Discriminating Between Apoptosis, Necrosis, Necroptosis, and Ferroptosis by Microscopy and Flow Cytometry
Aoife Costigan, Aoife Costigan, Emilie Hollville, Emilie Hollville, Seamus J. Martin, Seamus J. Martin
apoptosis
caspases
cell morphology
ferroptosis
flow cytometry
microscopy
necroptosis
necrosis
programmed necrosis
Abstract
Apoptosis is a mode of programmed cell death that plays important roles in tissue sculpting during development, in the maintenance of tissue homeostasis in the adult, and in the eradication of injured or infected cells during pathological processes. Numerous physiological as well as pathological stimuli trigger apoptosis, such as engagement of plasma-membrane-associated Fas, TRAIL, or TNF receptors, growth factor deprivation, hypoxia, radiation, and exposure to diverse cytotoxic drugs. Apoptosis is coordinated by members of the caspase family of cysteine proteases, which, upon activation, trigger a series of dramatic morphological and biochemical changes including retraction from the substratum, cell shrinkage, extensive and protracted plasma membrane blebbing, chromatin condensation, DNA hydrolysis, nuclear fragmentation, and proteolytic cleavage of numerous caspase substrates. These dramatic structural and biochemical alterations result not only in the controlled dismantling of the cell, but also in the rapid recognition and removal of apoptotic cells by phagocytes through the cell surface display of phagocytotic triggers such as phosphatidylserine. Necrosis, which is typically nonprogrammed or imposed upon the cell by overwhelming membrane or organelle damage, is characterized by high-amplitude cell swelling, followed by rapid plasma membrane rupture and release of cellular contents into the extracellular space. Necrosis is often provoked by infectious agents or severe departure from physiological conditions due to toxins, temperature extremes, or physical injury. However, forms of programmed necrosis (necroptosis, pyroptosis, ferroptosis) can also occur in specific circumstances. Nonprogrammed and programmed necrosis can be distinguished from apoptosis by morphological features, based on the rapid uptake of vital dyes, and through the application of specific inhibitors of key molecules associated with the latter modes of cell death. This unit describes protocols for the measurement of apoptosis and necrosis and for distinguishing apoptosis from programmed as well as conventional necrosis. