for subjecting the healthy and diabetic organoids from Basic Protocol 2 to immunofluorescence using confocal microscopy. Immunofluorescence analysis can help to identify key cellular markers within the organoids and to investigate the effects of pregestational diabetes on structural composition and morphology in the developing human heart organoids, such as myocardial and epicardial tissue formation.
In this assay, the organoids are fixed using paraformaldehyde (PFA), and primary antibodies are used to detect tissue-specific proteins (e.g., TNNT2 for cardiomyocytes and WT1 for epicardial cells) within the organoids. Secondary antibodies, which are conjugates with fluorescent labels, are then used to visualize the location of the primary antibodies under confocal microscopy. This technique allows us to determine the tissue composition of the organoids and visualize the structural organization of major heart tissues in relation to one another. The epicardium, for example, plays a crucial role in the developmental stages of the heart, and gives rise to other cardiac cell types (J. Cao & Poss, 2018; Y. Cao, Duca, & J. Cao, 2020), while the myocardium makes up the muscle tissue and forms the heart chambers (Keung et al., 2019; Zhao et al., 2020). The formation of these heart tissues is critical for establishing a recapitulative human heart model in vitro. Visualizing the development of these tissues in the organoids using immunofluorescence can provide essential information on the health of the heart organoids in response to treatments or toxicity screening.
We also briefly comment on how to prepare the organoids for other assays not described here, including live assays or RNA-based assays such as RT-qPCR or RNA-seq. We describe how to transfer and prepare the organoids for these analyses.
Day 15 cardiac organoids (from Basic Protocol 2) are used here as an example for analyses; however, organoids can be analyzed at any time point of interest throughout the differentiation protocol, as well as after long-term culture beyond 15 days.
For assays that require transferring the organoids out of the culture plate to a secondary location, it is advised to follow this protocol with care, to avoid damaging the organoids in the transfer process.
The tools described here can be used to investigate the mechanisms underlying the effects of hyperglycemia and hyperinsulinemia on the developing human heart and explore potential therapeutic avenues in a high-throughput systematic screening manner.
Materials
-
Healthy and diabetic cardiac organoids generated in Basic Protocol 2
-
RNAprotect Tissue Reagent (Qiagen, cat. no. 76104)
-
Extraction kit (RNEasy Mini Kit from Qiagen)
-
Paraformaldehyde (MP Biomedicals, cat. no. IC15014601)
-
1× PBS (Gibco, cat. no. 10010049)
-
Glycine (Millipore Sigma, cat. no. 410225)
-
Immunofluorescence blocking solution (see recipe)
-
Immunofluorescence antibody solution (see recipe)
-
Anti-Cardiac Troponin T antibody (Abcam, cat. no. ab8295)
-
Recombinant Anti-Wilms Tumor Protein antibody (Abcam, cat. no. ab89901)
-
Donkey anti-Rabbit I Secondary Antibody, Alexa Fluor™ 488 (ThermoFisher Scientific, cat. no. A-21206)
-
Donkey anti-Mouse Secondary Antibody, Alexa Fluor™ 594 (ThermoFisher Scientific, cat. no. A-21203)
-
Polybead Microspheres, 90-µm (Polysciences, Inc., cat. no. 73155)
-
Clearing mounting medium (see recipe)
-
Top Coat Nail Varnish (transparent)
-
P-200 micropipette andP-200 pipette tips
-
1.5-ml microcentrifuge tubes (Fisher Scientific, cat. no. 02682002)
-
Thermoshaker (ThermoFisher Scientific, cat. no. 13-687-711PM)
-
Kimwipes (Kimberly-Clark Professional, cat. no. 06-666)
-
Microscope slides (Fisherbrand, cat. no. 1255015)
-
Micro cover glasses (VWR, cat. no. 48393-241)
-
Nikon Instruments A1 Confocal Laser Microscope (Nikon, cat. no. HD25)
Organoid collection and transfer
1.Cut off the pointed end of a p200 pipette tip about 10 mm from the bottom so that the opening of the tip is about 3 mm in diameter.
2.Remove the day 15 organoids plate (from Basic Protocol 2) from the incubator. Using the p200 pipette with a cut tip (step 1), slowly collect one organoid from a well by collecting 100-200 µl of medium with the organoid. To do this, hold the pipette straight up and then lower it to the bottom of the well as you slowly extract the medium along with the organoid.
3.Slowly dispense the collected organoid and medium into a 1.5-ml microcentrifuge tube. Collect the necessary number of organoids per condition into this same tube, but without exceeding 8 in a single tube; larger amounts of organoids per tube are not recommended for some downstream analyses. If performing immunofluorescence, proceed to step 6.
4.For live assays, transfer organoids in tube with medium to the secondary location for the live assays (e.g., calcium transient imaging, see note below), and analyze immediately.
5.For gene expression analyses, including RT-qPCR and RNA sequencing, remove all medium from the tube without disturbing the organoids, add 100 µl of RNAprotect Tissue Reagent, place at 4°C for 24 hr, and then transfer to −20°C for long-term storage. Proceed to RNA extraction using an extraction kit such as RNEasy Mini Kit from Qiagen, per the manufacturer's instructions. Subject the RNA to the analysis of interest, for instance, (see Drakhlis et al., 2021; Lewis-Israeli et al., 2021; Manivannan et al., 2022).
Whole-mount immunofluorescence analysis
Tubes should not contain more than 8 organoids per tube; higher numbers will reduce the effectiveness of the fixing process.
6.Remove as much medium from the tube of organoids as possible without disturbing the organoids at the bottom.
7.Prepare 4% (w/v) paraformaldehyde (PFA) in PBS and add 400 µl to each tube. Incubate for 30-45 min at room temperature.
8.Prepare a stock solution of 20 mM glycine in PBS by adding 150 mg of glycine to 100 ml of 1× PBS (store stock solution at 4°C for up to 1 month). Remove PFA without disturbing the organoids at the bottom of the tube, and wash three times with 400 µl PBS supplemented with 20 mM glycine, each time for 5 min.
9.Remove PBS-glycine from each tube without disturbing the organoids. Add 100 µl of immunofluorescence blocking solution and place on a thermoshaker at room temperature overnight, shaking at 300 × g.
10.Remove immunofluorescence blocking solution from each tube without disturbing the organoids. Wash once with 100 µl of PBS.
11.Prepare antibody solution (see Reagents and Solutions) with added primary antibodies according to the manufacturer's recommended antibody dilution [e.g., Anti-Cardiac Troponin T antibody (Abcam, ab8295) at a 1:200 dilution, and Recombinant Anti-Wilms Tumor Protein antibody (Abcam, ab89901) at a 1:100 dilution, diluted in immunofluorescence antibody solution]. Remove PBS from the tube without disturbing the organoids and add 100 µl of immunofluorescence antibody solution containing the desired primary antibodies. Place on a thermoshaker at 4°C set to 300 × g , for 24 hr.
12.Remove immunofluorescence antibody solution without disturbing the organoids, and wash with 100 µl PBS three times, for 5 min each. Keep the tubes on the thermoshaker at 300 × g during the washes.
