Enhancing iPSC-CM Maturation Using a Matrigel-Coated Micropatterned PDMS Substrate
Eric N. Jimenez-Vazquez, Eric N. Jimenez-Vazquez, Abhilasha Jain, Abhilasha Jain, David K. Jones, David K. Jones
Abstract
Cardiac myocytes isolated from adult heart tissue have a rod-like shape with highly organized intracellular structures. Cardiomyocytes derived from human pluripotent stem cells (iPSC-CMs), on the other hand, exhibit disorganized structure and contractile mechanics, reflecting their pronounced immaturity. These characteristics hamper research using iPSC-CMs. The protocol described here enhances iPSC-CM maturity and function by controlling the cellular shape and environment of the cultured cells. Microstructured silicone membranes function as a cell culture substrate that promotes cellular alignment. iPSC-CMs cultured on micropatterned membranes display an in-vivo-like rod-shaped morphology. This physiological cellular morphology along with the soft biocompatible silicone substrate, which has similar stiffness to the native cardiac matrix, promotes maturation of contractile function, calcium handling, and electrophysiology. Incorporating this technique for enhanced iPSC-CM maturation will help bridge the gap between animal models and clinical care, and ultimately improve personalized medicine for cardiovascular diseases. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Cardiomyocyte differentiation of iPSCs
Basic Protocol 2 : Purification of differentiated iPSC-CMs using MACS negative selection
Basic Protocol 3 : Micropatterning on PDMS
INTRODUCTION
Animal models have aided in the understanding of a variety of cardiovascular diseases and, consequently, in the development of numerous cardiovascular therapies (Chorro, Such-Belenguer, & Lopez-Merino, 2009; Oh, Kho, Hajjar, & Ishikawa, 2019). However, animal models are not free of limitations. For example, the ion channel kinetics and functional effects of drug compounds in animals can vary widely from those observed in human pathophysiology.
Human induced pluripotent stem (iPS) cells circumvent the issues associated with interspecies comparisons. And because they also retain the genetic background of the individual from whom they are derived, human iPS cells have ushered in a new era of patient-specific research (Takahashi & Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). A comprehensive understanding of the human cellular phenotype remains elusive, largely because of a lack of adequate human cellular models. As a result, there is a great deal of interest in using iPSC-CMs to study the effects of mutations and their potential treatments in a patient-specific manner, as well as to supplement existing studies.
iPSC-derived cardiomyocytes have the potential to be excellent in vitro models of cardiac health and disease, but they differ from mature cardiomyocytes. They exhibit, for example, poorly organized excitation-contraction coupling machinery, they lack clear t-tubules, and they produce immature mitochondria (Gaspar et al., 2014; Li, Chen, & Li, 2013; Lieu et al., 2009; Rana, Anson, Engle, & Will, 2012). They also generally have a flat, circular shape along with isotropic filament organization, disorganized contractility patterns, and low expression of cytoskeletal proteins and ion channels (Ribeiro et al., 2015; van den Berg et al., 2015; Yang, Pabon, & Murry, 2014).
In 2016 it was demonstrated that culturing iPSC-CMs monolayers on soft polydimethylsiloxane (PDMS) membranes coated with Matrigel promotes their maturation, resulting in cardiomyocyte hypertrophy and expression of key mature sarcolemma (SCN5A, Kir2.1, and Cx43) and myofilament markers (cardiac troponin I), matured action potential profiles, and faster conduction velocities (Herron et al., 2016). These PDMS-cultured cells still displayed many immature morphological characteristics. This is important because cell shape is known to have important effects on cardiomyocyte properties including cytoskeletal structure, growth, and differentiation (Chen, Mrksich, Huang, Whitesides, & Ingber, 1997, 1998; Dike et al., 1999). Cell shape is also critical in determining contractile performance of single iPSC-cardiomyocytes and neonatal cardiomyocytes by regulating intracellular structure, increasing contractile activity, and promoting a more mature electrophysiology phenotype (faster upstroke and conduction velocities; Helms et al., 2020; Jimenez-Vazquez et al., 2022; Kuo et al., 2012; Ribeiro et al., 2015; Tsan et al., 2021).
A certain degree of iPSC-CM maturation is required for effective disease modeling or for regenerative purposes. iPSC-CMs with characteristics that closely resemble those of adult cardiomyocytes are a more effective model of a patient's disease. Matured human iPSC-CMs more closely reproduce the electrophysiological and pharmacological properties of both healthy and diseased adult cardiomyocytes (Davis et al., 2012; Itzhaki et al., 2011; Ma et al., 2011; Novak et al., 2012; Sun et al., 2012). The following protocols describe the generation of matured iPSC-CMs and their subsequent seeding on a Matrigel-coated micropatterned PDMS membrane. These methods generate a large number (∼8640 cells/stamp) of thick, cylindrical-shaped cardiomyocytes with well-defined sarcomeres. These rod-shaped iPSC-CMs can be used in a variety of techniques, including immunostaining, stretch assays, optical mapping, western blotting, and patch clamping. Furthermore, they can be adapted to create a variety of cell patterns on either hard or elastic membranes.
Basic Protocol 1: CARDIOMYOCYTE DIFFERENTIATION FROM iPSCs
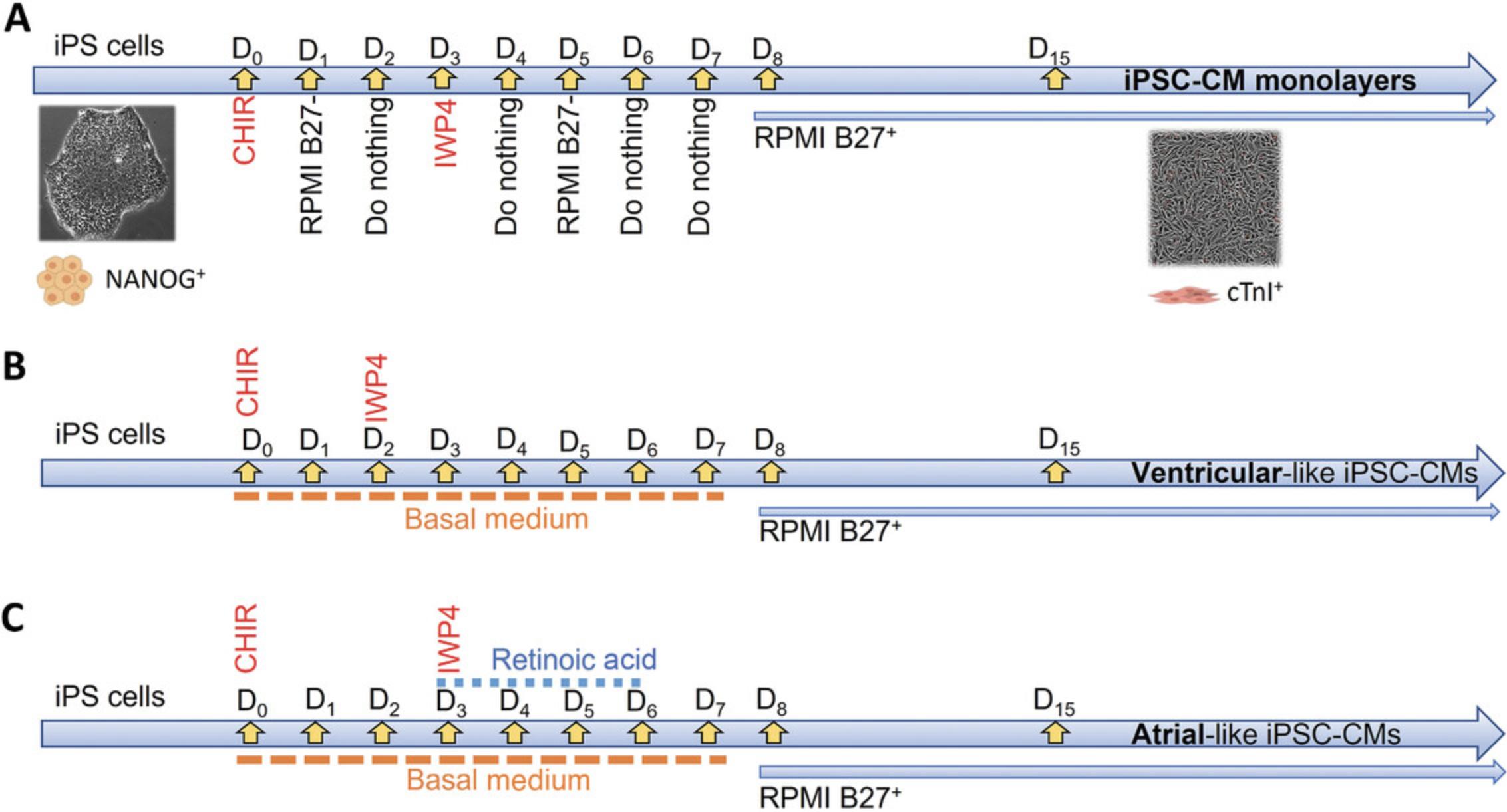
Years ago, Lian and colleagues developed a protocol for differentiating iPS cells into cardiomyocytes by modulating canonical Wnt signaling pathways with small molecules (Lian et al., 2013). Here, we have adapted that protocol from Lian et al. (2013) and Herron et al. (2016) to obtain differentiated cardiomyocytes from iPS cells (Fig. 1). The following protocol yields mostly ventricular-like cardiomyocytes, with a small proportion of atrial and nodal-like cells as well (Fig. 1A). Minor variations in the protocol can allow differentiation of only ventricular-like or atrial-like cardiomyocytes (Fig. 1B and 1C).