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Analysis of cell morphology by phase-contrast microscopy
Alternative Protocol 1 : Assessment of morphological changes using eosin-methylene blue staining
Alternative Protocol 2 : Analysis of nuclear morphology by fluorescence microscopy
Support Protocol : Preparation of cytospins
Basic Protocol 2 : Measurement of plasma membrane composition with annexin V and propidium iodide
Basic Protocol 3 : Measurement of DNA fragmentation by flow cytometry
Alternative Protocol 3 : Analysis of DNA fragmentation by the TUNEL assay
Basic Protocol 4 : Measurement of caspase activation by flow cytometry
Basic Protocol 5 : Discriminating between apoptosis, necrosis, necroptosis, and ferroptosis
INTRODUCTION
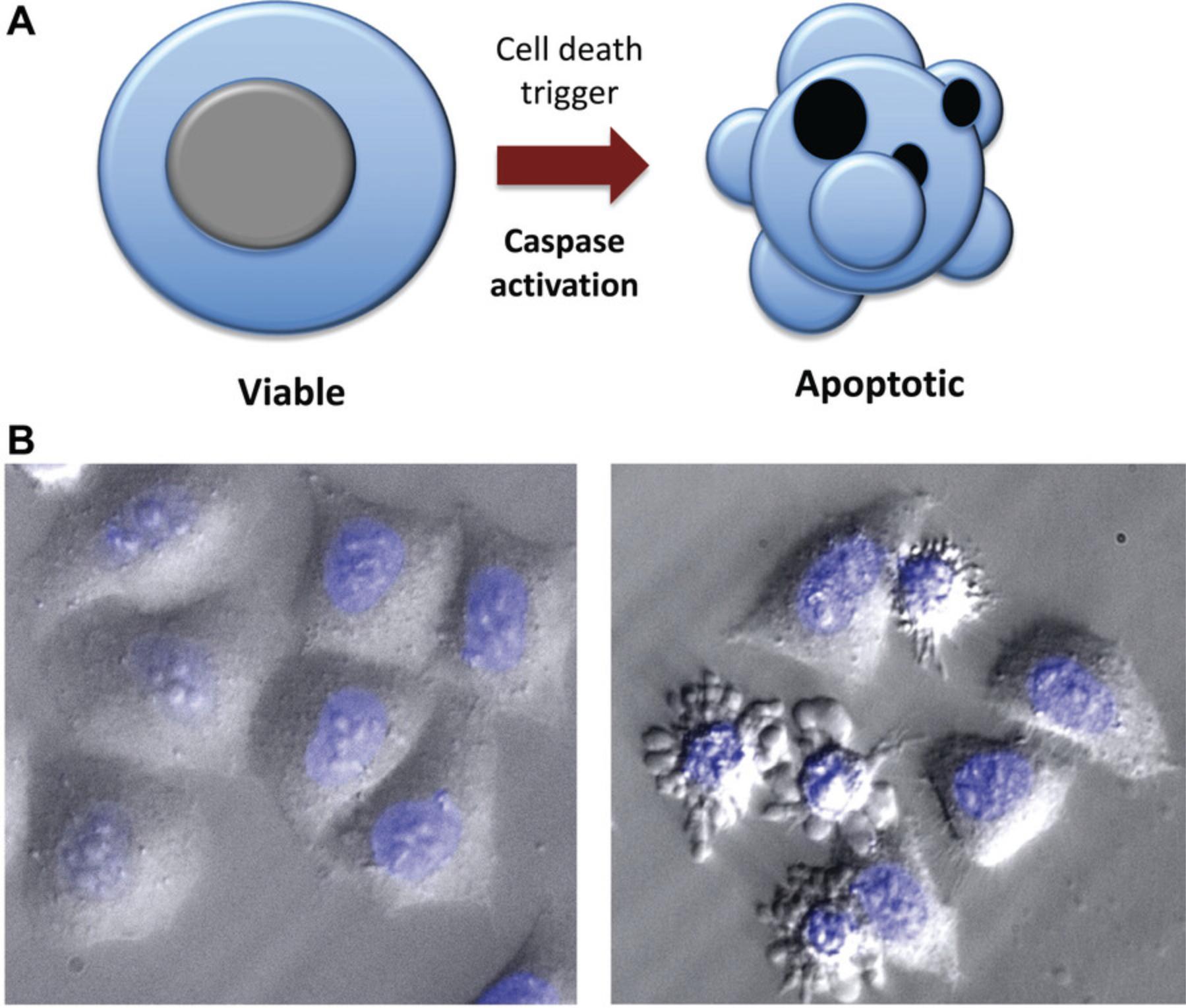
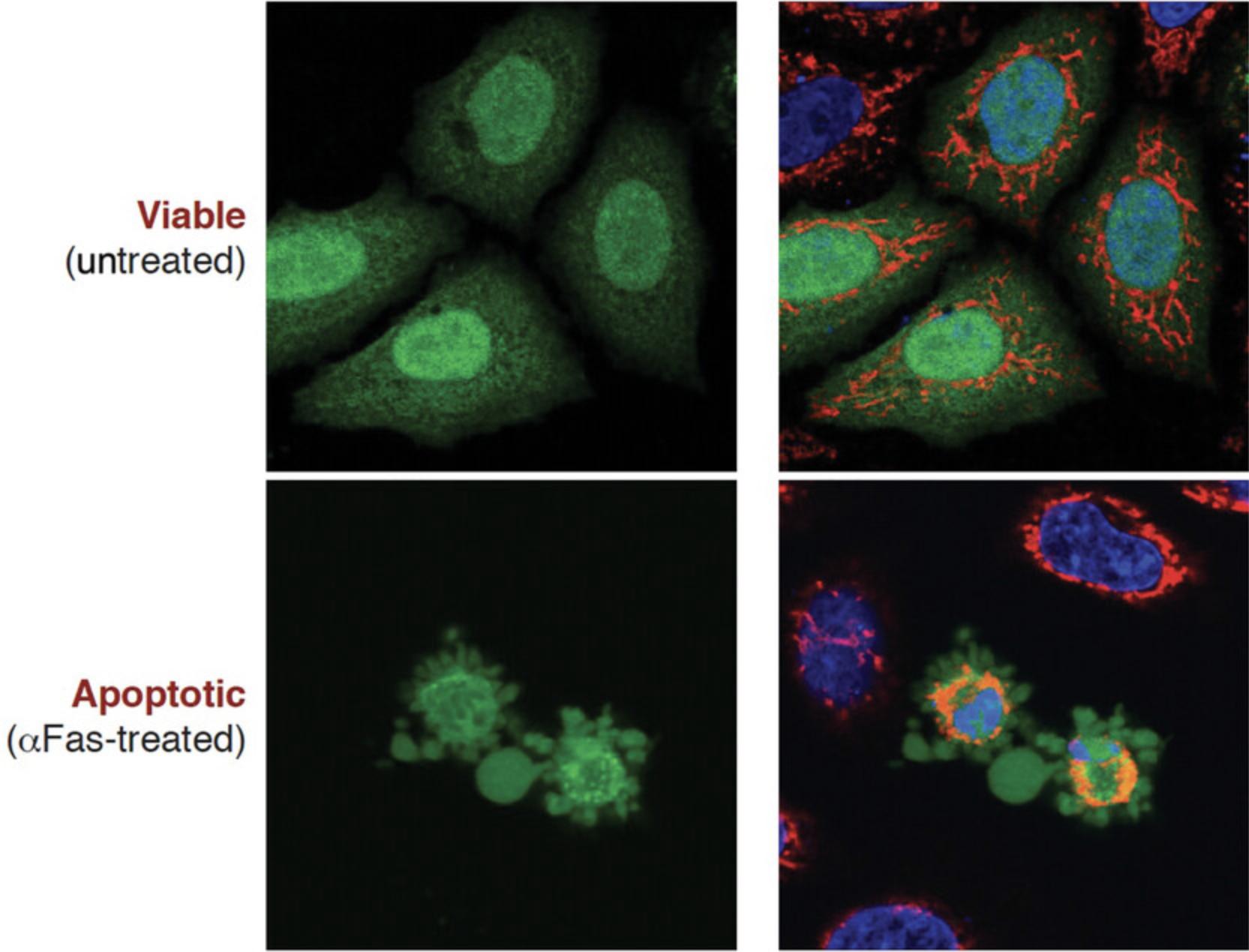
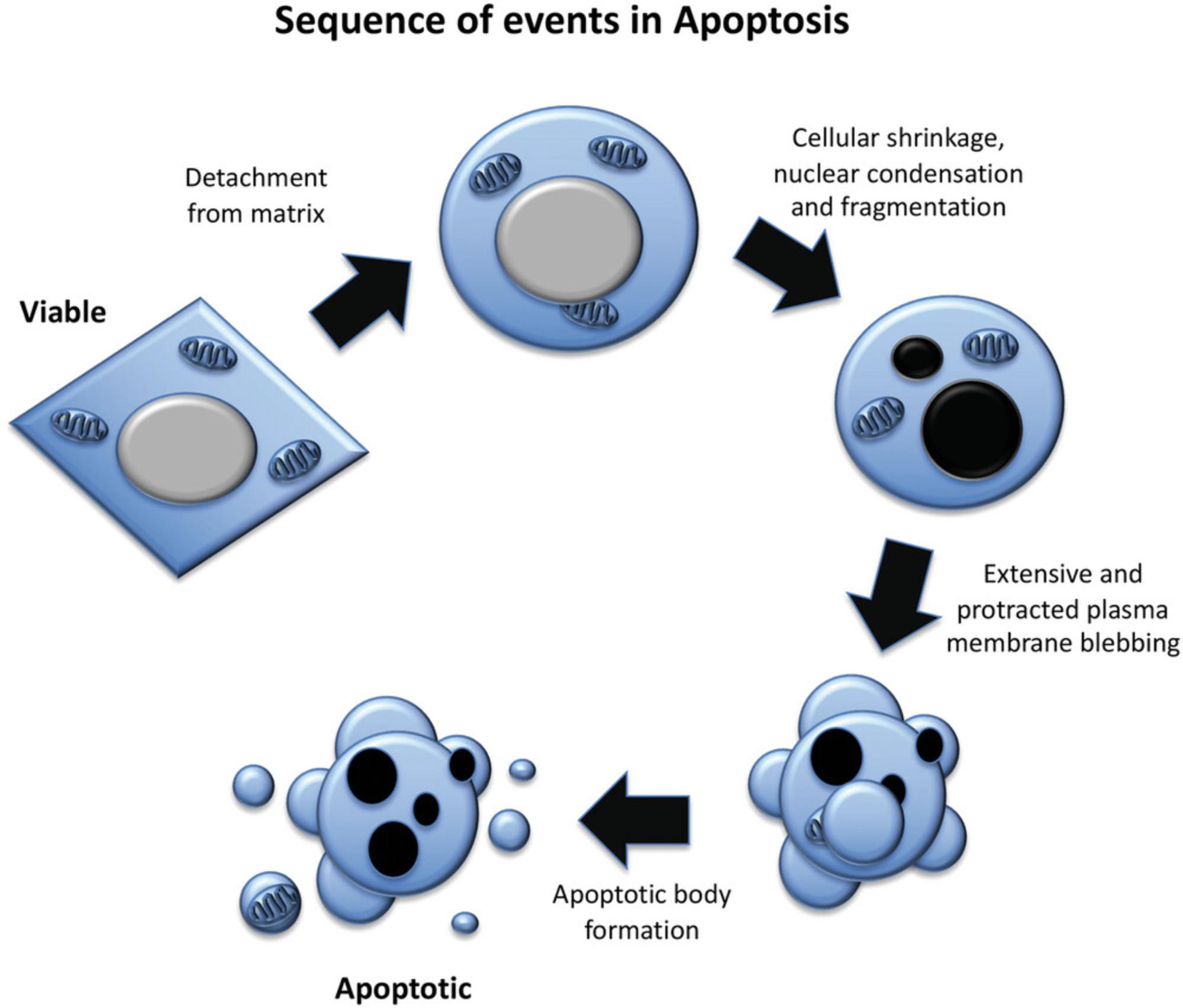
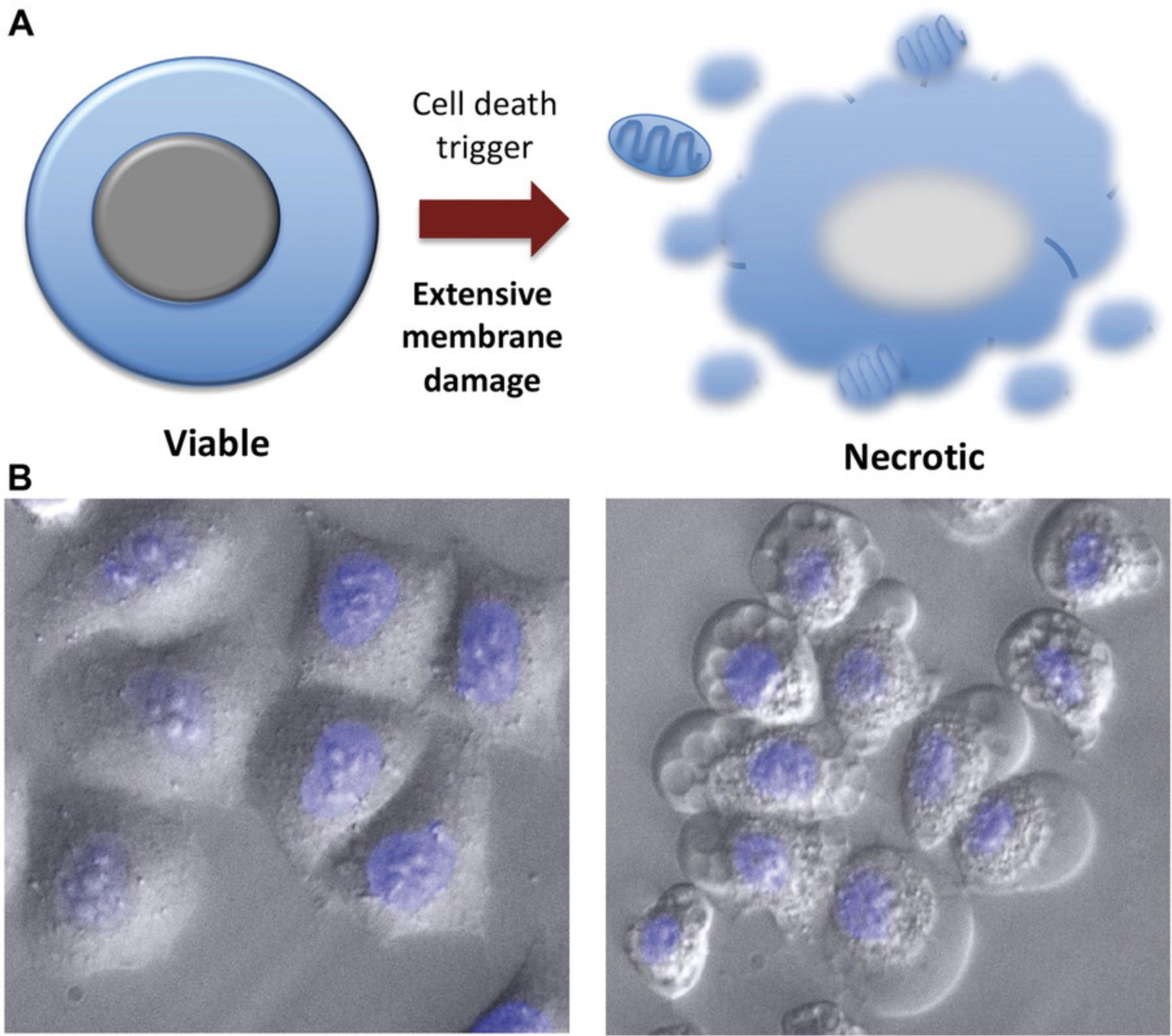
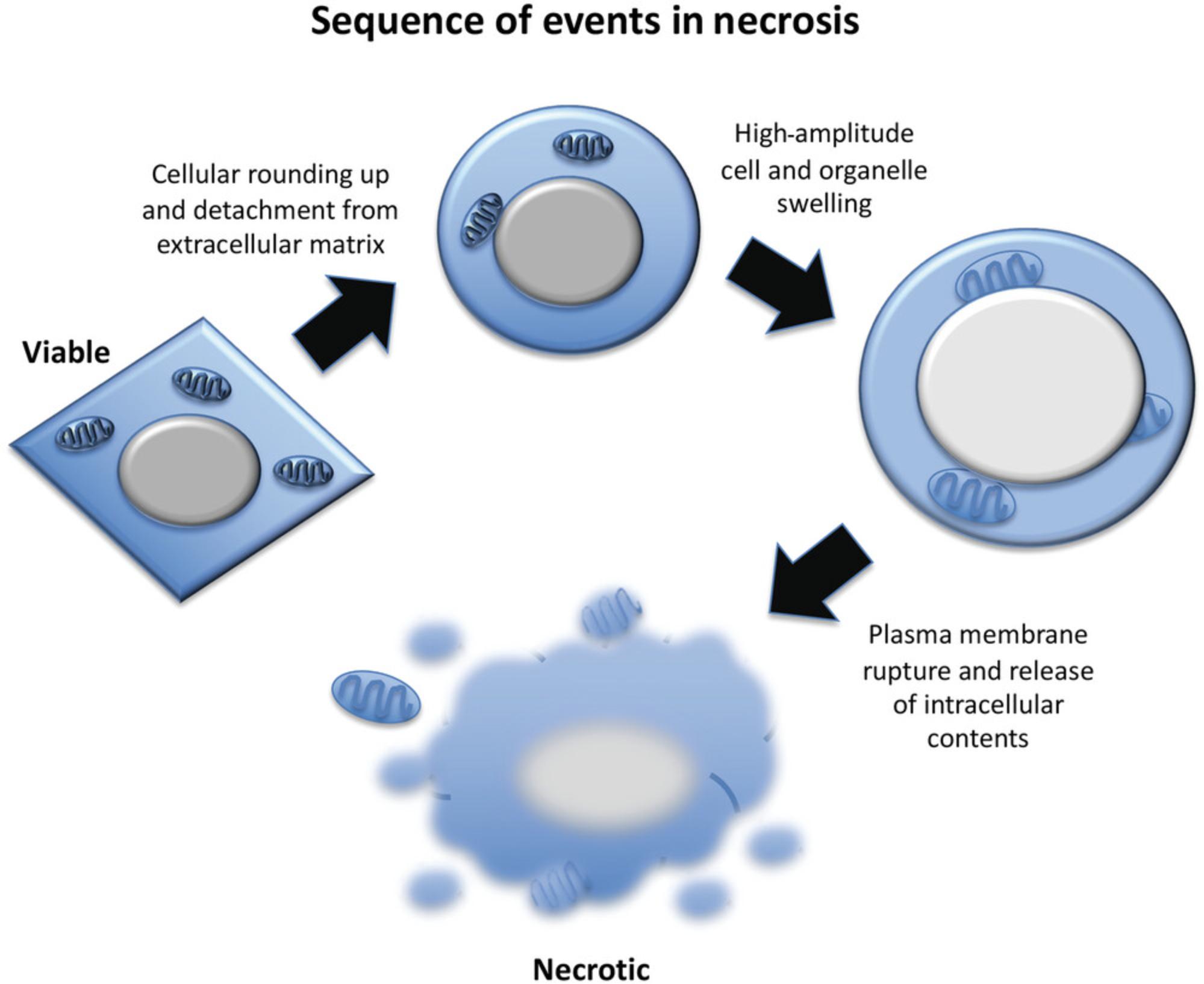
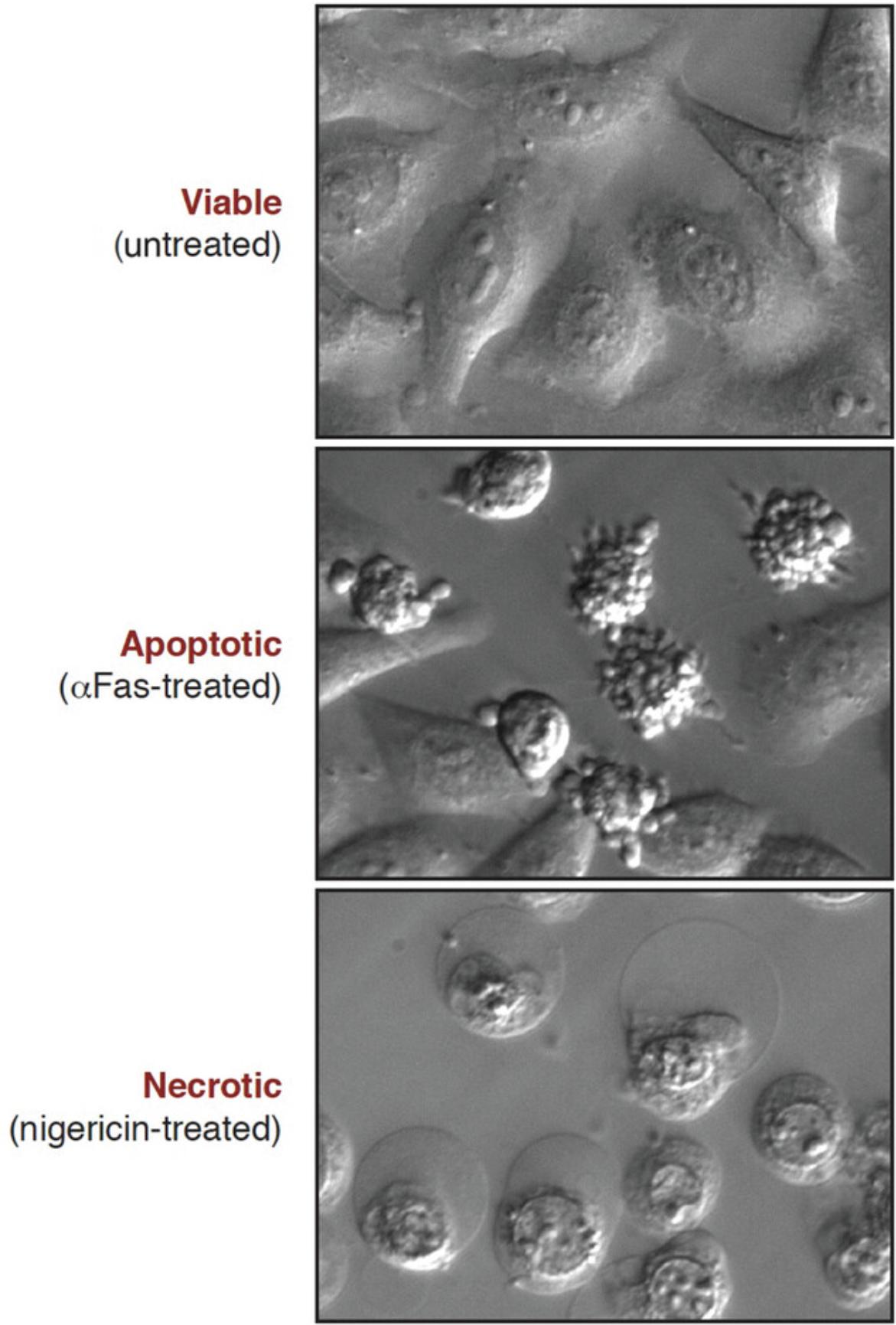
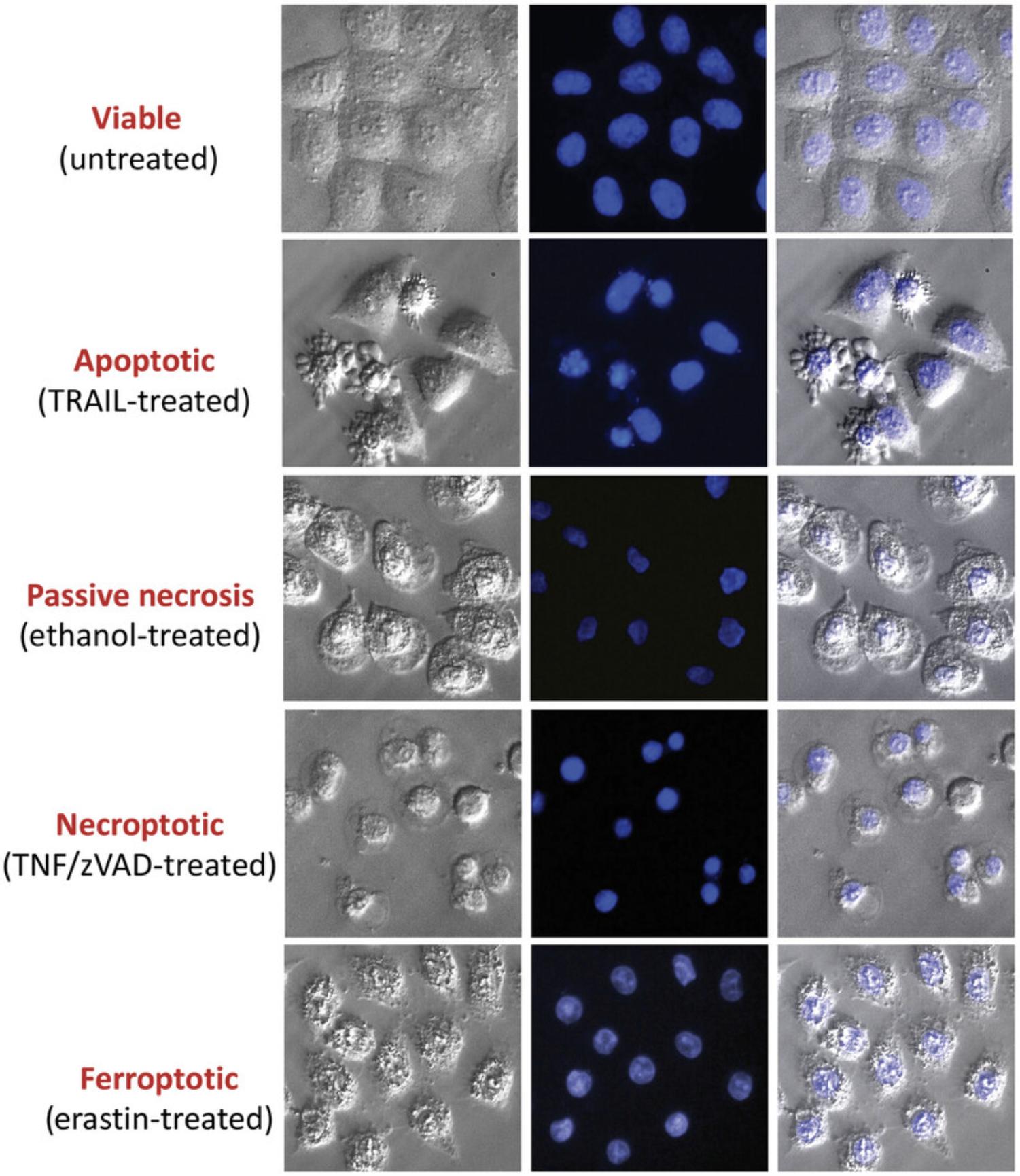
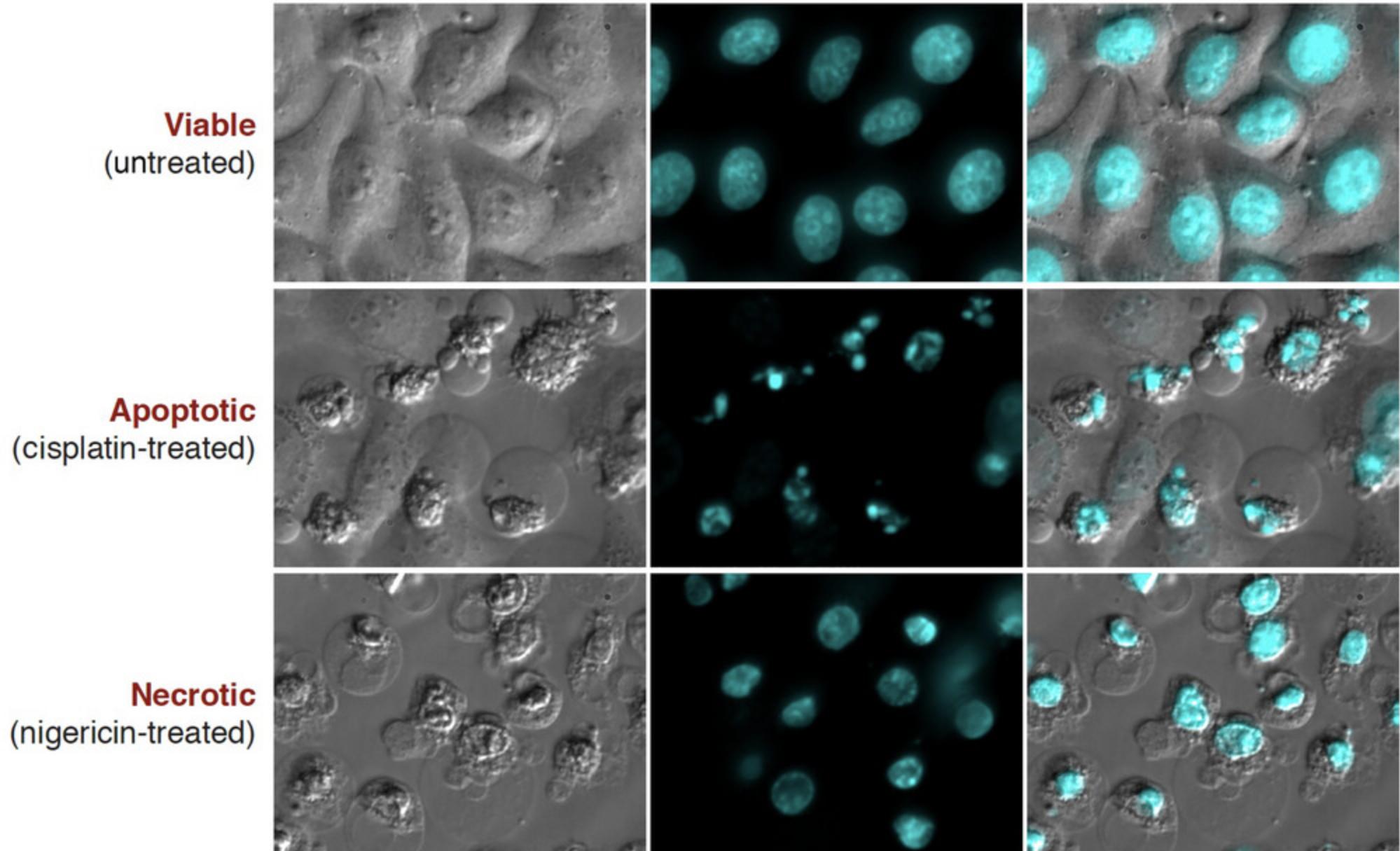
Microscopy is a simple yet powerful approach to identify the morphological alterations associated with apoptosis or necrosis (whether classical or programmed) and provides a rapid and unambiguous way to analyze and quantify cell death. Several characteristic features of apoptosis are readily observed by microscopy: the extensive membrane blebbing that frequently gives apoptotic cells an appearance similar to a bunch of grapes and the highly unusual chromatin compaction and nuclear fragmentation that occur during this mode of cell death (Figs. 1-3). These striking events are easy to quantify under phase-contrast (membrane blebbing) or fluorescence microscopy (DNA condensation and nuclear fragmentation). Similarly, necrotic cells are also relatively easy to identify from the striking morphological changes that sets them apart from healthy cells. In