13.Prepare antibody solution (see Reagents and Solutions) with secondary antibodies added according to the manufacturer's recommended antibody dilution [e.g., Donkey anti-Rabbit I Secondary Antibody, Alexa Fluor™ 488 (Thermo, A-21206) at a 1:200 dilution, and Donkey anti-Mouse Secondary Antibody, Alexa Fluor™ 594 (Thermo, A-21203) at a 1:200 dilution, diluted in immunofluorescence antibody solution]. Remove PBS from each tube without disturbing the organoids and add 100 µl immunofluorescence antibody solution containing the secondary antibodies corresponding to the primary antibodies used in the previous step. Place on a thermoshaker at 4°C set to 300 × g , for 24 hr. Cover thermoshaker with aluminum foil to provide a dark environment for the tubes.
14.Remove immunofluorescence antibody solution without disturbing the organoids, and wash with 100 µl PBS three times, each time for 5 min. Work with minimal light exposure, to minimize the effect on the fluorophores of the secondary antibodies.
15.Prepare slides by pipetting a drop of 90-µm Polybeads around the inside corners of where the cover glass will be. Dry the Polybeads using a Kimwipe. Place a drop of clearing mounting medium over the Polybeads so that they remain in place.
16.Using a cut p200 pipette tip (see step 1), transfer the stained organoids to the slide, positioning them between the Polybeads. Dry excess PBS around the organoids using the tip of a Kimwipe. Space out the organoids so that they do not touch each other and therefore interfere with the imaging process. Do not exceed 8 organoids per slide, for ease of imaging.
17.Place approximately 120 µl of clearing mounting medium on top of the organoids on the slide, ensuring that no bubbles are formed. Use a pipette tip to reposition organoids that may have moved when the mounting medium was added. Cover with a cover glass.
18.Seal the perimeter of the cover glass with transparent nail varnish, and store in the dark overnight at room temperature before imaging. Store in the dark at 4°C for long-term storage (up to 6 months).
19.Proceed to imaging using a confocal microscope (e.g., Nikon Instruments A1 Confocal Laser Microscope). Use a 10× objective for full organoid structure analysis, as shown in Figure 4, or a 60× objective for cellular-level imaging analysis, as previously described (Lewis-Israeli et al., 2021).
REAGENTS AND SOLUTIONS
Clearing mounting medium (Dekkers et al., 2019)
- Distilled H2O containing:
- 60% (v/v) glycerol (Millipore Sigma, cat. no. G5516)
- 2.5 M d-(−)-fructose (Millipore Sigma, cat. no. F0127)
- Store at 4°C for up to 3 months
Diabetes differentiation medium
- RPMI 1640 Medium, no glucose (Thermo Fisher Scientific, cat. no. 11879020)
- 2% (v/v) B-27™ Supplement, minus insulin (Thermo Fisher Scientific, cat. no. A1895601)
- 1% (v/v) penicillin-streptomycin (Gibco, cat. no. 15140122)
- 11.1 mM glucose (Thermo Fisher Scientific, cat. no. A2494001)
- Store at 4°C for up to 1 month
Diabetes maintenance medium
- RPMI 1640 Medium, no glucose (Thermo Fisher Scientific, cat. no. 11879020)
- 2% (v/v) B-27™ Supplement, minus insulin (Thermo Fisher Scientific, cat. no. A1895601)
- 1% (v/v) penicillin-streptomycin (Gibco, cat. no. 15140122)
- 5.5 mM glucose (Thermo Fisher Scientific, cat. no. A2494001)
- 1.14 nM insulin, human recombinant (Millipore Sigma, cat. no. 91077C)
- Store at 4°C for up to 1 month
Healthy differentiation medium
- RPMI 1640 Medium, no glucose (Thermo Fisher Scientific, cat. no. 11879020)
- 2% (v/v) B-27™ Supplement, minus insulin (Thermo Fisher Scientific, cat. no. A1895601)
- 1% (v/v) penicillin-streptomycin (Gibco, cat. no. 15140122)
- 5.5 mM glucose (Thermo Fisher Scientific, cat. no. A2494001)
- Store at 4°C for up to 1 month
Healthy maintenance medium
- RPMI 1640 Medium, no glucose (Thermo Fisher Scientific, cat. no. 11879020)
- 2% (v/v) B-27™ Supplement, minus insulin (Thermo Fisher Scientific, cat. no. A1895601)
- 1% (v/v) penicillin-streptomycin (Gibco, cat. no. 15140122)
- 5.5 mM glucose (Thermo Fisher Scientific, cat. no. A2494001)
- 170 pM insulin, human recombinant (Millipore Sigma, cat. no. 91077C)
- Store at 4°C for up to 1 month
Immunofluorescence antibody solution
- PBS (Gibco, cat. no. 10010049)
- 1% (v/v) donkey serum (Millipore Sigma, cat. no. S30-M)
- 0.5% (w/v) bovine serum albumin (Fisher Scientific, cat. no. 50253966)
- 0.5% (v/v) Triton™ X-100 (Millipore Sigma, cat. no. T8787)
- Store at 4°C for up to 1 month, or at −20°C for long-term storage (up to 1 year)
Immunofluorescence blocking solution
- PBS (Gibco, cat. no. 10010049)
- 10% (v/v) donkey serum (Millipore Sigma, cat. no. S30-M)
- 0.5% (w/v) bovine serum albumin (Fisher Scientific, cat. no. 50253966)
- 0.5% (v/v) Triton™ X-100 (Millipore Sigma, cat. no. T8787)
- Store at 4°C for up to 1 month, or at −20°C for long-term storage (up to 1 year)
COMMENTARY
Background Information
Congenital heart defects (CHDs) are the most common structural birth defect, occurring in 1%-5% of live births worldwide, making them the leading cause of death in the first year of life (Gilboa et al., 2016; Pierpont et al., 2007). The prevalence of CHDs has been rapidly increasing (Marelli et al., 2014), and it is estimated that ∼2.4 million Americans, including 1 million children, are living with a congenital malformation of the heart (Gilboa et al., 2016). The prevalence of diabetes mellitus (DM) around the world ranges from 2%-17% (Hunt & Schuller, 2007). This increasingly prevalent disease carries a high prenatal disease and mortality risk. Numerous retrospective case studies have reported a clear correlation between maternal DM during pregnancy and an increase in the risk of CHDs (Correa, 2016; Geurtsen et al., 2019; Øyen et al., 2016). In addition, in vivo studies in mice have investigated the symptoms and potential mechanisms in which pregestational diabetes affects fetal development. Many studies have demonstrated that maternal diabetes alters the expression of some genes involved in the development of the heart, including developmental morphogens and transcription factors, such as Bmp4, Msx1, Pax3 (Kumar, Dheen, & Tay, 2007), Gata4, Tbx5, Nkx2.5, Gata5, Bmp10, and Notch1 (Engineer et al., 2018), leading to congenital heart defects. These studies have also alluded to the involvement of increased oxidative stress during development (Engineer et al., 2018; Kumar et al., 2012; Moazzen et al., 2014). Importantly, in vitro studies using 2D monolayers of cardiomyocytes derived from human induced pluripotent stem cells (hiPSCs) have been used to investigate the effects of diabetic conditions in human cells (Drawnel et al., 2014; Geraets et al., 2018). In these reports, and to create a diabetogenic environment, hiPSC-derived cardiomyocytes were exposed to a high glucose (HG) environment. HG-treated cardiomyocytes were reported to be 5 times larger than normal glucose-treated cardiomyocytes. In line with the observed hypertrophic changes, high glucose treatment resulted in a significant increase in the expression of the cardiac hypertrophy markers ACTA1, FHL, and MLC2A (Ng et al., 2018).