NOTE : Perform all cell culture manipulations in a laminar flow hood.
Materials
- iPS cell lines
- Matrigel (cat. no. 345277, Corning)
- StemMACS iPS-Brew XF (with supplements; cat. no. 130-107-086, Miltenyi Biotec)
- Versene solution (cat. no. 15040066, Gibco)
- ROCK inhibitor (cat. no. Y-27632, Gibco)
- Hank's Balanced Salt Solution with Ca2+ and Mg2+ added (HBSS++; cat. no. 14025-092, Gibco)
- RPMI medium (with ʟ-glutamine; cat. no. 10-040-CV, Corning)
- B27 supplement minus insulin (cat. no. A1895602, Invitrogen)
- CHIR99021 (cat. no. C-6556, LC labs)
- IWP-4 (cat. no. 04-0036, Reprocell)
- B27 supplement with insulin (B27+; cat. no. 17504004, Invitrogen)
- Laminar flow hood
- 37°C, 5% CO2 incubator
1.Seed ∼5 million iPS cells into a Matrigel-coated six-well plate in 2 ml of StemMACS iPSC Brew XF medium per well. Culture in a 37°C, 5% CO2 incubator for 5-6 days, until confluent.
2.Aspirate the medium and then dissociate iPS cells using 1 ml/well of Versene solution at 37°C for 7 min. Reseed as monolayers on Matrigel-coated (100 μg/ml) 12-well plates at a density of 8.5 × 106 cells/well in 2 ml StemMACS iPSC Brew XF medium supplemented with 5 μmol/L ROCK inhibitor.
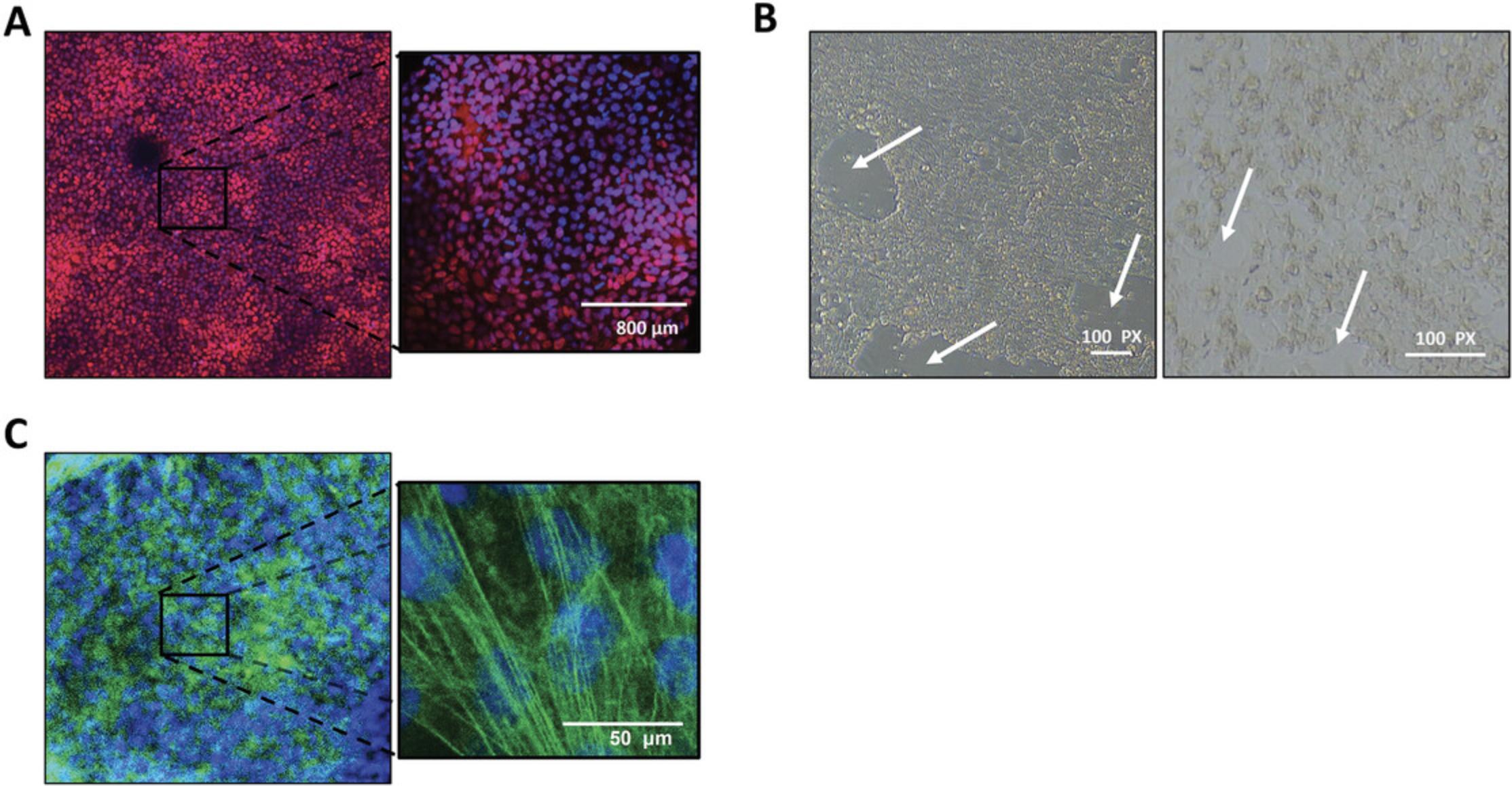
3.After 2 days, or when the monolayers reach 90%-100% confluence (Fig. 2A), remove medium, wash the stem cells with 1 ml/well HBSS++, and then replace with 3 ml RPMI supplemented with B27 minus insulin (RPMI/B27–) containing 10 μmol/L CHIR99021.
4.Day 1: Remove CHIR99021 by replacing the medium with 2 ml RPMI/B27–.
5.Day 2: Do nothing.
6.Day 3: Remove 1.5 ml medium from each well and add 1.5 ml RPMI/B27–. Add 1 μl/ml IWP4 (10 μmol/L); you will have a total volume of 3 ml per well.
7.Day 4: Do nothing.
8.Day 5: Aspirate the RPMI/IWP4 solution and add 2 ml RPMI/B27– per well.
9.Day 6: Do nothing.
10.Day 7: Aspirate the medium and add 2 ml of RPMI supplemented with B27 containing insulin (RPMI/B27+) per well.
11.From day 8 onward, change the medium every day until the cells are ready for purification.
Basic Protocol 2: PURIFICATION OF DIFFERENTIATED hiPSC-CMs USING MACS NEGATIVE SELECTION
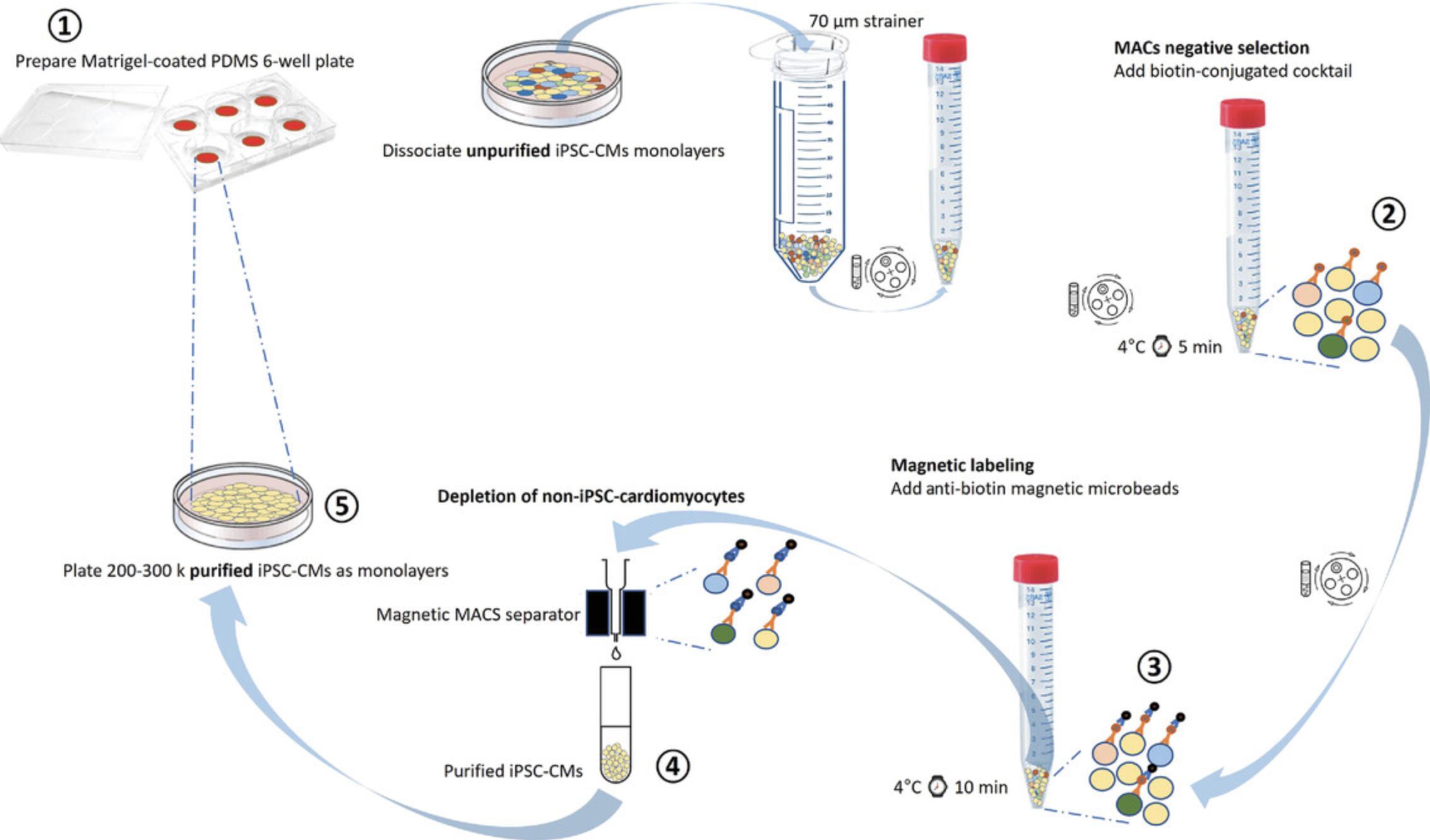
The directed differentiation method used here (adapted from Herron, Monteiro da Rocha, & Campbell, 2017, and Pekkanen-Mattila et al., 2019) should generate a cell population that is 20%-80% cardiomyocytes. For most downstream applications, the cardiac monolayers need to be purified. Currently, the most widely used method for purifying iPSC-CMs is based on THE significant differences in glucose and lactate metabolism between CMs and non-CMs (Tohyama et al., 2013). Although this means of iPSC-CM purification has been widely adopted and is a highly efficient method for large-scale purification, the same medium composition has also been used in the past to simulate ischemia preconditioning in cultured adult CMs (Diaz & Wilson, 2006). In other studies, a similar glucose-free solution was used as “injury solution” to cause localized ischemic injury in cardiac monolayers (Arutunyan, Webster, Swift, & Sarvazyan, 2001; Vanden Hoek et al., 1996). On the other hand, using the magnetic bead purification protocol that follows (Fig. 3), we obtain a large number of iPSC-CMs without an ischemic-injury-like phenotype that are appropriate for variety of studies (Block et al., 2020; da Rocha, Creech, Thonn, Mironov, & Herron, 2020; Davis et al., 2021; Herron et al., 2017; Jimenez-Vazquez et al., 2022; Ponce-Balbuena et al., 2018). This method has been also described and validated for use in proarrhythmia cardiotoxicity assays (da Rocha et al., 2017).