contrast to apoptotic cells, necrotic cells typically undergo rapid and high-amplitude swelling, with a swollen or “ballooned” morphology under phase-contrast microscopy (Figs. 4-6), that usually culminates in rupture of the plasma membrane within a few hours of exposure to the necrosis-inducing stimulus. By contrast, apoptosis usually occurs 8-20 hr after to exposure to a pro-apoptotic stimulus. Cells undergoing programmed necrosis (necroptosis, ferroptosis, pyroptosis) exhibit morphological changes essentially identical to those of cells undergoing nonprogrammed necrosis (Fig. 7), with the major difference being that programmed necrosis can be prevented through pharmacological inhibition or genetic inactivation of specific molecular pathways.
Flow cytometry is another widely used method to quantify cell death using a number of features of apoptotic or necrotic cells. The main approach involves the use of flow cytometry to evaluate the binding of fluorescently labeled annexin V, a probe for the plasma membrane phospholipid phosphatidylserine (PS), in tandem with propidium iodide (PI), which enters cells with damaged membranes. Because the redistribution of plasma membrane PS from the internal onto the external leaflet of the plasma membrane occurs early in apoptosis, before plasma membrane permeabilization, binding of annexin V to apoptotic cells typically precedes PI uptake, although PI uptake does eventually occur late in apoptosis after prolonged incubation when cells enter secondary necrosis. In sharp contrast, necrotic cells, which do not actively externalize PS, exhibit annexin V binding and PI uptake simultaneously due to rapid permeabilization of the plasma membrane during all forms of necrosis. In another flow cytometry approach, PI staining of permeabilized cells is used to measure DNA content on a per-cell basis, which permits the detection of the characteristic DNA fragmentation that occurs during apoptosis but not necrosis, leading to a distinct sub-G0/G1 DNA peak that is detectable in apoptotic cells. Although some limited DNA fragmentation is observed during necrosis, generally necrotic cells have a very similar DNA content to viable cells.