Human pluripotent stem cells (hPSCs), including embryonic stem cells (hESCs) and induced PSCs (hiPSCs), have the capability to self-organize into three-dimensional (3D) structures called organoids, resembling embryo-like tissue patterns in vitro. Although organoid models have been described for a broad range of tissues, progress in the cardiovascular field has been limited. The field of engineered heart tissues has traditionally relied on direct assembly approaches, where different cardiac cell types (obtained from animal tissues or, more recently, differentiated from PSCs) are co-cultured into a 3D aggregate using various approaches such as layering, surrounding pillars, embedding in hydrogels, or seeding in patterned molds, to name a few (Lewis-Israeli et al., 2021). More recently, self-assembling heart organoid protocols have emerged. Self-assembling organoids better mimic the physiological steps of heart development and give rise to complex organoids with sophisticated internal organization. These protocols, however, are often derived from non-human stem cells (Andersen et al., 2018; Lee et al., 2020; Rossi et al., 2020), giving rise to non-cardiac cells such as those from the foregut (Drakhlis et al., 2021), or rely on multiple protocols for co-culture approaches (Hofbauer et al., 2021). Here, we describe a protocol to create sophisticated, self-assembling human PSC–derived heart organoids (hHOs) in a simple, three step Wnt pathway modulation workflow in a single protocol. In a previous study, we employed the protocol described here and demonstrated the capacity of hiPSC-derived cardiac organoids to model the effects of pregestational diabetes on the developing human heart, and we were also able to recapitulate several observable symptoms, including enlargement of tissue, disorganization of cell types, and arrythmias in electrophysiology recordings (Lewis-Israeli et al., 2021). These organoids were extensively characterized, and exhibited high tissue complexity, containing all major cardiac cell types, including cardiomyocytes, epicardial cells, endocardial cells, endothelial cells, and cardiac fibroblasts (Lewis-Israeli et al., 2021). We further demonstrated the similarity of the hHOs to human fetal heart tissue at the transcriptome level. The hHOs also contain complex internal structures, including chambers and a sophisticated vascular network. We also reported a high level of reproducibility and control over the differentiation of the hHOs. Lastly, using a transgenic cell line with a fluorescent marker for apoptosis, we observed the absence of a necrotic core, and using doxorubicin as a positive control, we showed apoptotic responses and demonstrated the capacity of this hHO platform to model cardiotoxicity (Lewis-Israeli et al., 2021).
This methodology was optimized to simplify the required steps and minimize the addition of complex growth factor mixtures. In contrast to previous differentiation protocols, the concentrations of glucose and insulin in the medium were modified to reflect healthy physiological concentrations in humans as well as physiologically relevant elevated glucose and insulin levels to model pregestational diabetes.
Human organoid cultures have a number of advantages over animal models: first and foremost, they constitute a human model, and are more readily accessible than mouse fetuses, as PSCs are easy to procure, can be passaged numerous times, and yield hundreds of organoids at a time. Further, organoids can be easily followed over time in vitro , and culture conditions can be tailored to specific purposes. They can also be produced in large quantities in a systematic manner for high-throughput analyses or high-content screenings (Kim, Koo, & Knoblich, 2020). Heart organoids have been used for a variety of studies, including drug screenings (Mills et al., 2019; Skardal, Shupe, & Atala, 2016), disease development (Lewis-Israeli et al., 2021; Miyamoto, Nam, Kannan, & Kwon, 2021), and toxicity research (Hoang et al., 2021; Richards et al., 2020). Indeed, PSC-derived heart organoids provide an ideal platform to study the cardiotoxicity of therapeutic agents. Toxicity studies have previously relied on animals, but differences in species morphology, anatomy, and function of various tissues have limited our understanding of the mechanisms of toxicity in humans (Van Norman, 2020). For example, recent studies explored the effects of cardiotoxins on heart organoids, studying the effects of relative toxicant concentrations on cellular damage (Richards et al., 2017), as well as the effects of the chemotherapy drug doxorubicin (Richards et al., 2020). These demonstrate the ability to investigate cardiotoxicity with respect to therapeutic aid effects and drug discovery in human tissue.
Some limitations of organoid work, particularly for longer-term cultures, include a microenvironment that is lacking some critical features (e.g., lack of blood circulation and absence of neighboring organs that might provide additional cues), particularly as the organoids age. Over time, this phenomenon leads to a general loss of complex developmental or physiological cues, which result in an increasing deviation from the physiological environment meant to be modeled, thus reducing the value of the organoid model to address the specific problem at hand. Additionally, protocols for organoid generation and quality control are not globally standardized, affecting reproducibility, and their production and maintenance are relatively expensive compared to traditional cell lines or animal models. Finally, studies of organoids become increasingly difficult owing to the growing cellular and structural complexity required, and in many instances necessitate new technology development (Kim et al., 2020; Ming et al., 2022).
The protocol described here paves the path to studying the etiology of congenital heart disease associated with pregestational diabetes, and allows access to stages of human fetal heart development that are otherwise inaccessible in research. It provides a relatively low-cost and high-throughput platform to investigate heart development and disease in the context of drug screening and toxicology studies.
Critical Parameters
Stem cell culture
Diligent stem cell culture technique is crucial for the success of the protocols listed here. Stem cells must remain pluripotent until the directed differentiation protocol begins in Basic Protocol 2. Successful maintenance of stem cells is line dependent. hPSCs should be cultured and passaged at least twice after thawing, before embryoid body generation.
The use of defined medium is crucial for the success of this protocol. PSCs cultured in serum-based media are susceptible to variability and may result in suboptimal organoids or unsuccessful differentiation of cardiac lineages.
Embryoid body generation
This protocol utilizes round-bottom ultra-low-attachment plates to help shape the embryoid bodies into spheres. This results in the 3D spherical embryoid bodies sitting at the bottom of the well, at the lowest point, but not being attached to the plate itself. The embryoid bodies are very delicate, especially on the day after aggregation, and can very easily come apart. For this reason, the very first medium change after aggregation (Basic Protocol 1, Cell aggregation into embryoid bodies, step 14) should be executed slowly and with care not to cause movement of the EBs, which may result in their breakage. Broken EBs may lead to unsuccessful differentiation or in multiple smaller organoids in the same well that will not be reproducible or representative of the rest of the plate. This is true for both medium removal and medium addition.
Organoid differentiation
For successful differentiation into functional cardiac organoids, it is recommended that all medium changes take place at the same time of day, within a 20-min window. This allows differentiation to occur as intended and minimizes variability between plates.
The differentiation and maintenance media should be warmed to 37°C prior to medium changes, to minimize any temperature shocks to the organoids during the protocol (this is especially important in the early days of differentiation).