NOTE : Perform all cell culture manipulations in a laminar flow hood.
Materials
- PDMS (cat. no. SM21045730, Sigma Millipore)
- Matrigel solution: Matrigel (cat. no. 345277, Corning) diluted 1:100 in DMEM/F12 (cat. no. 11320-033, Gibco)
- Hank's Balanced Salt Solution (HBSS; cat. no. 14175-095, Gibco)
- 0.25% trypsin/EDTA (cat. no. 25200-056, Gibco)
- EB20 medium (see recipe)
- Trypan blue
- Hank's Balanced Salt Solution with Ca2+ and Mg2+ added (HBSS++; cat. no. 14025-092, Gibco)
- MACS separation buffer (cat. no. 130-091-221, Miltenyi Biotech), ice cold
- Cardiomyocyte purification kit (includes non-cardiomyocyte depletion cocktail and anti-biotin magnetic microbeads; cat. no. 130-110-188, Miltenyi Biotech)
- RPMI medium (with ʟ-glutamine; cat. no. 10-040-CV, Corning)
- B27+ supplement (cat. no. 17504004, Invitrogen)
- Gasket hole punch kit
- Laminar flow hood
- 37°C, 5% CO2 incubator
- 50-ml conical tube (sterile)
- 70-µm strainers (sterile; cat. no. 352350, Corning)
- Hemocytometer
- Benchtop centrifuge
- Ice bucket
- Magnetic MACS separator (cat. no. 130-090-976, Miltenyi Biotech)
- LS columns (cat. no. 130-042-401, Miltenyi Biotech)
- 30-µm pre-separation filters (cat. no. 130-041-407, Miltenyi Biotech)
- Penicillin-Streptomycin-Amphotericin B solution (PSA; cat. no. 15070063, Gibco)
Cell collection
1.Prepare a six-well plate containing a PDMS membrane at the bottom. For monolayer plating, pipet 500 μl ice-cold diluted Matrigel onto the PDMS in a large droplet. Allow the droplet to gel at room temperature for at least 30 min.
2.After ∼30 days in culture (see Basic Protocol 1, step 11), wash unpurified iPSC-CMs with HBSS and dissociate them by adding 1 ml of 0.25% trypsin/EDTA per well and then incubating the plates for 3-5 min at 37°C, 5% CO2.
3.Add 2 ml EB20 medium per well of dissociated cells to inactivate the trypsin. Triturate each well up to 10 times with a 10-ml serological pipet and transfer the cells into a sterile 50-ml conical tube fitted with a 70-µm strainer. Wash the strainer with 3 ml EB20 and add that to the tube as well.
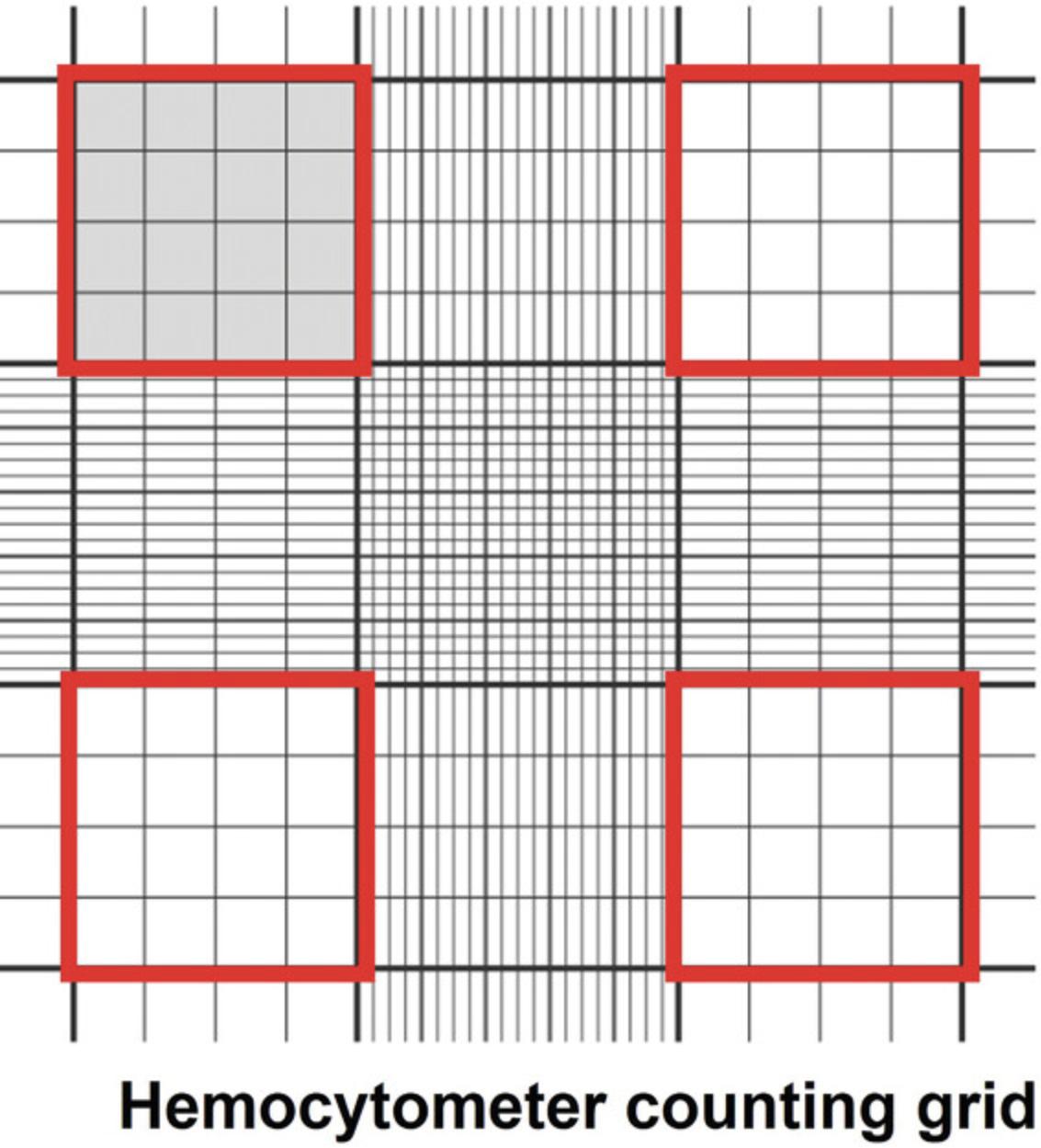
4.Count the cells as follows: Transfer 5 μl of the trypsinized cell suspension to a 1-ml centrifuge tube and mix with 45 μl trypan blue. Transfer 15-20 μl of the cell suspension between the hemocytometer and cover glass using a 20-μl (P-20) micropipettor. Count the number of cells in all four outer squares (Fig. 4, red squares) and divide by four (the mean number of cells/square). The number of cells per square × 104 × dilution (10) = the number of cells/ml of suspension.