Aside from the basic quantification of apoptosis or necrosis based on cell morphology by phase-contrast microscopy (Basic Protocol 1), two additional microscopy methods are also presented. Alternate Protocol 1 describes how standard light microscopy can be used to measure the classical apoptotic or necrotic features using standard histological staining of fixed samples. Alternate Protocol 2 details a procedure for staining fixed samples to facilitate the observation of the typical nuclear changes associated with apoptosis or necrosis by fluorescence (UV) microscopy. In addition, a protocol for the preparation of fixed samples (Support Protocol) is also provided.
Microscopy and flow cytometry can also be used to measure biochemical properties of dying cells. Basic Protocol 2 uses flow cytometry to measure early plasma membrane alterations (PS externalization) associated with apoptosis, as well as the membrane permeabilization (as assessed by uptake of cell-impermeant fluorescent dyes such as propidium iodide) associated with necrosis, or very late apoptosis (i.e., secondary necrosis). This protocol can be used to discriminate between apoptosis and all forms of necrosis.
The characteristic hydrolysis of DNA into ∼200-bp multiples and the nuclear compaction and fragmentation that is observed during apoptosis can also be assessed by microscopy and flow cytometry. Necrotic cells do not manifest significant DNA hydrolysis, nuclear compaction, or fragmentation and so can be readily discriminated from apoptotic cells on this basis. Two protocols based on DNA content analysis (Basic Protocol 3) and DNA strand break measurement (Alternate Protocol 3) of fixed samples using flow cytometry are presented. Although more complex than Basic Protocol 3, Alternate Protocol 3 is more accurate and can also be used to stain cells for analysis by fluorescence microscopy.
Basic Protocol 4 describes a method to quantify cells containing activated caspases, proteases that are specifically activated during apoptosis but not classical necrosis, necroptosis, or ferroptosis. Fixed samples stained according to this procedure can be analyzed by flow cytometry, as outlined, or by fluorescence microscopy.