For organoid cultures over 15 days, the user should note that a slight decrease in the size of the organoids after day 13 is to be expected, as the organoids become more compact. Organoids achieve human-fetal-like heart features and, therefore, studies into development and diseases of the adult/mature human heart may need to incorporate additional maturation steps that are outside the scope of this protocol, but that have been explored in recently published work (Ergir et al., 2022; Silva et al., 2021; Yang et al., 2019).
Organoid analysis and testing
When transferring organoids from the 96-well culture plate to a secondary location for live analysis (e.g., calcium imaging, life fluorescence), fixation, or molecular biology assays, we strongly recommend cutting the tip off a p200 tip to create a wide opening, to avoid any damage or stress to the organoids (see Fig. 1).
When fixing organoids using 4% PFA, we recommend a fixing time of 30-45 min, as shorter times would fail to properly fix the internal regions of the organoids, and longer fixing times may require an additional antigen retrieval step.
We also recommend not surpassing 8 organoids per tube for fixation and immunostaining, as we have experienced diminished immunofluorescence quality under these conditions.
Troubleshooting
Please see Table 1 for a list of common problems with the protocols, their causes, and potential solutions.
| Protocol | Problem | Possible cause | Solution |
|---|---|---|---|
| Basic Protocol 1 | Visible differentiation during stem cell culture prior to dissociation (steps 1-2) | Stress-inducing passage or thawing | Mark differentiated areas using an object marker under a microscope and scratch differentiated cells using a pipette tip before proceeding to another passage |
| Stem cell colonies are growing in 3D (yellowish center appearing on colonies) prior to dissociation (steps 1-2) | Colonies were too large or too confluent when last passaged | Passage cells at a lower confluency and ensure colonies are small when dissociated | |
| Stem cells are not dissociating to single cells with Accutase | Accutase not at room temperature, or is expired/inactive |
Make sure to allow enough time for Accutase to reach room temperature before proceeding with dissociation. Never place Accutase at temperatures above room temperature. Check expiration date. |
|
| EBs did not form the day after plate was centrifuged |
Medium was cold or centrifuge was not at room temperature TZV was not added |
Make sure that the cell culture medium was at room temperature or 37°C and contained TZV. Make sure that the centrifuge was set to room temperature, if it has temperature control. | |
| More than one EB has formed in a single well | Medium change after aggregation was too aggressive, resulting in the EBs splitting | Change the medium with extra care, gently removing and slowly adding the medium | |
| Basic Protocol 2 | Organoids are not beating by day 10 |
Unsuccessful differentiation due to the use of serum-based stem cell culture medium Unsuccessful differentiation due to nonadherence to protocol |
This protocol was optimized with PSCs grown in E8-based medium; less defined media will result in inferior differentiation results It is crucial to follow this protocol exactly as written, especially when it comes to concentrations and medium changing times |
| Organoids are broken, or have broken pieces | Organoid was in contact with pipette tip, or medium was changed too aggressively | The pipette tips should never be in contact with the organoid. Medium changes should be conducted with care, with minimal disturbance to the organoids. | |
| Basic Protocol 3 | Unable to collect organoid into the pipette tip | Users are being too gentle with the pipette suction, or organoids have been lost |
While it is important to collect the organoids with care, it is recommended to pipette up slightly fast when collecting organoids compared to regular medium changes Ensure that there is an organoid in the well; organoids should be visible to the naked eye when looking at the plate from the bottom |
| Organoids keep breaking during collection | The collection tip opening is not large enough, or organoids are extra delicate |
Make sure the tip is cut about 10 mm from the bottom so that the opening is 3 times larger than an organoid Depending on additional treatments to the organoids, they may become more delicate than usual, in which case the user should proceed with extra care or consider analyzing or fixing the organoids in the culture plate before transferring them (if possible) |
|
| No or little RNA was extracted from the organoids | Organoids were not pooled before extraction or were not submerged in RNAprotect reagent |
Make sure to pool at least 3-4 organoids for sufficient RNA Ensure that the organoids are completely submerged in the RNAprotect reagent, to preserve the RNA |
|
| Immunofluorescence images only stain the surface or the organoids | Immunofluorescence antibody solution was not well prepared or mixed |
It is crucial that the antibody solution be prepared according to the recipe provided and that the Triton X-100 detergent be at 0.5% (v/v) and mixed well so that the detergent is no longer visible and the solution in uniform and transparent If the problem persists, consider a longer incubation time with antibodies, and ensure that enough antibodies are used |
Understanding Results
The generation of functional human heart organoids under healthy and diabetic conditions, as described in this article, is a delicate process that requires some practice and knowledge of basic cell culture techniques. To achieve the desired results, the user must start off with a good culture of pluripotent stem cells, and follow the dissociation and aggregation steps described in the protocol. The top panel of Figure 1 illustrates the steps that should be followed to obtain acceptable embryoid bodies that are amenable to the differentiation protocol described in Basic Protocol 2. The differentiation protocol depicted by the center panel of Figure 1 demonstrates the steps for the generation of healthy and diabetic human heart organoids, which can be subsequently analyzed in the steps described in Basic Protocol 3 (Fig. 1, bottom panel).