5.Transfer cells into a 15-ml centrifuge conical tube and centrifuge 5 min at 1000 RPM (112 × g), 4°C.
6.Remove the supernatant and add 1 ml HBSS++ to wash the pellet.
7.Centrifuge the cells 5 min at 1000 RPM (112 × g), 4°C. Immediately aspirate the supernatant and add 100 μl ice-cold MACS Buffer per 5 × 106 cells to resuspend the pellet in the 15-ml conical tube.
iPSC-CM purification
8.Add 20 μl of non-cardiomyocyte depletion cocktail (biotin conjugated) per 5 × 106 cells, flick five times to mix, and incubate on ice for 5 min.
9.Add 1 ml ice-cold MACS Buffer per 15-ml conical tube. Triturate the cells gently and centrifuge 5 min at 1000 RPM (112 × g), 4°C.
10.Aspirate the supernatant and resuspend the pellet in 100 μl ice-cold MACS Buffer per 5 × 106 cells.
11.Add 20 μl of anti-biotin magnetic microbeads per 5 × 106 cells, flick five times to mix, and incubate on ice for 10 min.
12.Mix the cells gently with 8 ml MACS Buffer.
13.Add the cell suspension to the pre-separating filter on top of the LS flowing column on the Magnetic MACS Separator while continuously collecting the total flowthrough.
14.Collect 11 ml of flowthrough and centrifuge it for 5 min at 1000 RPM (112 × g), 4°C.
15.Discard the supernatant and gently triturate the iPSC-CM pellet with 1 ml EB20 medium.
iPSC-CM monolayer plating
16.Resuspend the purified iPSC-CM fractions in EB20 medium with 5 µM ROCK inhibitor to 200,000-300,000 cells/200-300 μl volume. Plate cells as monolayers on the Matrigel-coated 22 × 22-mm PDMS membranes adhered to the bottom of a six-well culture dish (from step 1).
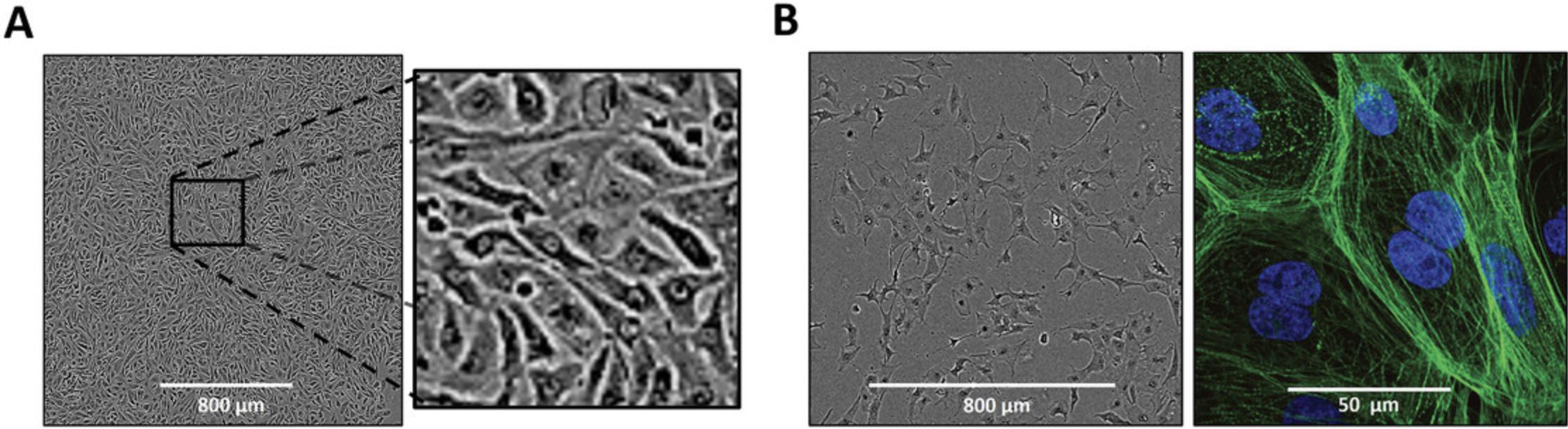
17.Transfer the plate to the incubator at 37°C and 5% CO2 and leave it there for at least 4 hr to allow attachment of iPSC-CMs to the Matrigel.
18.After ≥4 hr, add 3 ml EB20 medium with ROCK inhibitor to each well.
19.After 2 days, wash the cells with 3 ml HBSS++, aspirate, and replace with 3 ml RPMI medium containing B27+ supplement (RPMI/B27+).
20.Culture the highly purified iPSC-CMs monolayer on Matrigel-PDMS for at least 7 days before re-plating onto micropatterned PDMS (Fig. 5A).
Basic Protocol 3: MICROPATTERNING ON PDMS
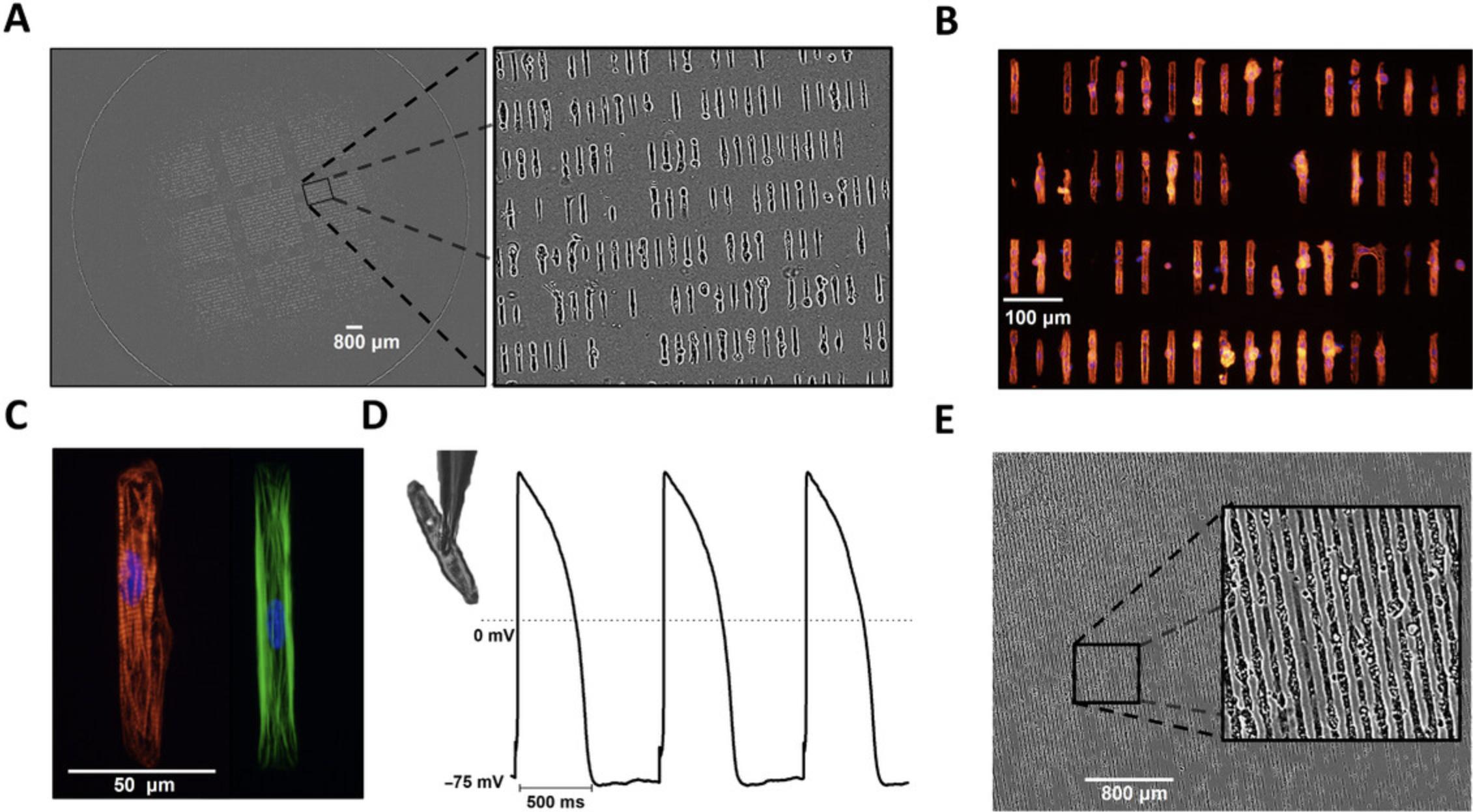
Standard cell culture techniques produce confluent but unstructured monolayers. Unlike native adult cardiomyocytes, iPSC-CMs do not retain many key structural and functional properties that are essential for many applications in cardiac research. The lack of myocyte alignment in iPSC-CMs is a significant deviation from the phenotype of adult cardiomyocytes in vivo, as evidenced by the irregular sarcomere alignment of iPSC-CMs plated on traditional planar substrates (Fig. 5B). Importantly, there are several reports indicating that cell shape and substrate stiffness improve contractile activity and facilitate maturation of iPSC-CMs (Chen, Mrksich, Huang, Whitesides, & Ingber, 1998; da Rocha et al., 2017; Dike et al., 1999; Herron et al., 2016; Kuo et al., 2012; Ribeiro et al., 2015). Micropatterning provides a structural framework that promotes the formation of rod-shaped iPSC-CMs that more accurately reproduce adult cardiomyocyte structure and function (e.g., increased expression of structural genes, greater sarcomere organization, matured mitochondria function, binucleation, etc.). The micropatterning platform detailed below (Fig. 6), adapted from Kuo et al. (2012), produces contracting single cells (Fig. 7) through the use of a reusable micropatterned silicone stamp that is compatible with a variety of matrix protein solutions. The stamp will leave a patterned growth substrate on the PDMS membrane that will provide the spatial resolution and structure for appropriate iPSC-CM anisotropy. The cell seeding pattern is also easily adaptable using different types of micropatterned silicone stamps. iPSC-CMs should be cultured as a monolayer for at least 7 days to induce maturation (see Basic Protocol 2, step 20) before the cardiomyocytes are re-plated onto the micropatterned PDMS.