Finally, Basic Protocol 5 describes a method to discriminate between apoptosis, conventional necrosis, necroptosis, and ferroptosis using pharmacological inhibitors that can block or delay apoptosis, necroptosis, or ferroptosis but not nonprogrammed necrosis.
Basic Protocol 1: ANALYSIS OF CELL MORPHOLOGY BY PHASE-CONTRAST MICROSCOPY
Morphological changes to the cellular architecture are the most striking features of apoptosis and necrosis (Figs. 1-5). Membrane blebbing and cell shrinkage (associated with apoptosis) or membrane swelling and ballooning (associated with necrosis) can easily be detected by observing cells with a phase-contrast microscope (Figs. 6 and 7). Stimuli that initiate apoptosis will typically take 8-20 hr to provoke the latter changes in cells, whereas stimuli that provoke necrosis often elicit morphological alterations to cells within 2-3 hr. Quantification of cell death can be done directly from the culture dish without any processing of the sample. As a result, microscopy is probably the simplest and most accurate method to assess cell death and to discriminate between live, apoptotic, and necrotic cells. However, this approach does require knowledge of the distinct features displayed by apoptotic versus necrotic cells and involves manual cell counting, which can be time consuming. That said, this is the preferred method for rapidly and accurately quantifying apoptosis and necrosis in many laboratories, including our own. Numerous cytotoxic drugs and other stimuli can induce apoptosis, and it is useful to include a well-known pro-apoptotic agent as a positive control (Table 1), as the specific morphological features of apoptosis can vary between cell types. Because necrosis is largely a physical process due to plasma membrane rupture, followed by high-amplitude cellular and organelle swelling, the features of necrotic cells tend to be broadly similar even among diverse cell types (Fig. 7).
| Pro-apoptotic compound | Mechanism(s) of action | Concentration range | Source |
|---|---|---|---|
| TRAIL ligand | Activator of death receptor pathway (TRAIL receptors 1/2, DR4/5) | 10-200 ng/ml | Adipogen: Superkiller TRAIL AG-40T-0002-C020 |
| Fas ligand | Activator of death receptor pathway (Fas/CD95 receptor) | 50-200 ng/ml | Merck: anti-Fas antibody (human, activating), clone CH11 |
| Cisplatin | Inducer of DNA damage by DNA crosslinking | 10-200 μM | Merck: cisplatin PHR1624 |
| Etoposide | Inducer of DNA damage by topoisomerase 2 inhibition | 2-150 μM | Enzo Life Sciences: etoposide BML-GR307 |
| Daunorubicin | Inducer of DNA damage by intercalation between DNA base pairs | 5-10 μM | Enzo Life Sciences: daunorubicin hydrochloride, ALX-380-043-M010 |
| Doxorubicin | Inducer of DNA damage by intercalation between base pairs and inhibition of topoisomerase activity | 5-10 μM | Merck: doxorubicin hydrochloride, D1515 |
| Staurosporine | General protein kinase inhibitor | 0.2-1 μM | Merck: staurosporine from Streptomyces sp. S5921 |
| Actinomycin D | Inhibitor of RNA transcription | 5-10 μM | Merck: actinomycin D, A9415 |
| Cycloheximide | Inhibitor of protein translation | 5-100 μg/ml | Merck: cycloheximide, C1988 |
| Paclitaxel (Taxol) | Freezes microtubules during mitosis | 25-100 μM | Merck: paclitaxel, T1912 |
| Bortezomib | Proteasome inhibitor | 0.05-5 μM | Selleckchem: bortezomib (PS-341), S1013 |
| Carfilzomib | Proteasome inhibitor | 0.05-5 μM | Selleckchem: carfilzomib (S2853) PR-171 |
| MG-132 | Proteasome inhibitor | 10-50 μM | Merck: MG-132 474790 |
| ABT-737 | Inhibitor of anti-apoptotic BCL2 + BCLXL | 1-10 μM | Merck: BCL-2 inhibitor VI, ABT 737 197333 |
Materials
-
Cells of interest
-
Positive control pro-apoptotic agent (optional; Table 1)
-
Phase-contrast microscope
-
High-resolution camera (optional)
-
Hemacytometer (optional)
1.Count live, apoptotic, or necrotic cells (i.e., those that display extensive plasma membrane blebbing or extensive membrane swelling, as illustrated in Figs. 1, 4, 6, and 7) among 3 × 100 cells from at least three separate fields.
2.Calculate the percentage of apoptotic or necrotic cells as follows:
3.Use a high-resolution camera to image cells.
Alternate Protocol 1: ASSESSMENT OF MORPHOLOGICAL CHANGES USING EOSIN-METHYLENE BLUE STAINING
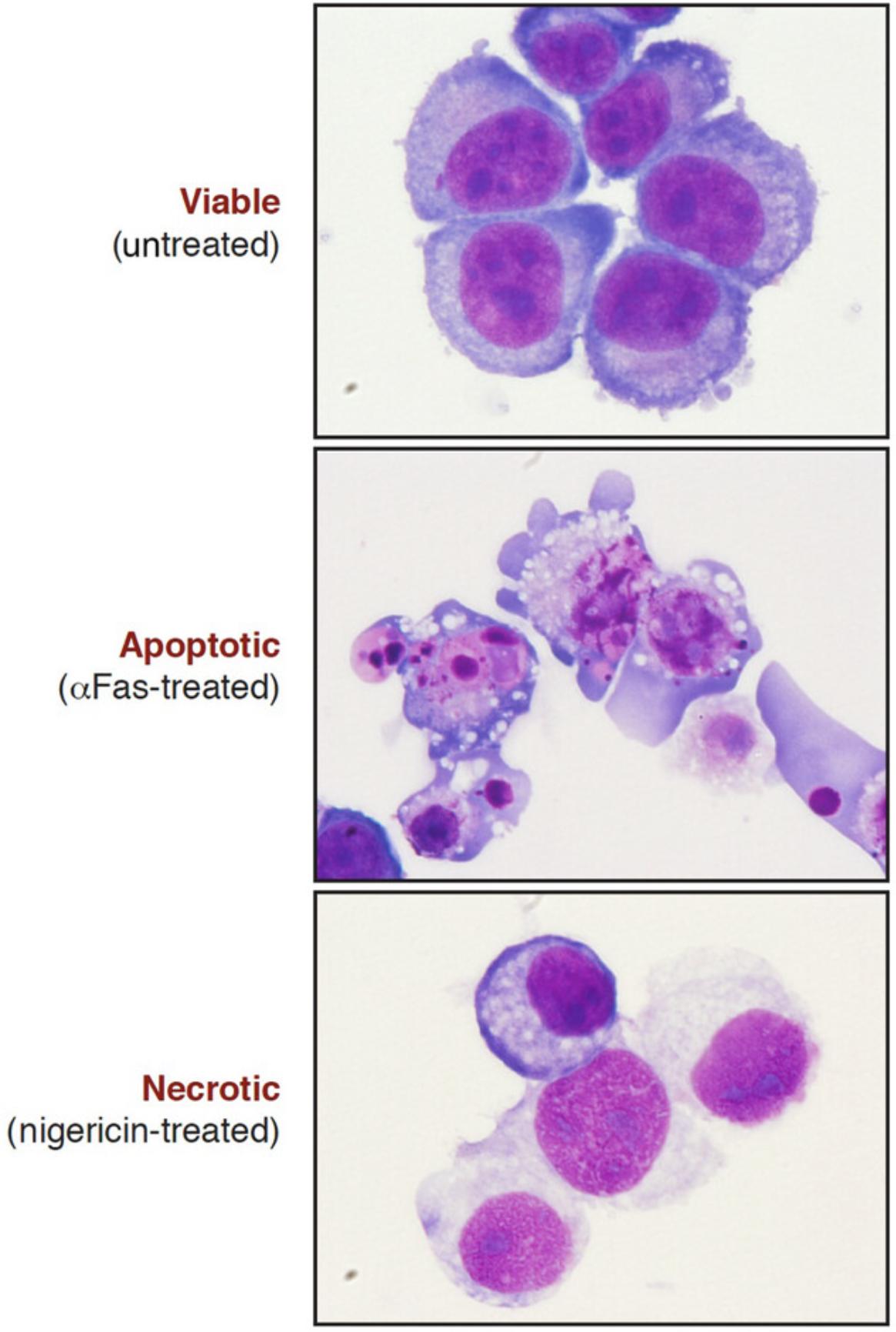
Differential staining of the nucleus and cytoplasm facilitates the high-power analysis of cellular morphology by light microscopy (Fig. 8). This can be achieved by eosin and methylene blue staining of cytospun and fixed cell preparations. The cell cytoplasm is stained purple/blue and the nucleus becomes pink after eosin and methylene blue staining. This method is more involved than direct cell counting from tissue culture but allows a more detailed inspection of nuclear morphology; and, because cells are fixed and mounted, slides can be stored and re-counted at a later date.
Materials
-
Cytospins (see Support Protocol)
-
100% methanol
-
1% (w/v) eosin Y staining solution (see recipe)
-
1% (w/v) methylene blue staining solution (see recipe)
-
DPX mounting medium
-
Coplin staining jar
-
Coverslips, no. 1.5 thickness
-
Phase-contrast microscope equipped with a high-resolution color camera
1.Fix cytospins with 100% methanol for 5 min at room temperature.
2.Transfer slides to a Coplin jar containing eosin Y solution. Stain for 30 s.
3.Drain excess eosin solution from the slides, and rinse gently in distilled water with a wash bottle.