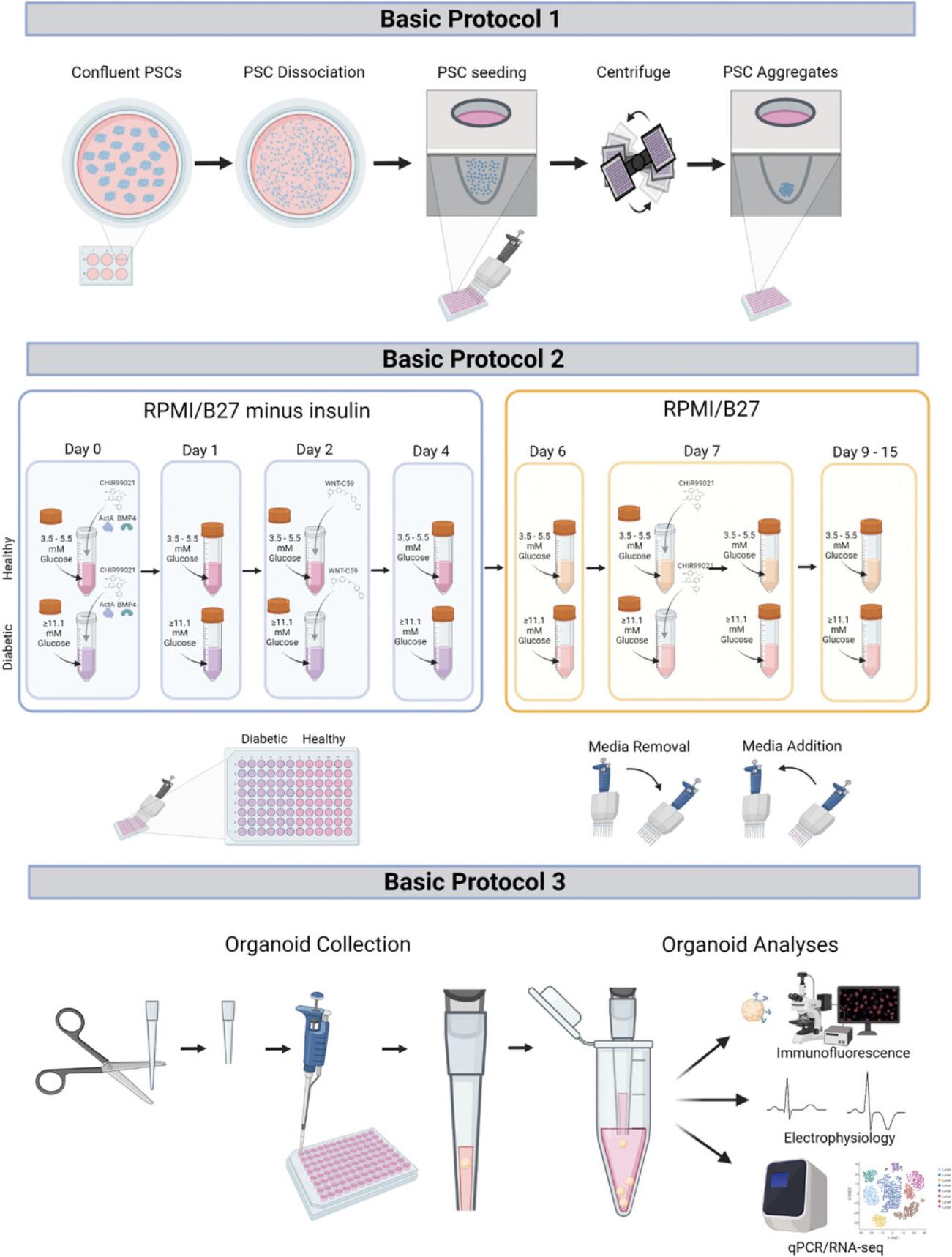
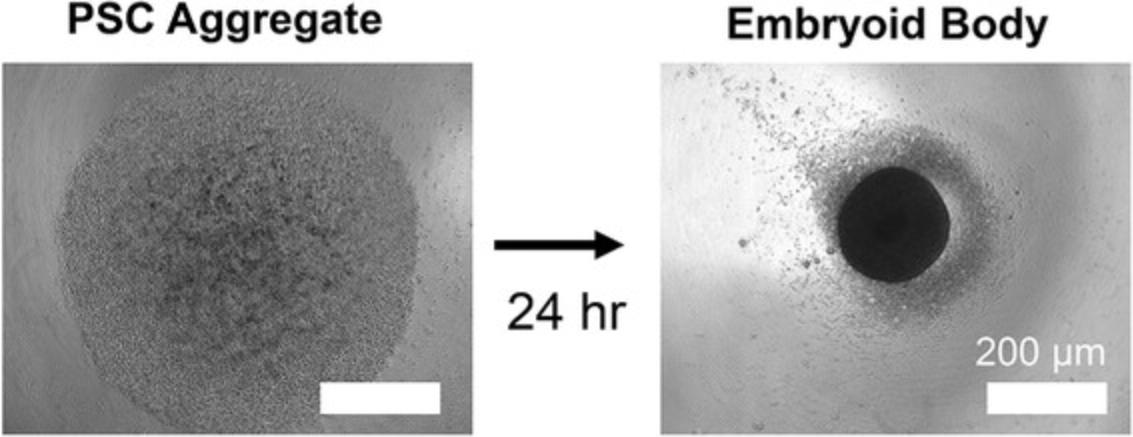
Figure 2 shows images under a light microscope of a PSC aggregate after centrifugation (left) and subsequent formation of EB (right) 24 hr later, illuminated from below. These should be used as a guide of what Basic Protocol 1 should yield, and if the results differ, the user should consult the Troubleshooting section for potential solutions. Once differentiation begins, the EBs will noticeably grow to be about 2-3 times their size in the first 4 days, and will maintain that size for the remainder of the protocol. At day 15, healthy organoids should be 600-800 µm in diameter, while diabetic organoids should be 800–1200 µm in diameter (Fig. 3). Organoids may begin beating as early as day 6, with all organoids expected to beat by day 10. Healthy organoids often begin to beat earlier, and diabetic organoids often beat more vigorously.
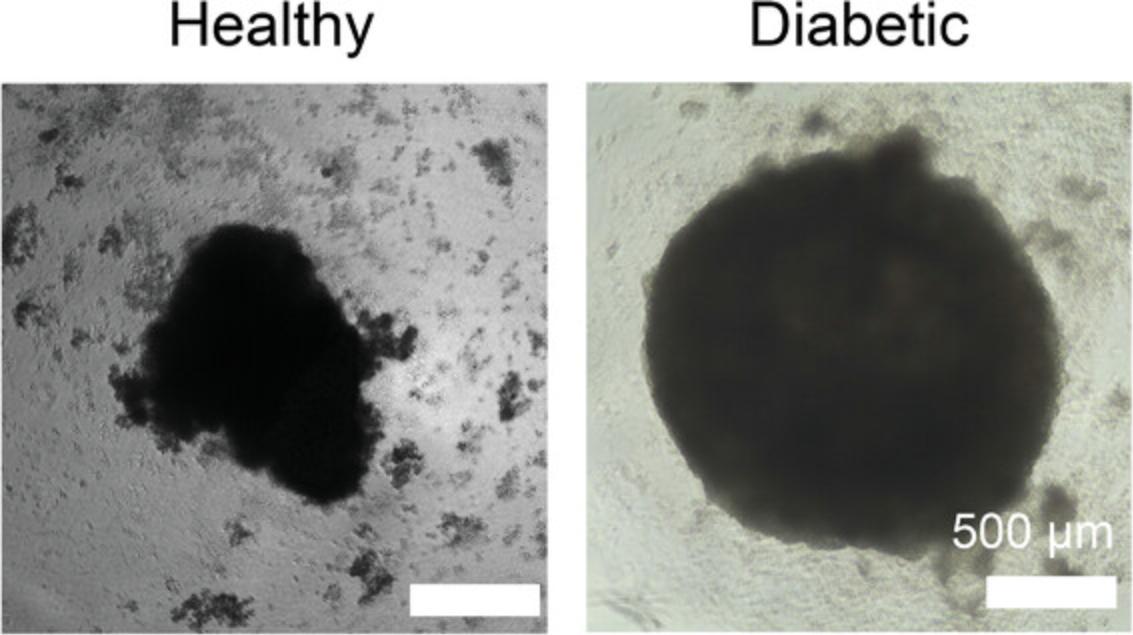
Immunofluorescence imaging should be clear throughout most of the organoid, and is limited by the depth penetration of confocal microscopy. For this reason, 90-µm Polybeads are used between the slide and the coverslip, so that the entire organoid can be imaged throughout without losing all the 3D structure this heart model provides. It is important to keep in mind that the organoids are still flattened on the slide and will, therefore, appear larger in a confocal image and will lose some 3D information (Fig. 4). Immunofluorescence imaging of cardiomyocyte marker TNNT2 and epicardial cell marker WT1 (shown in red and green, respectively, in Fig. 4) show the formation of myocardial and epicardial tissue, and allow observing the notable disparity in tissue organization between the healthy organoids (5.5 mM glucose) and diabetic organoids (11.1 mM glucose). These disparities include an enlarged myocardial area in the diabetic organoids, suggesting hypertrophy, and a disorganized and scattered epicardial tissue, suggesting morphological abnormalities under diabetes conditions.