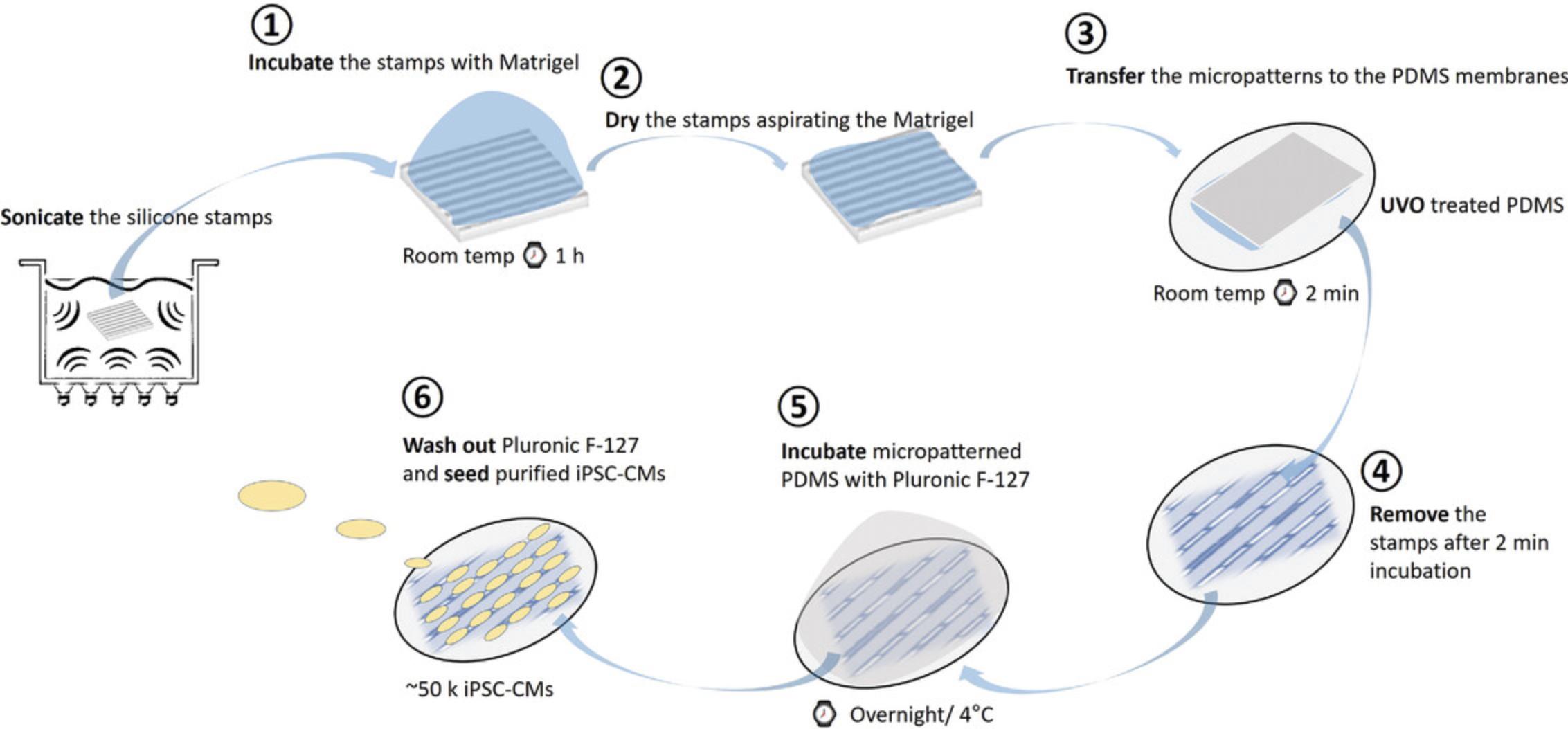
NOTE : Perform all cell culture manipulations in a laminar flow hood.
Materials
- 70% (v/v) ethanol in Milli-Q water
- Matrigel (cat. no. 345277, Corning)
- Pluronic F-127 (cat. no. P2443, Sigma)
- PDMS (cat. no. SM21045730, Sigma Millipore)
- Phosphate-buffered saline (PBS)
- Penicillin-Streptomycin-Amphotericin B solution (PSA; cat. no. 15070063, Gibco)
- 0.25% trypsin/EDTA (cat. no. 25200-056, Gibco)
- RPMI medium (with ʟ-glutamine; cat. no. 10-040-CV, Corning)
- Fetal bovine serum
- Replating medium: RPMI with 2% fetal bovine serum (FBS), B27+ supplement (cat. no. 17504004, Invitrogen), and 5 μM ROCK inhibitor (cat. no. Y-27632, Cayman Chemical), prewarmed to 37°C
- Silicone stamps
- Scotch tape
- Sonicator
- Laminar flow hood
- Gasket hole punch kit
- UV/ozone (UVO) machine
- 37°C, 5% CO2 incubator
- 50-ml conical tube (sterile)
- 70-µm strainers (sterile; cat. no. 352350, Corning)
- 15-ml centrifuge tube
- Benchtop centrifuge
- Ice bucket
- Trypan blue
- Hemocytometer
1.Carefully clean the surface of the silicone stamps with Scotch tape by placing it onto the stamp surface and slowly removing it (this is to remove residues from Matrigel used before). After that, sonicate the stamps in 70% ethanol/Milli-Q water for at least 20 min.
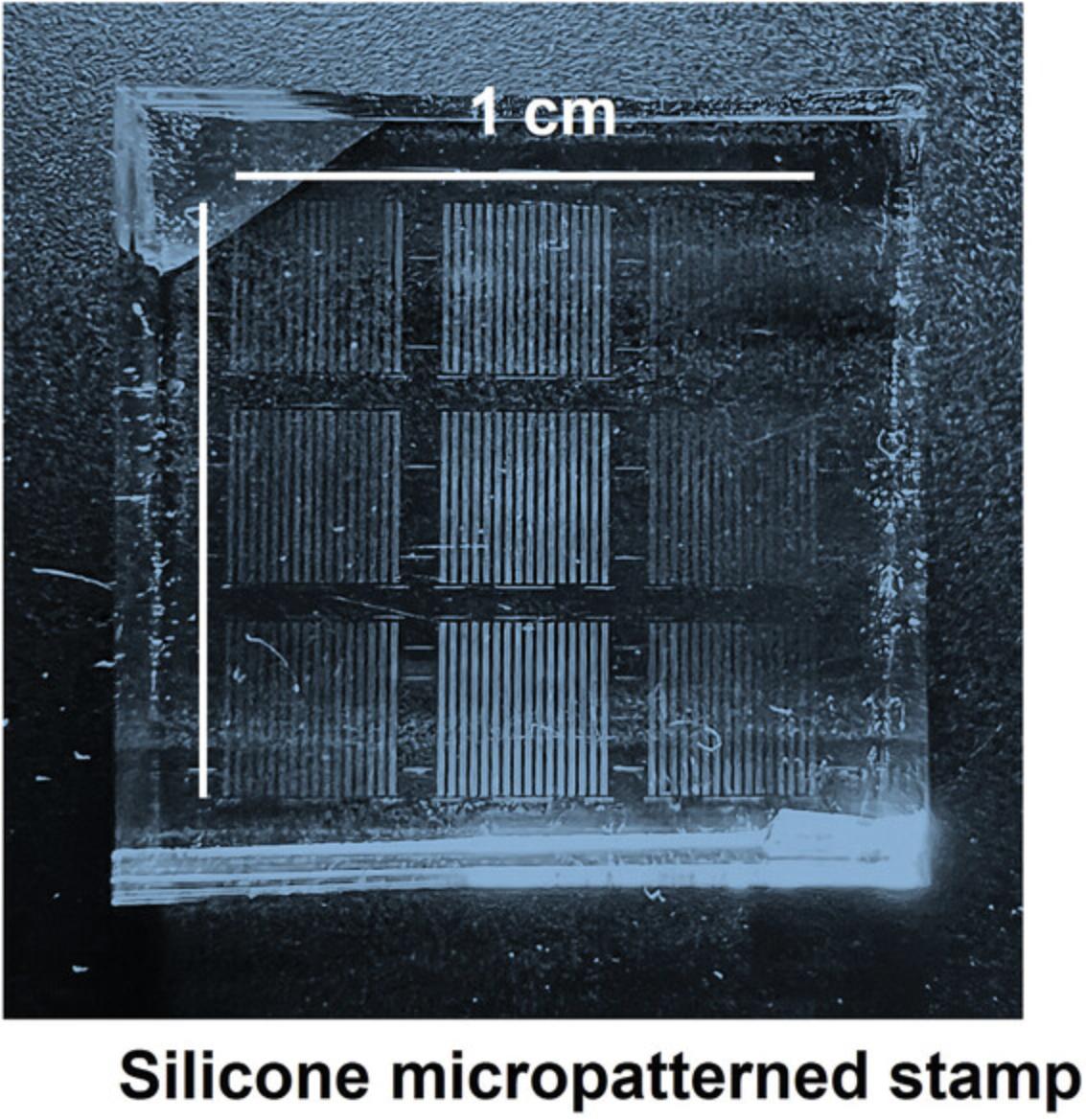
2.Dry the stamps in a sterile hood and then apply 250 μl Matrigel diluted 1:100 in di-water. Incubate the stamps in the diluted Matrigel at room temperature for at least 1 hr.