4.Transfer slides to a Coplin jar containing methylene blue solution. Stain for 30 s.
5.Drain excess methylene blue solution from the slides, and rinse in distilled water.
6.Air-dry the slides.
7.Place a drop of DPX mounting medium on a coverslip and mount on the stained slide. Keep slides protected from light.
8.Analyze the slides with a phase-contrast microscope.
-
Count live, apoptotic or necrotic cells (i.e., cells with hypercondensed and fragmented nuclei [apoptosis] or swollen and decondensed nuclei [necrosis]) among 3 × 100 cells from at least three separate fields.
-
Take representative photographs using an eyepiece-mounted camera or equivalent.
Alternate Protocol 2: ANALYSIS OF NUCLEAR MORPHOLOGY BY FLUORESCENCE MICROSCOPY
The condensation and fragmentation of nuclei that occurs during apoptosis can easily be detected with fluorescent DNA-binding dyes such as Hoechst 33342 or propidium iodide (PI). This protocol describes the use of the cell-permeable dye Hoechst 33342 to stain fixed cells that have been grown on coverslips. Hoechst 33342 can be excited with a UV laser or a xenon or a mercury lamp of a fluorescence microscope and emits blue fluorescence. Cells stained in this way can then be counted, with apoptotic nuclei displaying characteristic hypercondensation and fragmentation into distinct globules (Fig. 9). In contrast, necrotic, necroptotic, or ferroptotic cells display a swollen, decondensed morphology that is very distinct from the features exhibited by apoptotic nuclei (Fig. 7). Hoechst 33342 can also be used to stain live as well as dead cells directly in the culture dish.
Materials
- Cells grown on coverslips (no. 1.5 thickness) or cytospins (see Support Protocol)
- Phosphate-buffered saline (PBS)
- 3.5% (w/v) paraformaldehyde (see recipe)
- 0.15% (v/v) Triton X-100 (see recipe)
- 2 μg/ml Hoechst 33342 staining solution (see recipe)
- Antifade fluorescence mounting medium (e.g., SlowFade or equivalent)
- Clear nail varnish
- Fluorescence (or confocal) microscope with a xenon or mercury lamp and a blue filter
1.Wash the cells grown on coverslips with 2 ml PBS.
2.Fix cells grown on coverslips (or cytospins) with 3.5% paraformaldehyde for 10 min at room temperature.
3.Wash cells with PBS for 5 min.
4.Repeat the washing step twice more.
5.Permeabilize cells with 0.15% Triton X-100 for 15 min at room temperature.
6.Wash cells with PBS for 5 min.
7.Incubate cells with 2 μg/ml Hoechst 33342 staining solution for 10 min.
8.Drain excess Hoechst solution.
9.Mount coverslip with a drop of antifade mounting medium. Seal the edges of the coverslip with a clear nail varnish.
10.Analyze under a fluorescence microscope using a blue filter or, alternatively, with a confocal microscope. Count live versus apoptotic (i.e., cells with highly compacted and fragmented nuclei) or necrotic (i.e., cells with swollen or decondensed nuclei) cells among 3 × 100 cells from at least three separate fields. Take representative photographs if desired.
Support Protocol: PREPARATION OF CYTOSPINS
Cytocentrifugation is a very useful method of adhering cells to slides (cytospins) for staining as well as for long-term storage. In this protocol, cells are spun at low speed onto glass slides while the liquid from the cell suspension is absorbed by a filter paper. After cytocentrifugation, cells should appear as a flat monolayer on the microscope slide. Cytospin preparations can be used for eosin and methylene blue staining (see Alternate Protocol 1) or classical immunofluorescence staining.
Materials
-
Single-cell suspension sample
-
Phosphate-buffered saline (PBS)
-
Hemacytometer
-
Centrifuge
-
Cytocentrifuge (e.g., ThermoShandon)
-
Cytospin slides (e.g., Sigma)
-
Cytospin filters (e.g., ThermoShandon)
-
Cytospin chambers (e.g., ThermoShandon)
Harvest cells
1.Harvest the cells and estimate their concentration using a hemacytometer.
2.Centrifuge 3 min at 400 × g , room temperature.
3.Resuspend the pellet with PBS at a concentration of 5 × 105 cells/ml.
Prepare cytospins
4.Remove the rotor from the cytocentrifuge.
5.Place the slides and filters into appropriate cytospin chambers with the filters on the top of the slides, facing the rotor.
6.Load 200 μl cell suspension into each cytospin chamber.
7.Place the rotor in the cytocentrifuge.
8.Centrifuge 3 min at 10 × g (250 rpm on a ThermoShandon Cytospin 4), room temperature.
9.Air dry the slides prior to staining.
Basic Protocol 2: MEASUREMENT OF PLASMA MEMBRANE COMPOSITION WITH ANNEXIN V AND PROPIDIUM IODIDE
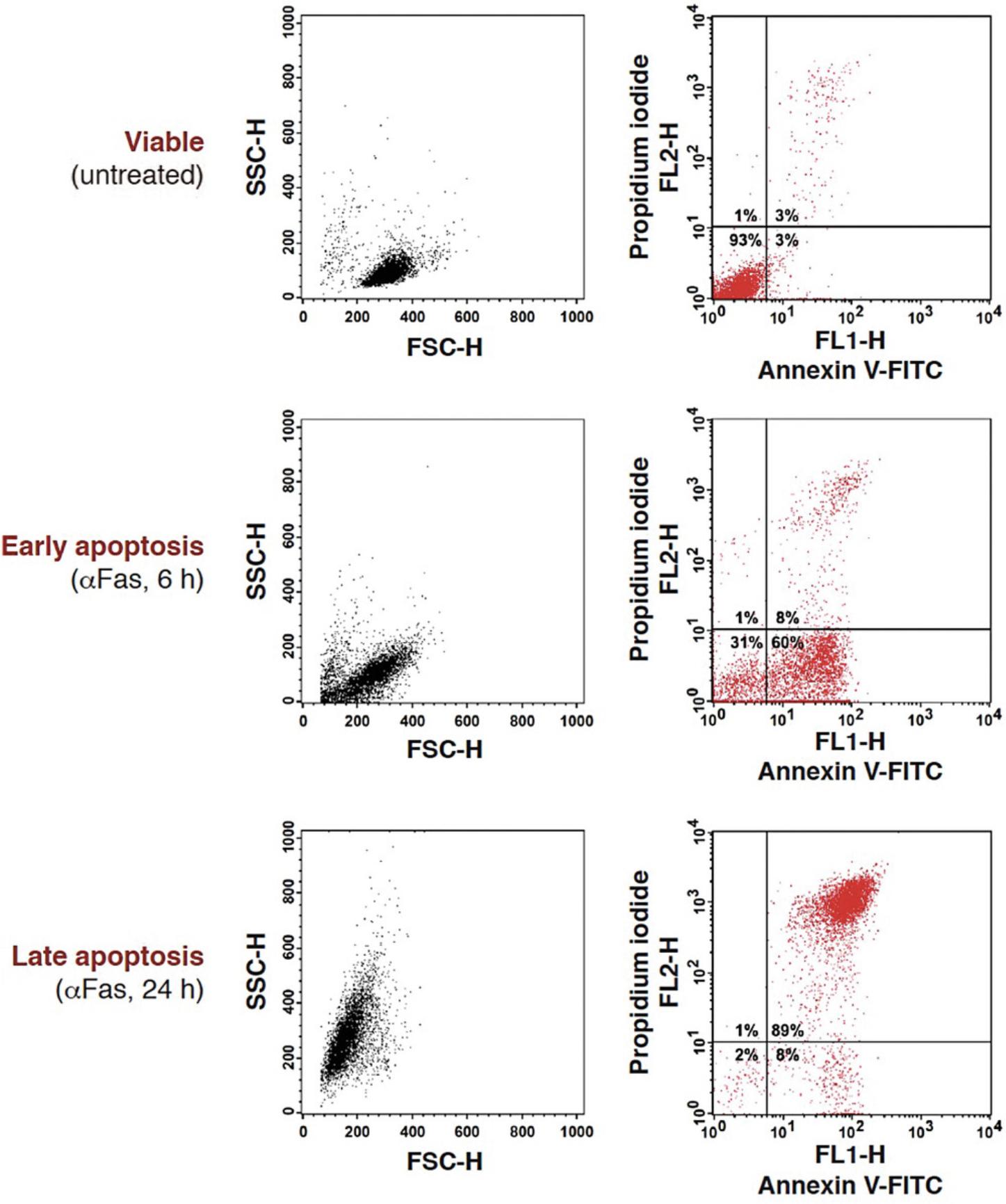
The annexin V binding assay is one of the most sensitive and widely used techniques to detect and distinguish between early and late apoptosis, as well as between apoptosis and necrosis. However, it is important to emphasize that annexin V staining does not discriminate between apoptotic and necrotic cells when plasma membrane permeabilization has occurred. In this assay, FITC-conjugated annexin V binds to phosphatidylserine (PS), a phospholipid normally maintained on the inner leaflet of the plasma membrane that is rapidly externalized on the plasma membrane of apoptotic cells. Because plasma membrane integrity is maintained in the early stages of apoptosis, the use of a vital dye such as propidium iodide (PI) enables discrimination between live cells (annexin V-negative and PI-negative), early apoptotic cells (annexin V-positive and PI-negative), and late apoptotic or necrotic cells (annexin V-positive and PI-positive; Fig. 10). This protocol can also be adapted for the analysis of annexin V-positive cells by fluorescence microscopy using cells grown on coverslips or suspension cell populations.