Time Considerations
Allow at least two passages of PSCs after thawing, which can take 1-2 weeks depending on the cell line. Basic Protocol 1 can be initiated as soon as healthy and pluripotent stem cells reach the required confluency. The dissociation of the stem cells and subsequent aggregation should take about 1-2 hr, depending on user experience. EBs are ready for differentiation 2 days after aggregation. The differentiation protocol (Basic Protocol 2) is 15 days long, unless further culture is desired for other specific applications. Allow for at least 1 hr on every medium change day for medium preparation and slow and careful medium changes. Changing the medium on a single 96-well plate should take anywhere from 10 to 30 min, depending on user experience. Organoid analysis time varies depending on the assay. Immunofluorescence (Basic Protocol 3) takes 4 days, allowing a day for the fixing and blocking step, a day for primary antibodies, a day for secondary antibodies, and a final day for imaging. Analyzing the organoids for live imaging or for RNA analyses can take between 1 and 4 hr, depending on the user and the protocol used. Fixed organoids can be stored in PBS at 4°C for up to 2 weeks before starting immunofluorescence. Organoid transfer from culture to a microcentrifuge tube using a cut pipette tip can take between 5 and 20 min, depending on how many organoids are retrieved at any one time.
Acknowledgements
We wish to thank all members of the Aguirre Lab for their valuable comments and advice. Work in Dr. Aguirre's laboratory was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award numbers K01HL135464 and R01HL151505, by the American Heart Association under award number 19IPLOI34660342, and by the Spectrum-MSU Foundation.
Author Contributions
Yonatan R. Lewis-Israeli : conceptualization, data curation, formal analysis, investigation, methodology, resources, software, validation, visualization, writing original draft; Mishref Abdelhamid : data curation, formal analysis, investigation, methodology, resources, software, validation, visualization, writing original draft; Isoken Olomu : conceptualization, supervision, writing original draft, writing review and editing; Aitor Aguirre : conceptualization, funding acquisition, investigation, methodology, project administration, supervision, validation, writing original draft, writing review and editing.
Conflict of Interest
The authors have no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
Literature Cited
- Andersen, P., Tampakakis, E., Jimenez, D. V., Kannan, S., Miyamoto, M., Shin, H. K., … Kwon, C. (2018). Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nature Communications , 9(1), 1–13. doi: 10.1038/s41467-018-05604-8.
- Bao, X., Lian, X., Hacker, T. A., Schmuck, E. G., Qian, T., Bhute, V. J., … Palecek, S. P. (2017). Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nature Biomedical Engineering , 1(1), 1–12. doi: 10.1038/s41551-016-0003.
- Basu, M., & Garg, V. (2018). Maternal hyperglycemia and fetal cardiac development: Clinical impact and underlying mechanisms. Birth Defects Research , 110(20), 1504–1516. doi: 10.1002/bdr2.1435.
- Bredenoord, A. L., Clevers, H., & Knoblich, J. A. (2017). Human tissues in a dish: The research and ethical implications of organoid technology. Science , 355(6322), eaaf9414–eaaf9414. doi: 10.1126/science.aaf9414.
- Buchanan, T., & Kitzmiller, J. (1994). Metabolic interactions of diabetes and pregnancy. Annual Review of Medicine , 45, 245–260.
- Cao, J., & Poss, K. D. (2018). The epicardium as a hub for heart regeneration. Nature Reviews Cardiology , 15(10), 631–647. doi: 10.1038/s41569-018-0046-4.
- Cao, Y., Duca, S., & Cao, J. (2020). Epicardium in heart development. Cold Spring Harbor Perspectives in Biology , 12(2), 1–18. doi: 10.1101/cshperspect.a037192.
- Centers for Disease Control and Prevention. (2020). National Diabetes Statistics Report. Retrieved from https://www.cdc.gov/diabetes/data/statistics-report/index.html.
- Chen, G., Gulbranson, D. R., Hou, Z., Bolin, J. M., Ruotti, V., Probasco, M. D., … Thomson, J. A. (2011). Chemically defined conditions for human iPSC derivation and culture. Nature Methods , 8(5), 424–429. doi: 10.1038/nmeth.1593.
- Correa, A. (2016). Pregestational diabetes mellitus and congenital heart defects. Circulation , 133(23), 2219–2221. doi: 10.1161/CIRCULATIONAHA.116.022960.
- Dekkers, J. F., Alieva, M., Wellens, L. M., Ariese, H. C. R., Jamieson, P. R., Vonk, A. M., … Rios, A. C. (2019). High-resolution 3D imaging of fixed and cleared organoids. Nature Protocols , 14(6), 1756–1771. doi: 10.1038/s41596-019-0160-8.
- Drakhlis, L., Biswanath, S., Farr, C.-M., Lupanow, V., Teske, J., Ritzenhoff, K., … Zweigerdt, R. (2021). Human heart-forming organoids recapitulate early heart and foregut development. Nature Biotechnology , 39(6), 737–746. doi: 10.1038/s41587-021-00815-9.
- Drawnel, F. M. M., Boccardo, S., Prummer, M., Delobel, F., Graff, A., Weber, M., … Iacone, R. (2014). Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Reports , 9(3), 810–820. doi: 10.1016/j.celrep.2014.09.055.
- Engineer, A., Saiyin, T., Lu, X., Kucey, A. S., Urquhart, B. L., Drysdale, T. A., … Feng, Q. (2018). Sapropterin treatment prevents congenital heart defects induced by pregestational diabetes mellitus in mice. Journal of the American Heart Association , 7(21), e009624. doi: 10.1161/JAHA.118.009624.
- Ergir, E., La Cruz, J. O.-D., Fernandes, S., Cassani, M., Niro, F., Sousa, D., … Forte, G. (2022). Generation and maturation of human iPSC-derived cardiac organoids in long term culture [Preprint]. bioRxiv. doi: 10.1101/2022.03.07.483273.
- Geraets, I. M. E., Chanda, D., van Tienen, F. H. J., van den Wijngaard, A., Kamps, R., Neumann, D., … Nabben, M. (2018). Human embryonic stem cell-derived cardiomyocytes as an in vitro model to study cardiac insulin resistance. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease , 1864(5), 1960–1967. doi: 10.1016/j.bbadis.2017.12.025.
- Geurtsen, M. L., van Soest, E. E. L. L., Voerman, E., Steegers, E. A. P. P., Jaddoe, V. W. V. V., & Gaillard, R. (2019). High maternal early-pregnancy blood glucose levels are associated with altered fetal growth and increased risk of adverse birth outcomes. Diabetologia , 62(10), 1880–1890. doi: 10.1007/s00125-019-4957-3.