3.Using a punch tool, cut 18-mm PDMS circles from a sheet of PDMS and sonicate them in 70% ethanol for 20 min.
4.Shake excess ethanol off the 18-mm PDMS circles and transfer them to a six-well plate.
5.When ready for microprinting, treat the PDMS culture dish with UVO (UV light/ozone) for 9 min with lid off.
6.While UVO is performed on PDMS circles, aspirate the Matrigel solution from the PDMS stamps.
7.After UVO is completed, invert the dried stamps onto each PDMS circle, and remove one by one after ∼2 min.
8.Incubate the micropatterned PDMS plate with 1% Pluronic F-127 overnight at room temperature.
9.Before re-plating iPSC-CMs, clean the micropatterned plates for 1 hr with 3× PSA diluted in PBS, aspirate the PSA, and expose them to UV light for 15 min.
10.Dissociate the iPSC-CMs monolayers using 1 ml of 0.25% trypsin/EDTA and incubate the cardiomyocytes for 8-10 min at 37°C, 5% CO2.
11.After dissociation, add 1 ml of RPMI medium containing 10% FBS to inactivate the trypsin.
12.Transfer dissociated iPSC-CMs through a 70-μm filter into a 50-ml conical tube.
13.Collect the iPSC-CMs into a 15-ml conical centrifuge tube and centrifuge for 3 min at (112 × g), room temperature.
14.Resuspend the iPSC-CMs in 1 ml re-plating medium.
15.Count (see Basic Protocol 2, step 4) and plate ∼30,000 iPSC-CMs in 350 μl re-plating medium in the center of the micropatterned area.
16.After ∼5 hr, gently add 2 ml re-plating medium.
17.Return the plate to the incubator
18.Change the medium on days 1 and 3 after re-plating.
REAGENTS AND SOLUTION
EB20 medium
- 80% DMEM/F12 (cat. no. 11320-033, Gibco)
- 0.1 mM non-essential amino acids (cat. no. 11140-050, Gibco)
- 1 mM ʟ-glutamine (cat. no. 25030-081, Gibco)
- 0.1 mM 2-mercaptoethanol (cat. no. M6250, Sigma)
- 20% fetal bovine serum (FBS; cat. no. F0926, Sigma)
- 10 µM blebbistatin (cat. no. 13186, Cayman Chemical)
- 5 µM ROCK inhibitor (cat. no. Y-27632, Cayman Chemical)
- If properly covered from light, EB20 medium will remain stable at 4°C for at least 1 month.
COMMENTARY
Background Information
Animal models have been and will continue to be useful in defining mechanisms of cardiovascular development, physiology, and disease (Chorro et al., 2009; Houser et al., 2012; Oh et al., 2019). However, differences between humans and animals often limit the translation of findings from animal models into human therapies (Doncheva et al., 2021). Patient-specific in vitro models can help to fill the gap between animal models and the clinic to facilitate the development of novel therapies (Kim, 2014). Human-based models are especially important for cardiovascular research because the cardiac physiology of many animal models differs dramatically from that of humans. These differences include beating rates and calcium handling, as well as ion channel types and their expression levels (Liu, Laksman, & Backx, 2016; Milani-Nejad & Janssen, 2014; Zhao et al., 2018). And, although the physiological differences in large animal models are less than those observed in murine models (Dixon & Spinale, 2009), there is still a need for a human model that can replicate the cellular and genetic environment of the human cardiomyocyte.
Models that recapitulate individual patient disease at the molecular and cellular levels are obvious candidates for improving the understanding of disease pathogenesis and progression and predicting the responses of individual patients to specific treatments. The development of iPS cell technology (Takahashi & Yamanaka, 2006; Takahashi et al., 2007), and the improved ability to differentiate iPS cells into disease-relevant cell types such as cardiomyocytes (Burridge, Keller, Gold, & Wu, 2012), have created an extraordinary opportunity for generating human patient-specific cell lines for use in disease modeling, personalized drug screening, and regenerative approaches to precision medicine (Kim, 2014; Liu et al., 2016; Matsa, Burridge, & Wu, 2014; Moretti et al., 2010).
Many barriers remain in the field of iPS cell differentiation. Even though human iPS cell technology has advanced rapidly since 2007, iPS cell clones can exhibit differences in differentiation efficiency and in phenotype, including between clones derived from the same person. Thus, line-to-line variation often complicates data interpretation (Shi, Inoue, Wu, & Yamanaka, 2017). Another barrier is the variability in differentiated cell maturation across lines (Ivashchenko et al., 2013; Zhu, Santana, & Laflamme, 2009). Modeling diseases with iPS cells has been further hindered by the fetal-like properties of iPS-cell-derived cell lines (Studer, Vera, & Cornacchia, 2015). Standardized human iPS cell protocols reduce technical variability and enable more reliable identification of true biological phenotypes. Concerning the variations in maturation, the micropatterning technique has been shown to be useful in controlling cell size, shape, and maturation state (Helms et al., 2020; Jimenez-Vazquez et al., 2022; Kuo et al., 2012; Ribeiro et al., 2015; Tsan et al., 2021). Plating myocytes or other cell types (e.g., BHK cells, MDCK cells, etc.) in controlled two-dimensional arrangements has previously been shown to also improve cell differentiation, functionality, and longevity (Clark, Connolly, Curtis, Dow, & Wilkinson, 1991; Motlagh, Hartman, Desai, & Russell, 2003; Motlagh, Senyo, Desai, & Russell, 2003).
A variety of cell patterns can be generated using the Matrigel-based micropatterning protocol. They can be used with a variety of techniques and studies, including optical mapping, immunostaining, high-throughput compound testing, stretching analysis, and patch clamping. This method creates a culture environment that combines a flexible platform with a microenvironment that promotes iPSC-CM cell alignment, growth, connection, and maturation.
This protocol cannot reproduce the three-dimensional multilayer structure of the native myocardium. This is significant because the electromechanical function in the heart is closely related to the three-dimensional spatiotemporal gradients that are essential to cardiac structure and function (Burton et al., 2006; Vadakkumpadan et al., 2010). As a result, although this protocol produces iPSC-CMs with morphologies that are similar to those of adult cardiomyocytes, it does not accurately reflect the in situ environment. Despite these limitations, iPSC-CMs seeded on micropatterned surfaces clearly mimic important structural and functional aspects of native ventricular myocardium.
Critical Parameters and Troubleshooting
The micropatterning technique is a valuable tool for improving several functional aspects of iPSC-CMs. However, its success is dependent upon several critical parameters and on an understanding of the limitations associated with this technology. Table 1 discusses the main troubleshooting strategies.
| Possible problems | Troubleshooting strategies |
|---|---|
| No single cells are found on the micropatterns |
Pipet the iPSC-CM monolayers up and down more times to better dissociate them. Plate no more than 50,000 iPSC-CMs per micropatterned area. |
| Micropatterns are not transferred from the stamp to the PDMS sheet |
Incubate the PDMS plate under UVO for a longer time. Stiffness of the PDMS is important. Thicker PDMS usually needs longer exposure time to UVO. Before transferring the micropatterns to the PDMS, remove any excess Matrigel from the stamp. |
| When transferring the Matrigel micropatterns, there is only smearing | You are pressing the silicone stamp too firmly against the PDMS sheet. When transferring the micropattern to the PDMS, apply minimal pressure to the stamp. |
| Micropatterns do not last long enough on the PDMS sheets, or iPSC-CMs adhere poorly to the Matrigel micropatterns |
Matrigel degrades quickly. Use a proper dilution of ≥1:100. Do not leave the micropatterned PDMS plate under UV light for >20 min, as it will damage the Matrigel micropatterns. |
| iPSC-CMs do not survive after re-plating on micropatterns |
When dissociating the iPSC-CM monolayers, be gentle. Allow the cells to incubate with trypsin for no more than 10 min. Because Pluronic F-127 is cytotoxic, rinse the Matrigel-coated micropatterned PDMS. plate thoroughly after incubation. |
Critical steps
- The patterned region of the silicone stamp should never be touched by anything other than the matrix protein solution, and the Scotch tape used when cleaning it.
- When preparing the PDMS plates, make sure all air bubbles are removed. Otherwise, they will be incorporated into the final micropattern, resulting in micropattern defects.
- Place the silicone stamp covered in Matrigel carefully onto the PDMS to avoid smearing the transferred micropatterns.
- Pluronic F-127 is cytotoxic. Make sure to wash it carefully at least five times with PBS before plating the iPSC-CMs.
- Do not expose the micropatterned PDMS plate to UV light for >20 min. This may damage the micropatterns.
- Before plating the iPSC-CMs, ensure that there are no clumps of cells to avoid forming small monolayers or cell clusters.