Materials
-
Single-cell suspension at a density of 106 cells/ml
-
Appropriate flow cytometry compensation control samples (see step 8)
-
1 μg/ml annexin V-FITC staining solution (see recipe)
-
1 mg/ml propidium iodide (PI) stock solution (see recipe)
-
Flow cytometry tubes
-
Centrifuge
-
Flow cytometer with a 488-nm argon-ion laser
Annexin V-FITC and propidium iodide staining
1.Harvest a sample of the cells to be analyzed (∼2 × 105 cells), and transfer to flow cytometry tubes.
2.Pellet the cells by centrifugation for 5 min at 400 × g , room temperature.
3.Resuspend the cell pellets with 300 μl of 1 μg/ml annexin V-FITC staining solution.
4.Incubate 15 min at room temperature.
5.Add PI stock solution to the cell suspension to achieve a final concentration of 10 μg/ml immediately before analysis.
Analysis by flow cytometry
6.Set up a flow cytometer for detection of the green fluorescence of annexin V-FITC with the FL1 detection channel and for detection of the red fluorescence of PI with the FL2 detection channel. Set both FL1 and FL2 channels in logarithmic (LOG) mode.
7.Prepare the following plot: single-parameter histogram (FL1-H versus cell counts) or two-parameter dot plots (FL1-H versus FL2-H) in logarithmic scale.
8.For dot plot analysis of annexin V-FITC and PI double-stained samples, adjust compensation to minimize spectral overlap between the green and the red channel.
9.Acquire ≥10,000 cells.
Basic Protocol 3: MEASUREMENT OF DNA FRAGMENTATION BY FLOW CYTOMETRY
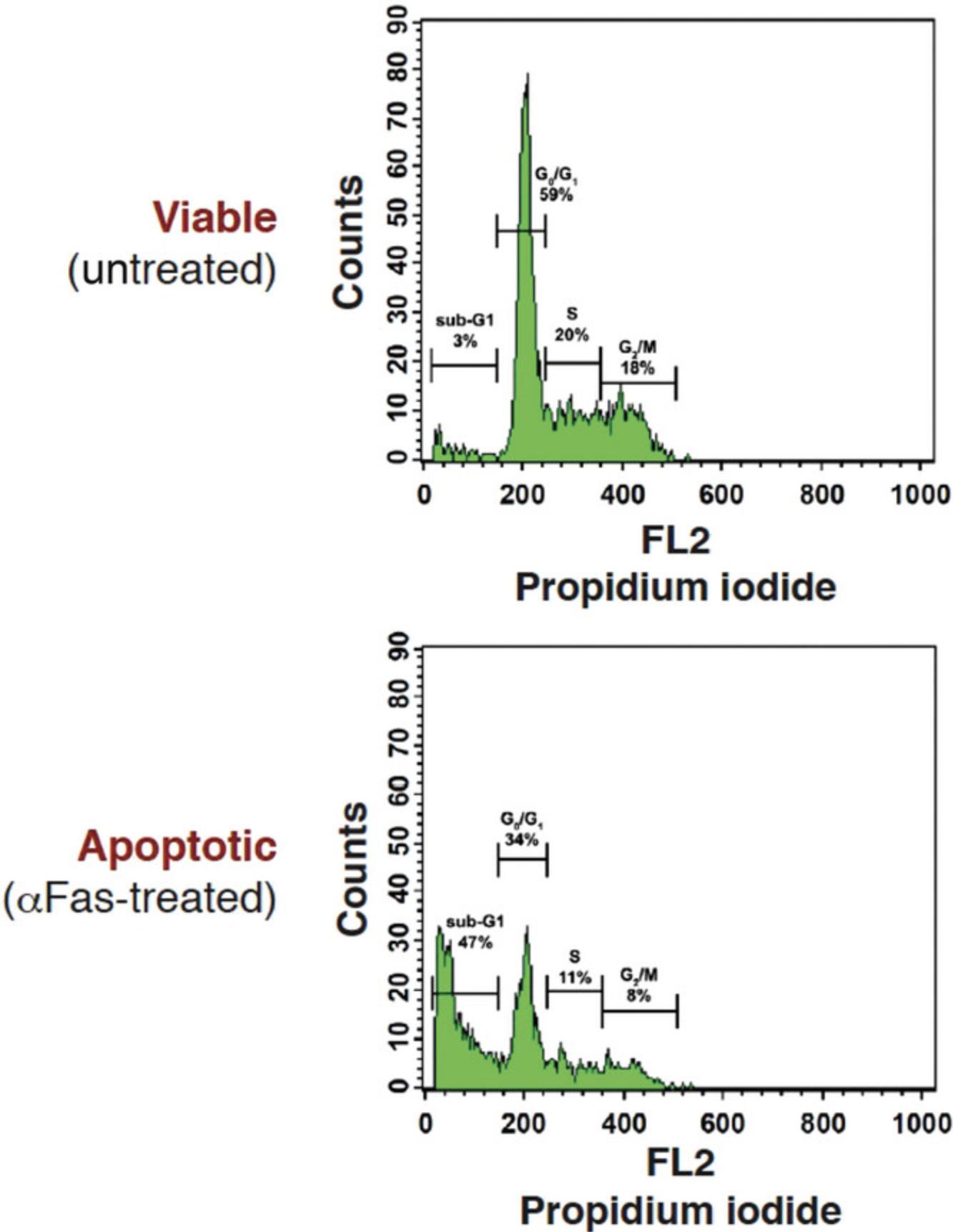
Intranucleosomal DNA fragmentation is a major hallmark of apoptosis, whereas necrotic cells exhibit little DNA fragmentation by comparison. Activation of caspase-activated endonucleases (CAD) during apoptosis results in the generation of low-molecular-weight (∼200 bp multiples) DNA fragments. In this protocol, the small DNA fragments of apoptotic cells are passively released with a phosphate-citrate buffer after fixation/permeabilization of the cells with ethanol. As a result, apoptotic cells display a markedly reduced DNA content (i.e., apoptotic cells appear below the typical 2 N DNA peak observed for the majority of viable cells in G1 phase of the cell cycle) upon analysis by flow cytometry. This can readily be measured by flow cytometry after staining with the DNA-intercalating dye propidium iodide (PI), which binds stoichiometrically to nucleic acids, resulting in a red fluorescence emission proportional to the DNA content of the cell. Using this assay method, apoptotic cells appear below the G1 peak on the DNA histogram and are therefore called sub-G1 (Fig. 11).
Materials
-
Single-cell suspension
-
Phosphate-buffered saline (PBS)
-
100% ethanol, ice cold
-
Phosphate-citrate wash buffer (see recipe)
-
50 μg/ml propidium iodide (PI) staining solution containing 100 μg/ml DNase-free RNase A (see recipe)
-
Centrifuge
-
1.5-ml microcentrifuge tubes Ice
-
Flow cytometry tubes
-
Flow cytometer with a 488-nm argon-ion laser
Propidium iodide staining
1.Harvest a sample (106 cells) of the cells to be analyzed.
2.Centrifuge 5 min at 400 × g , room temperature.
3.Wash cells with 1 ml PBS, and transfer to 1.5-ml microcentrifuge tubes.
4.Pellet the cells by centrifugation for 5 min at 400 × g , room temperature.
5.Gently resuspend the pellet with 300 μl PBS and then add 700 μl ice-cold absolute ethanol (i.e., 70% final concentration). Mix well by inverting tubes, and fix for at least 2 hr on ice.
6.Pellet the cells by centrifugation for 5 min at 400 × g , room temperature.
7.Gently resuspend the pellet with 1 ml phosphate-citrate wash buffer and incubate at room temperature for 10 min to allow DNA fragments to diffuse out of apoptotic cells. Pellet cells by centrifugation for 5 min at 400 × g , room temperature, and wash pellet once with 1 ml PBS.
8.Pellet the cells by centrifugation for 5 min at 400 × g , room temperature, and then resuspend with 500 μl PI staining solution containing RNase A. Incubate 30 min at room temperature or 15 min at 37°C.
9.Transfer the cells to flow cytometry tubes.
Analysis by flow cytometry
10.Set up the flow cytometer for detection of the red fluorescence of PI with the FL2 channel in linear (LIN) mode. Gate out cell aggregates by creating a gate on an FSC-H versus SSC-H dot plot.
11.Prepare the following plots: FSC-H versus SSC-H dot plot and FL2 histogram.
12.Gate out the doublets, cell debris, and cell aggregates from the analysis by creating a gate on the FSC-H versus SSC-H dot plot.
13.Plot the gated single cells on an FL2 histogram and measure the percentage of cells forming the sub-G1 peak that display a reduced fluorescence intensity compared to the G0/G1 peak.
14.Acquire ≥10,000 gated cells.
Alternate Protocol 3: ANALYSIS OF DNA FRAGMENTATION BY THE TUNEL ASSAY
Although detection of the sub-G1 cell population is a relatively easy way to determine whether cells have undergone apoptosis, this technique is usually considered a semi-qualitative method for apoptosis detection. This protocol provides an alternate method allowing the quantitative detection of cells displaying DNA fragmentation. The TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling) assay is more complex and significantly more expensive than the sub-G1 assay. In this protocol, cleaved ends of genomic DNA are labeled with 5-bromodeoxyuridine (BrdU) by the terminal deoxynucleotidyl transferase (TdT) enzyme. BrdU-labeled DNA breaks are subsequently detected with a FITC-conjugated anti-BrdU antibody. Here, BrdU is preferred to the traditional dUTP-biotin because it is more sensitive, cheaper, and easier to use.