- Gilboa, S. M., Devine, O. J., Kucik, J. E., Oster, M. E., Riehle-Colarusso, T., Nembhard, W. N., … Marelli, A. J. (2016). Congenital heart defects in the United States. Circulation , 134(2), 101–109. doi: 10.1161/CIRCULATIONAHA.115.019307.
- Gupta, A., Chacko, V. P., Schär, M., Akki, A., & Weiss, R. G. (2011). Impaired ATP kinetics in failing in vivo mouse heart. Circulation: Cardiovascular Imaging , 4(1), 42–50. doi: 10.1161/CIRCIMAGING.110.959320.
- Hamlin, R. L., & Altschuld, R. A. (2011). Extrapolation from mouse to man. Circulation: Cardiovascular Imaging , 4(1), 2–4. doi: 10.1161/CIRCIMAGING.110.961979.
- Hoang, P., Kowalczewski, A., Sun, S., Winston, T. S., Archilla, A. M., Lemus, S. M., … Ma, Z. (2021). Engineering spatial-organized cardiac organoids for developmental toxicity testing. Stem Cell Reports , 16(5), 1228–1244. doi: 10.1016/j.stemcr.2021.03.013.
- Hofbauer, P., Jahnel, S. M., Papai, N., Giesshammer, M., Deyett, A., Schmidt, C., … Mendjan, S. (2021). Cardioids reveal self-organizing principles of human cardiogenesis. Cell , 184(12), 3299–3317.e22. doi: 10.1016/j.cell.2021.04.034.
- Hunt, K. J., & Schuller, K. L. (2007). The increasing prevalence of diabetes in pregnancy. Obstetrics and Gynecology Clinics of North America , 34(2), 173–199. doi: 10.1016/j.ogc.2007.03.002.
- Keung, W., Chan, P. K. W., Backeris, P. C., Lee, E. K., Wong, N., Wong, A. O. T., … Li, R. A. (2019). Human cardiac ventricular-like organoid chambers and tissue strips from pluripotent stem cells as a two-tiered assay for inotropic responses. Clinical Pharmacology and Therapeutics , 106(2), 402–414. doi: 10.1002/cpt.1385.
- Kim, J., Koo, B.-K., & Knoblich, J. A. (2020). Human organoids: Model systems for human biology and medicine. Nature Reviews Molecular Cell Biology , 21, 571–584. doi: 10.1038/s41580-020-0259-3.
- Kretzschmar, K., & Clevers, H. (2016). Organoids: modeling development and the stem cell niche in a dish. Developmental Cell , 38(6), 590–600. doi: 10.1016/j.devcel.2016.08.014.
- Kumar, S. D., Dheen, S. T., & Tay, S. S. W. (2007). Maternal diabetes induces congenital heart defects in mice by altering the expression of genes involved in cardiovascular development. Cardiovascular Diabetology , 6(1), 34–34. doi: 10.1186/1475-2840-6-34.
- Kumar, S. D., Vijaya, M., Samy, R. P., Dheen, S. T., Ren, M., Watt, F., … Tay, S. S. W. (2012). Zinc supplementation prevents cardiomyocyte apoptosis and congenital heart defects in embryos of diabetic mice. Free Radical Biology and Medicine , 53(8), 1595–1606. doi: 10.1016/j.freeradbiomed.2012.07.008.
- Lee, J., Sutani, A., Kaneko, R., Takeuchi, J., Sasano, T., Kohda, T., … Ishino, F. (2020). In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nature Communications , 11(1), 4283–4283. doi: 10.1038/s41467-020-18031-5.
- Lehtoranta, L., Koskinen, A., Vuolteenaho, O., Laine, J., Kytö, V., Soukka, H., … Räsänen, J. (2017). Gestational hyperglycemia reprograms cardiac gene expression in rat offspring. Pediatric Research , 82(2), 356–361. doi: 10.1038/pr.2017.42.
- Lehtoranta, L., Vuolteenaho, O., Laine, V. J., Koskinen, A., Soukka, H., Kytö, V., … Räsänen, J. (2013). Maternal hyperglycemia leads to fetal cardiac hyperplasia and dysfunction in a rat model. American Journal of Physiology—Endocrinology and Metabolism , 305(5), E611–E619. doi: 10.1152/ajpendo.00043.2013.
- Lewis-Israeli, Y. R., Volmert, B. D., Gabalski, M. A., Huang, A. R., & Aguirre, A. (2021). Generating self-assembling human heart organoids derived from pluripotent stem cells. Journal of Visualized Experiments , 175, 10.3791/63097. doi: 10.3791/63097.
- Lewis-Israeli, Y. R., Wasserman, A. H., & Aguirre, A. (2021). Heart organoids and engineered heart tissues: Novel tools for modeling human cardiac biology and disease. Biomolecules , 11(9), 1277–1277. doi: 10.3390/biom11091277.
- Lewis-Israeli, Y. R., Wasserman, A. H., Gabalski, M. A., Volmert, B. D., Ming, Y., Ball, K. A., … Aguirre, A. (2021). Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nature Communications , 12(1), 5142–5142. doi: 10.1038/s41467-021-25329-5.
- Lian, X., Hsiao, C., Wilson, G., Zhu, K., Hazeltine, L. B., Azarin, S. M., … Palecek, S. P. (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America , 109(27), E1848–E1857. doi: 10.1073/pnas.1200250109.
- Loiselle, D. S., & Gibbs, C. L. (1979). Species differences in cardiac energetics. American Journal of Physiology—Heart and Circulatory Physiology , 237(1), H90–H98. doi: 10.1152/ajpheart.1979.237.1.H90.
- Malik, N., & Rao, M. S. (2013). A review of the methods for human iPSC derivation. Methods in Molecular Biology , 997, 23–33. doi: 10.1007/978-1-62703-348-0_3.
- Manivannan, S., Mansfield, C., Zhang, X., Kodigepalli, K. M., Majumdar, U., Garg, V., & Basu, M. (2022). Single-cell transcriptomic profiling unveils cardiac cell-type specific response to maternal hyperglycemia underlying the risk of congenital heart defects. BioRxiv. doi: 10.1101/2021.05.28.446177.
- Marelli, A. J., Ionescu-Ittu, R., Mackie, A. S., Guo, L., Dendukuri, N., & Kaouache, M. (2014). Lifetime prevalence of congenital heart disease in the general population from 2000 to 2010. Circulation , 130(9), 749–756. doi: 10.1161/CIRCULATIONAHA.113.008396.
- Mills, R. J., Parker, B. L., Quaife-Ryan, G. A., Voges, H. K., Needham, E. J., Bornot, A., … Hudson, J. E. (2019). Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell , 24(6), 895–907. doi: 10.1016/j.stem.2019.03.009.
- Ming, Y., Hao, S., Wang, F., Lewis-Israeli, Y. R., Volmert, B. D., Xu, Z., … Zhou, C. (2022). Longitudinal morphological and functional characterization of human heart organoids using optical coherence tomography. Biosensors and Bioelectronics , 207, 114136. doi: 10.1016/j.bios.2022.114136.
- Miyamoto, M., Nam, L., Kannan, S., & Kwon, C. (2021). Heart organoids and tissue models for modeling development and disease. Seminars in Cell & Developmental Biology, 118, 119–128. doi: 10.1016/j.semcdb.2021.03.011.