- Rinse the PSA from the micropatterned PDMS plate thoroughly because antibiotics such as streptomycin block stretch-activated ion channels in isolated cells (Belus & White, 2003; Shen, Chou, & Chiu, 2003) and may affect the responses of the iPSC-CMs to stretch.
- Depending on the stiffness of the PDMS, the UVO incubation time may need to be adjusted.
Anticipated Results
This protocol describes the methods used to create in vitro models of iPSC-CMs that mimic important structural and mechanical parameters of adult cardiomyocytes. iPSC-CMs grown on Matrigel-coated micropatterned PDMS membranes develop in vivo-like cell morphology, including organized sarcomeres and hyperpolarized resting membrane potentials (Fig. 7A, Fig. 7B, Fig. 7C, Fig. 7D). Other silicone stamps can be used to produce strands of aligned iPSC-CMs that can be plated with fibroblasts to mimic the cellular spatial distribution of the native myocardium (Fig. 7E).
In addition to increased maturation, these micropatterns can be used to study mechanical, biophysical, and biochemical events from iPSC-CMs derived from healthy and diseased individuals. Importantly, the application of this technique is not limited to cardiomyocytes; controlling shape and size may be significant in a wide range of cells, including fibroblasts, smooth muscle cells, endothelial cells, and others.
Time Considerations
The generation of iPSC-cardiomyocytes as described in Basic Protocol 1 will take at least 15 days from the start of the differentiation process, which may vary depending on the iPS cell line. The purification protocol (Basic Protocol 2) takes only a few hours to complete, depending on the number of cells to be purified. However, the purified iPSC-CMs must then mature as monolayers for an additional 7 days before being seeded on micropatterns. The micropatterning process (Basic Protocol 3) is typically carried out in two stages: 1 day to prepare the Matrigel-coated micropatterned PDMS plates, and another day to perform the cell re-plating. Before any type of analysis is performed, iPSC-CMs must be on the micropatterns for at least 3 days. As a result, the entire process, from iPSC differentiation to their use in the desired assay following micropatterning, takes about a month. The processing and acquisition of the data following iPSC-CM micropatterning will determined by the type of study to be conducted.
Acknowledgments
This research was supported by US NIH/NHLBI R00HL133482 to DKJ.
Author Contributions
Eric N. Jimenez-Vazquez : Conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; Abhilasha Jain : Investigation, methodology; David K. Jones : Funding acquisition, project administration, resources, writing—original draft, writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
All data generated or analyzed in this study are included in this manuscript.
Literature Cited
- Arutunyan, A., Webster, D. R., Swift, L. M., & Sarvazyan, N. (2001). Localized injury in cardiomyocyte network: A new experimental model of ischemia-reperfusion arrhythmias. American Journal of Physiology. Heart and Circulatory Physiology , 280(4), H1905–1915. doi: 10.1152/ajpheart.2001.280.4.H1905
- Belus, A., & White, E. (2003). Streptomycin and intracellular calcium modulate the response of single guinea-pig ventricular myocytes to axial stretch. Journal of Physiology , 546(Pt 2), 501–509. doi: 10.1113/jphysiol.2002.027573
- Block, T., Creech, J., da Rocha, A. M., Marinkovic, M., Ponce-Balbuena, D., Jimenez- Vazquez, E. N., … Herron, T. J. (2020). Human perinatal stem cell derived extracellular matrix enables rapid maturation of hiPSC-CM structural and functional phenotypes. Science Reports , 10(1), 19071. doi: 10.1038/s41598-020-76052-y
- Burridge, P. W., Keller, G., Gold, J. D., & Wu, J. C. (2012). Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell , 10(1), 16–28. doi: 10.1016/j.stem.2011.12.013
- Burton, R. A., Plank, G., Schneider, J. E., Grau, V., Ahammer, H., Keeling, S. L., … Kohl, P. (2006). Three-dimensional models of individual cardiac histoanatomy: Tools and challenges. Annals of the New York Academy of Sciences , 1080, 301–319. doi: 10.1196/annals.1380.023
- Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M., & Ingber, D. E. (1997). Geometric control of cell life and death. Science , 276(5317), 1425–1428.
- Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M., & Ingber, D. E. (1998). Micropatterned surfaces for control of cell shape, position, and function. Biotechnology Progress , 14(3), 356–363. doi: 10.1021/bp980031m
- Chorro, F. J., Such-Belenguer, L., & Lopez-Merino, V. (2009). Animal models of cardiovascular disease. Revista Espanola de Cardiologia , 62(1), 69–84.
- Clark, P., Connolly, P., Curtis, A. S., Dow, J. A., & Wilkinson, C. D. (1991). Cell guidance by ultrafine topography in vitro. Journal of Cell Science , 99(Pt 1), 73–77. doi: 10.1242/jcs.99.1.73
- da Rocha, A. M., Campbell, K., Mironov, S., Jiang, J., Mundada, L., Guerrero-Serna, G., … Herron, T. J. (2017). hiPSC-CM monolayer maturation state determines drug responsiveness in high throughput pro-arrhythmia screen. Science Reports , 7(1), 13834. doi: 10.1038/s41598-017-13590-y
- da Rocha, A. M., Creech, J., Thonn, E., Mironov, S., & Herron, T. J. (2020). Detection of drug-induced Torsades de Pointes arrhythmia mechanisms using hiPSC-CM syncytial monolayers in a high-throughput screening voltage sensitive dye assay. Toxicological Sciences , 173(2), 402–415. doi: 10.1093/toxsci/kfz235
- Davis, J., Chouman, A., Creech, J., Monteiro da Rocha, A., Ponce-Balbuena, D., Jimenez Vazquez, E. N., … Herron, T. J. (2021). In vitro model of ischemic heart failure using human induced pluripotent stem cell-derived cardiomyocytes. JCI Insight , 6(10), e134368. doi: 10.1172/jci.insight.134368
- Davis, R. P., Casini, S., van den Berg, C. W., Hoekstra, M., Remme, C. A., Dambrot, C., … Mummery, C. L. (2012). Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation , 125(25), 3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092
- Diaz, R. J., & Wilson, G. J. (2006). Studying ischemic preconditioning in isolated cardiomyocyte models. Cardiovascular Research , 70(2), 286–296. doi: 10.1016/j.cardiores.2005.12.003
- Dike, L. E., Chen, C. S., Mrksich, M., Tien, J., Whitesides, G. M., & Ingber, D. E. (1999). Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. In Vitro Cellular & Developmental Biology Animal, 35(8), 441–448. doi: 10.1007/s11626-999-0050-4
- Dixon, J. A., & Spinale, F. G. (2009). Large animal models of heart failure: A critical link in the translation of basic science to clinical practice. Circulation. Heart Failure , 2(3), 262–271. doi: 10.1161/CIRCHEARTFAILURE.108.814459
- Doncheva, N. T., Palasca, O., Yarani, R., Litman, T., Anthon, C., Groenen, M. A. M., … Gorodkin, J. (2021). Human pathways in animal models: Possibilities and limitations. Nucleic Acids Research , 49(4), 1859–1871. doi: 10.1093/nar/gkab012
- Gaspar, J. A., Doss, M. X., Hengstler, J. G., Cadenas, C., Hescheler, J., & Sachinidis, A. (2014). Unique metabolic features of stem cells, cardiomyocytes, and their progenitors. Circulation Research , 114(8), 1346–1360. doi: 10.1161/CIRCRESAHA.113.302021
- Helms, A. S., Tang, V. T., O'Leary, T. S., Friedline, S., Wauchope, M., Arora, A., & Day, S. M. (2020). Effects of MYBPC3 loss-of-function mutations preceding hypertrophic cardiomyopathy. JCI Insight , 5(2), e133782. doi: 10.1172/jci.insight.133782
- Herron, T., Monteiro da Rocha, A., & Campbell, K. (2017). Cardiomyocyte purification from pluripotent stem cells. Retrieved from https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.601/attachments/App_note_PSC_derived_cardiomyocytes.pdf
- Herron, T. J., Rocha, A. M., Campbell, K. F., Ponce-Balbuena, D., Willis, B. C., Guerrero- Serna, G., … Jalife, J. (2016). Extracellular matrix-mediated maturation of human pluripotent stem cell-derived cardiac monolayer structure and electrophysiological function. Circulation. Arrhythmia and Electrophysiology , 9(4), e003638. doi: 10.1161/CIRCEP.113.003638
- Houser, S. R., Margulies, K. B., Murphy, A. M., Spinale, F. G., Francis, G. S., Prabhu, S. D., … Translational, B. (2012). Animal models of heart failure: A scientific statement from the American Heart Association. Circulation Research , 111(1), 131–150. doi: 10.1161/RES.0b013e3182582523
- Itzhaki, I., Maizels, L., Huber, I., Zwi-Dantsis, L., Caspi, O., Winterstern, A., … Gepstein, L. (2011). Modelling the long QT syndrome with induced pluripotent stem cells. Nature , 471(7337), 225–229. doi: 10.1038/nature09747