- Moazzen, H., Lu, X., Ma, N. L., Velenosi, T. J., Urquhart, B. L., Wisse, L. J., … Feng, Q. (2014). N-Acetylcysteine prevents congenital heart defects induced by pregestational diabetes. Cardiovascular Diabetology , 13, 46. doi: 10.1186/1475-2840-13-46.
- Ng, K.-M., Lau, Y.-M., Dhandhania, V., Cai, Z.-J., Lee, Y.-K., Lai, W.-H., … Siu, C.-W. (2018). Empagliflozin ammeliorates high glucose induced-cardiac dysfuntion in human iPSC-derived cardiomyocytes. Scientific Reports , 8(1), 14872–14872. doi: 10.1038/s41598-018-33293-2.
- Ornoy, A., Reece, E. A., Pavlinkova, G., Kappen, C., & Miller, R. K. (2015). Effect of maternal diabetes on the embryo, fetus, and children: Congenital anomalies, genetic and epigenetic changes and developmental outcomes. Birth Defects Research Part C: Embryo Today: Reviews , 105(1), 53–72. doi: 10.1002/bdrc.21090.
- Øyen, N., Diaz, L. J., Leirgul, E., Boyd, H. A., Priest, J., Mathiesen, E. R., … Melbye, M. (2016). Prepregnancy diabetes and offspring risk of congenital heart disease: A nationwide cohort study. Circulation , 133(23), 2243–2253. doi: 10.1161/CIRCULATIONAHA.115.017465.
- Panopoulos, A. D., D'Antonio, M., Benaglio, P., Williams, R., Hashem, S. I., Schuldt, B. M., … Frazer, K. A. (2017). IPSCORE: A resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports , 8(4), 1086–1100. doi: 10.1016/j.stemcr.2017.03.012.
- Paşca, S. P. (2019). Assembling human brain organoids. Science , 363(6423), 126–127. doi: 10.1126/science.aau5729.
- Piddington, R., Joyce, J., Dhanasekaran, P., & Baker, L. (1996). Diabetes mellitus affects prostaglandin E2 levels in mouse embryos during neurulation. Diabetologia , 39(8), 915–920. doi: 10.1007/BF00403910.
- Pierpont, M. E., Basson, C. T., Benson, D. W., Gelb, B. D., Giglia, T. M., Goldmuntz, E., … Webb, C. L. (2007). Genetic basis for congenital heart defects: Current knowledge. Circulation , 115(23), 3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056.
- Richards, D. J., Coyle, R. C., Tan, Y., Jia, J., Wong, K., Toomer, K., … Mei, Y. (2017). Inspiration from heart development: Biomimetic development of functional human cardiac organoids. Biomaterials , 142, 112–123. doi: 10.1016/j.biomaterials.2017.07.021.
- Richards, D. J., Li, Y., Kerr, C. M., Yao, J., Beeson, G. C., Coyle, R. C., … Mei, Y. (2020). Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nature Biomedical Engineering , 4(4), 446–462. doi: 10.1038/s41551-020-0539-4.
- Rossi, G., Broguiere, N., Miyamoto, M., Boni, A., Guiet, R., Girgin, M., … Lutolf, M. P. (2020). Capturing cardiogenesis in gastruloids. Cell Stem Cell , 28(2), 1–11. doi: 10.1016/j.stem.2020.10.013.
- Sasai, Y. (2013). Next-generation regenerative medicine: Organogenesis from stem cells in 3D culture. Cell Stem Cell , 12(5), 520–530. doi: 10.1016/j.stem.2013.04.009.
- Silva, A. C., Matthys, O. B., Joy, D. A., Kauss, M. A., Natarajan, V., Lai, M. H., … McDevitt, T. C. (2021). Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell , 28(12), 2137–2152.e6. doi: 10.1016/j.stem.2021.11.007.
- Skardal, A., Shupe, T., & Atala, A. (2016). Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discovery Today , 21(9), 1399–1411. doi: 10.1016/j.drudis.2016.07.003.
- Strober, W. (2015). Trypan blue exclusion test of cell viability. Current Protocols in Immunology , 111, A.3.1–A.3.5. doi: 10.1002/0471142735.ima03bs111.
- Tabib, A., Shirzad, N., Sheikhbahaei, S., Mohammadi, S., Qorbani, M., Haghpanah, V., … Baghaei-Tehrani, R. (2013). Cardiac malformations in fetuses of gestational and pre gestational diabetic mothers. Iranian Journal of Pediatrics , 23(6), 664–668.
- Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell , 126(4), 663–676. doi: 10.1016/j.cell.2006.07.024.
- Tuveson, D., & Clevers, H. (2019). Cancer modeling meets human organoid technology. Science , 364(6444), 952–955.
- Van Norman, G. A. (2020). Limitations of animal studies for predicting toxicity in clinical trials. JACC: Basic to Translational Science , 5(4), 387–397. doi: 10.1016/j.jacbts.2020.03.010.
- Weeber, F., Ooft, S. N., Dijkstra, K. K., & Voest, E. E. (2017). Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chemical Biology , 24(9), 1092–1100. doi: 10.1016/J.CHEMBIOL.2017.06.012.
- Yang, X., Rodriguez, M. L., Leonard, A., Sun, L., Fischer, K. A., Wang, Y., … Murry, C. E. (2019). Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports , 13(4), 657–668. doi: 10.1016/j.stemcr.2019.08.013.
- Zaidi, S., & Brueckner, M. (2017). Genetics and genomics of congenital heart disease. Circulation Research , 120(6), 923–940. doi: 10.1161/CIRCRESAHA.116.309140.
- Zhao, Y., Rafatian, N., Wang, E. Y., Wu, Q., Lai, B. F. L., Lu, R. X., … Radisic, M. (2020). Towards chamber specific heart-on-a-chip for drug testing applications. Advanced Drug Delivery Reviews , 165-166, 60–76. doi: 10.1016/j.addr.2019.12.002.
Citing Literature
Number of times cited according to CrossRef: 3
- Dosh Whye, Delaney Wood, Wardiya Afshar Saber, Erika M. Norabuena, Nina R. Makhortova, Mustafa Sahin, Elizabeth D. Buttermore, A Robust Pipeline for the Multi‐Stage Accelerated Differentiation of Functional 3D Cortical Organoids from Human Pluripotent Stem Cells, Current Protocols, 10.1002/cpz1.641, 3 , 1, (2023).
- Yingjuan Liu, Honglin Xu, Sabu Abraham, Xin Wang, Bernard D. Keavney, Progress of 3D Organoid Technology for Preclinical Investigations: Towards Human In Vitro Models, International Journal of Drug Discovery and Pharmacology, 10.53941/ijddp.v1i1.188, (9), (2022).
- Dosh Whye, Delaney Wood, Kristina H. Kim, Cidi Chen, Nina Makhortova, Mustafa Sahin, Elizabeth D. Buttermore, Dynamic 3D Combinatorial Generation of hPSC‐Derived Neuromesodermal Organoids With Diverse Regional and Cellular Identities, Current Protocols, 10.1002/cpz1.568, 2 , 10, (2022).