- Ivashchenko, C. Y., Pipes, G. C., Lozinskaya, I. M., Lin, Z., Xiaoping, X., Needle, S., … Willette, R. N. (2013). Human-induced pluripotent stem cell-derived cardiomyocytes exhibit temporal changes in phenotype. American Journal of Physiology. Heart and Circulatory Physiology , 305(6), H913–922. doi: 10.1152/ajpheart.00819.2012
- Jimenez-Vazquez, E. N., Arad, M., Macias, A., Vera-Pedrosa, M. L., Cruz, F. M., Gutierrez, L. K., … Jalife, J. (2022). SNTA1 gene rescues ion channel function and is antiarrhythmic in cardiomyocytes derived from induced pluripotent stem cells from muscular dystrophy patients. Elife , 11, e76576. doi: 10.7554/eLife.76576
- Kim, C. (2014). Disease modeling and cell based therapy with iPSC: Future therapeutic option with fast and safe application. Blood Research , 49(1), 7–14. doi: 10.5045/br.2014.49.1.7
- Kuo, P. L., Lee, H., Bray, M. A., Geisse, N. A., Huang, Y. T., Adams, W. J., … Parker, K. K. (2012). Myocyte shape regulates lateral registry of sarcomeres and contractility. American Journal of Pathology , 181(6), 2030–2037. doi: 10.1016/j.ajpath.2012.08.045
- Li, S., Chen, G., & Li, R. A. (2013). Calcium signalling of human pluripotent stem cell- derived cardiomyocytes. Journal of Physiology , 591(21), 5279–5290. doi: 10.1113/jphysiol.2013.256495
- Lian, X., Zhang, J., Azarin, S. M., Zhu, K., Hazeltine, L. B., Bao, X., … Palecek, S. P. (2013). Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nature Protocols , 8(1), 162–175. doi: 10.1038/nprot.2012.150
- Lieu, D. K., Liu, J., Siu, C. W., McNerney, G. P., Tse, H. F., Abu-Khalil, A., … Li, R. A. (2009). Absence of transverse tubules contributes to non-uniform Ca2+ wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells and Development , 18(10), 1493–1500. doi: 10.1089/scd.2009.0052
- Liu, J., Laksman, Z., & Backx, P. H. (2016). The electrophysiological development of cardiomyocytes. Advanced Drug Delivery Reviews , 96, 253–273. doi: 10.1016/j.addr.2015.12.023
- Ma, J., Guo, L., Fiene, S. J., Anson, B. D., Thomson, J. A., Kamp, T. J., & January, C. T. (2011). High purity human-induced pluripotent stem cell-derived cardiomyocytes: Electrophysiological properties of action potentials and ionic currents. American Journal of Physiology. Heart and Circulatory Physiology , 301(5), H2O06–2017. doi: 10.1152/ajpheart.00694.2011
- Matsa, E., Burridge, P. W., & Wu, J. C. (2014). Human stem cells for modeling heart disease and for drug discovery. Science Translational Medicine , 6(239), 239ps236. doi: 10.1126/scitranslmed.3008921
- Milani-Nejad, N., & Janssen, P. M. (2014). Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacology & Therapeutics, 141(3), 235–249. doi: 10.1016/j.pharmthera.2013.10.007
- Moretti, A., Bellin, M., Welling, A., Jung, C. B., Lam, J. T., Bott-Flugel, L., … Laugwitz, K. L. (2010). Patient-specific induced pluripotent stem-cell models for long-QT syndrome. New England Journal of Medicine , 363(15), 1397–1409. doi: 10.1056/NEJMoa0908679
- Motlagh, D., Hartman, T. J., Desai, T. A., & Russell, B. (2003). Microfabricated grooves recapitulate neonatal myocyte connexin43 and N-cadherin expression and localization. Journal of Biomedical Materials Research. Part A , 67(1), 148–157. doi: 10.1002/jbm.a.10083
- Motlagh, D., Senyo, S. E., Desai, T. A., & Russell, B. (2003). Microtextured substrata alter gene expression, protein localization and the shape of cardiac myocytes. Biomaterials , 24(14), 2463–2476. doi: 10.1016/s0142-9612(02)00644-0
- Novak, A., Barad, L., Zeevi-Levin, N., Shick, R., Shtrichman, R., Lorber, A., … Binah, O. (2012). Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to beta-adrenergic stimulation. Journal of Cellular and Molecular Medicine , 16(3), 468–482. doi: 10.1111/j.1582-4934.2011.01476.x
- Oh, J. G., Kho, C., Hajjar, R. J., & Ishikawa, K. (2019). Experimental models of cardiac physiology and pathology. Heart Failure Reviews , 24(4), 601–615. doi: 10.1007/s10741-019-09769-2
- Pekkanen-Mattila, M., Hakli, M., Polonen, R. P., Mansikkala, T., Junnila, A., Talvitie, E., … Aalto-Setala, K. (2019). Polyethylene terephthalate textiles enhance the structural maturation of human induced pluripotent stem cell-derived cardiomyocytes. Materials , 12(11), 1805. doi: 10.3390/ma12111805
- Ponce-Balbuena, D., Guerrero-Serna, G., Valdivia, C. R., Caballero, R., Diez-Guerra, F. J., Jiménez-Vázquez, E. N., … Campbell, K. F. (2018). Cardiac Kir2. 1 and NaV1. 5 channels traffic together to the sarcolemma to control excitability. Circulation Research , 122(11), 1501–1516.
- Rana, P., Anson, B., Engle, S., & Will, Y. (2012). Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: Bioenergetics and utilization in safety screening. Toxicological Sciences , 130(1), 117–131. doi: 10.1093/toxsci/kfs233
- Ribeiro, A. J., Ang, Y. S., Fu, J. D., Rivas, R. N., Mohamed, T. M., Higgs, G. C., … Pruitt, B. L. (2015). Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proceedings of the National Academy of Sciences of the United States of America , 112(41), 12705–12710. doi: 10.1073/pnas.1508073112
- Shen, M. R., Chou, C. Y., & Chiu, W. T. (2003). Streptomycin and its analogues are potent inhibitors of the hypotonicity-induced Ca2+ entry and Cl– channel activity. FEBS Letters , 554(3), 494–500. doi: 10.1016/s0014-5793(03)01231-6
- Shi, Y., Inoue, H., Wu, J. C., & Yamanaka, S. (2017). Induced pluripotent stem cell technology: A decade of progress. Nature Reviews Drug Discovery , 16(2), 115–130. doi: 10.1038/nrd.2016.245
- Studer, L., Vera, E., & Cornacchia, D. (2015). Programming and reprogramming cellular age in the era of induced pluripotency. Cell Stem Cell , 16(6), 591–600. doi: 10.1016/j.stem.2015.05.004
- Sun, N., Yazawa, M., Liu, J., Han, L., Sanchez-Freire, V., Abilez, O. J., … Wu, J. C. (2012). Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Science Translational Medicine , 4(130), 130ra147. doi: 10.1126/scitranslmed.3003552
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., & Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell , 131(5), 861–872. doi: 10.1016/j.cell.2007.11.019
- Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell , 126(4), 663–676. doi: 10.1016/j.cell.2006.07.024
- Tohyama, S., Hattori, F., Sano, M., Hishiki, T., Nagahata, Y., Matsuura, T., … Fukuda, K. (2013). Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell , 12(1), 127–137. doi: 10.1016/j.stem.2012.09.013
- Tsan, Y. C., DePalma, S. J., Zhao, Y. T., Capilnasiu, A., Wu, Y. W., Elder, B., … Helms, A. S. (2021). Physiologic biomechanics enhance reproducible contractile development in a stem cell derived cardiac muscle platform. Nature Communication , 12(1), 6167. doi: 10.1038/s41467-021-26496-1
- Vadakkumpadan, F., Arevalo, H., Prassl, A. J., Chen, J., Kickinger, F., Kohl, P., … Trayanova, N. (2010). Image-based models of cardiac structure in health and disease. Wiley Interdisciplinary Reviews: Systems Biology and Medicine , 2(4), 489–506. doi: 10.1002/wsbm.76
- van den Berg, C. W., Okawa, S., Chuva de Sousa Lopes, S. M., van Iperen, L., Passier, R., Braam, S. R., … Mummery, C. L. (2015). Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development , 142(18), 3231–3238. doi: 10.1242/dev.123810
- Vanden Hoek, T. L., Shao, Z., Li, C., Zak, R., Schumacker, P. T., & Becker, L. B. (1996). Reperfusion injury on cardiac myocytes after simulated ischemia. American Journal of Physiology , 270(4 Pt 2), H1334–1341. doi: 10.1152/ajpheart.1996.270.4.H1334
- Yang, X., Pabon, L., & Murry, C. E. (2014). Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circulation Research , 114(3), 511–523. doi: 10.1161/CIRCRESAHA.114.300558
- Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., … Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science , 318(5858), 1917–1920. doi: 10.1126/science.1151526
- Zhao, Z., Lan, H., El-Battrawy, I., Li, X., Buljubasic, F., Sattler, K., … Akin, I. (2018). Ion channel expression and characterization in human induced pluripotent stem cell-derived cardiomyocytes. Stem Cells International , 2018, 6067096. doi: 10.1155/2018/6067096
- Zhu, W. Z., Santana, L. F., & Laflamme, M. A. (2009). Local control of excitation- contraction coupling in human embryonic stem cell-derived cardiomyocytes. Plos One , 4(4), e5407. doi: 10.1371/journal.pone.0005407
Citing Literature
Number of times cited according to CrossRef: 1
- Kamil Elkhoury, Sacha Kodeih, Eduardo Enciso‐Martínez, Ali Maziz, Christian Bergaud, Advancing Cardiomyocyte Maturation: Current Strategies and Promising Conductive Polymer‐Based Approaches, Advanced Healthcare Materials, 10.1002/adhm.202303288, 13 , 13, (2024).