Unlike the sub-G1 assay, the TUNEL assay requires a cross-linking step to prevent the release of cleaved DNA fragments. This is achieved with the use of the cross-linking fixative paraformaldehyde. BrdU-labeled cells can be quantitatively measured by flow cytometry. Alternatively, TUNEL staining can be performed on cells grown on coverslips for fluorescence microscopy analysis. In this case, counterstaining of the nuclei with propidium iodide or Hoechst 33342 can be performed to facilitate counting of TUNEL-positive nuclei.
Materials
-
Single-cell suspension or cells grown on coverslips (no. 1.5 thickness)
-
Phosphate-buffered saline (PBS)
-
3.5% (w/v) paraformaldehyde (see recipe)
-
0.15% (v/v) Triton X-100 (see recipe)
-
TdT equilibration buffer (see recipe)
-
TdT reaction solution (see recipe)
-
FITC-conjugated anti-BrdU staining solution (see recipe)
-
Centrifuge
-
1.5-ml microcentrifuge tubes
-
Ice
-
37°C incubator
-
Flow cytometer with a 488-nm argon-ion laser
Cell fixation and permeabilization
1.Harvest a sample (106 cells) of the cells to be analyzed.
2.Centrifuge 5 min at 400 × g , room temperature.
3.Wash cells with 1 ml PBS and transfer the cells to 1.5-ml microcentrifuge tubes.
4.Pellet cells by centrifugation for 5 min at 400 × g , room temperature.
5.Gently resuspend the pellet and fix cells with 1 ml of 3.5% paraformaldehyde for 10 min on ice.
6.Centrifuge 5 min at 400 × g , 4°C, and wash with 1 ml PBS.
7.Centrifuge 5 min at 400 × g , 4°C, and permeabilize with 1 ml of 0.15% Triton X-100 for 15 min at room temperature.
8.Centrifuge 5 min at 400 × g , room temperature, and wash with 1 ml PBS. Repeat centrifugation to pellet the cells.
TUNEL staining
9.Resuspend the cell pellet with 50 μl TdT equilibration buffer and equilibrate for 10 min at room temperature.
10.Centrifuge 5 min at 400 × g , room temperature, and resuspend with 50 μl TdT reaction solution.
11.Incubate 45 min at 37°C.
12.Centrifuge 5 min at 400 × g , room temperature, and wash the cells with 1 ml PBS.
13.Resuspend the cells with 100 μl FITC-conjugated anti-BrdU staining solution.
14.Incubate 1 hr at room temperature, protected from light.
15.Add 1 ml PBS, and centrifuge 5 min at 400 × g , room temperature.
16.Resuspend the cells with 300 μl PBS.
Analysis by flow cytometry
17.Set up a flow cytometer for detection of the green fluorescence of FITC with the FL1 detection channel.
18.Prepare a single-parameter histogram (FL1-H versus cell counts) in logarithmic scale and acquire ≥10,000 cells.
Basic Protocol 4: MEASUREMENT OF CASPASE ACTIVATION BY FLOW CYTOMETRY
Several members of the caspase family of cysteine proteases, particularly caspase-3 and caspase-7, play key roles in the phenotypic changes to the cell that occur during apoptosis (Figs. 1 and 3). In contrast, caspase are not typically activated during passive necrosis, ferroptosis, or necroptosis. Indeed, necroptosis is typically observed upon engagement of death receptors, such as TNFR, when caspase activity is inhibited. Activation of other caspases (i.e., caspase-1, -4, and -5) is, however, associated with pyroptosis (a programmed mode of necrosis that is typically associated with caspase-1-mediated activation of the pore-forming gasdermin D protein), and pyroptotic cells are best discriminated from apoptotic cells on the basis of morphological criteria (see Basic Protocol 1). Therefore, measuring caspase activity is an easy way to discriminate between apoptosis and most forms of necrosis, with the exception of pyroptosis. This can be achieved with the use of FAM-VAD-FMK, a cell-permeable caspase substrate. The tripeptide VAD, which is able to bind the active site of all caspase family members, is coupled to a fluoromethylketone (FMK) moiety, which is responsible for the covalent binding of the probe to the active site of catalytically active caspases. This inhibitor is conjugated with a green fluorescent carboxyfluorescein moiety (FAM); therefore, cells with active caspases will stain green, whereas cells with inactive caspases will remain unstained. In this protocol, cells are also counterstained with propidium iodide (PI) to allow discrimination between viable and dead cells by flow cytometry. However, as an alternative, cells can be fixed for UV or confocal microscopy analysis.
Materials
-
Single-cell suspension
-
Cell culture medium appropriate for the cells of interest
-
300 μM FAM-VAD-FMK FLICA reagent (e.g., Molecular Probes or ImmunoChemistry Technologies) in phosphate-buffered saline (PBS)
-
1 mg/ml propidium iodide (PI) stock solution (see recipe)
-
Centrifuge
-
1.5-ml microcentrifuge tubes
-
37°C incubator
-
Flow cytometer with a 488-nm argon-ion laser
FLICA and propidium iodide staining
1.Harvest a sample (typically 1–3 × 105 cells) of the cells to be analyzed.
2.Centrifuge 5 min at 400 × g , room temperature.
3.Resuspend the cells with 290 μl of the appropriate cell culture medium and transfer the cells to 1.5-ml microcentrifuge tubes.
4.Add 10 μl of 300 μM FLICA reagent to achieve a 10 μM final concentration.
5.Incubate 30 min at 37°C.
6.Add 1 ml cell culture medium and pellet the cells by centrifugation for 5 min at 400 × g , room temperature.
7.Wash the cells with 1 ml cell culture medium for 10 min at room temperature.
8.Centrifuge 5 min at 400 × g , room temperature.
9.Resuspend the cells with 300 μl cell culture medium containing 1 mg/ml PI to achieve a 10 μg/ml final concentration immediately before analysis.
Analysis by flow cytometry
10.Set up a flow cytometer for detection of the green fluorescence of FAM-VAD-FMK with the FL1 detection channel and for detection of the red fluorescence of PI with the FL2 detection channel. Set both FL1 and FL2 channels in logarithmic (LOG) mode.
11.Prepare the following plot: single-parameter histogram (FL1-H versus cell counts) or two-parameter dot plots (FL1-H versus FL2-H) in logarithmic scale.
12.For dot plots analysis of FAM-VAD-FMK and PI double-stained samples, adjust compensation with appropriate controls: unstained cells and FAM-VAD-FMK positively stained cells (apoptotic cells).
13.Acquire ≥10,000 cells.
Basic Protocol 5: DISCRIMINATING BETWEEN APOPTOSIS, NECROSIS, NECROPTOSIS AND FERROPTOSIS
Because caspases play a key role in apoptosis, the effects of broad-spectrum caspase inhibitors (such as zVAD-FMK, Emricisan, or qVD-OPh) can be used to determine whether a specific agent is promoting apoptosis, necrosis, necroptosis, or ferroptosis. Whereas caspase inhibitors typically inhibit apoptosis, or radically alter the morphological features of cells undergoing apoptosis, such inhibitors typically have little effect on conventional necrosis or ferroptosis and can even enhance or accelerate necroptosis.
Necroptosis is a mode of programmed necrosis that is typically only observed when caspases are inhibited and can be suppressed using a RIP kinase 1 inhibitor called necrostatin. Ferroptosis is a mode of programmed necrosis typically seen in response to agents that promote extensive lipid peroxidation and is normally suppressed by the lipid hydroperoxidase glutathione peroxidase 4 (GPX4). Ferroptosis can be discriminated from apoptosis, necrosis, and necroptosis through use of the radical-trapping antioxidants ferrostatin-1 or liproxstatin-1, which block this mode of cell death. Thus, assessing the effects of poly-caspase inhibitors, necrostatin, ferrostatin-1, and liproxstatin-1 on the morphology and kinetics of cell death induced by a specific stimulus is a relatively straightforward way to discriminate between apoptosis, necrosis, necroptosis, and ferroptosis.
Materials
- Cells of interest
- Cytotoxic agent under investigation
- zVAD-FMK (or alternative poly-caspase inhibitor; Table 2)
- Necroptotic inhibitor (Table 2)
- Ferroptotic inhibitor (Table 2)
- Positive control pro-apoptotic agent (optional)
| Inhibitors | Mechanism(s) of action | Concentration range | Source |
|---|---|---|---|
| Apoptotic Inhibitors | |||
| Z-VAD-FMK | Poly-caspase inhibitor | 5-20 μM | Bachem: Z-Val-Ala-DL-Asp-fluoromethylketone, 4026865 |
| Q-VD-OPh | Poly-caspase inhibitor | 5-20 μM | ApexBio: Q-VD-OPh, A1901 |
| Emricasan | Poly-caspase inhibitor, caspase 8 inhibitor | 1-10 μM | Merck: emricasan, SML2227 |
| Necroptotic Inhibitors | |||
| Necrostatin-1 (Nec-1) | Inhibitor of RIPK1 | 10-100 μM | Merck: necrostatin-1, N9037 |
| Necrostatin-1 Stable (Nec-1s) | Specific inhibitor of RIPK1 | 10-100 μM | Selleck: necrostatin 2 racemate (Nec-1s), S8641 |
| Necrosulfonamide | Binds to MLKL | 1-20 μM | Selleck: necrosulfonamide, S8251 |
| Ferroptotic Inhibitors | |||
| Ferrostatin | Lipid ROS scavenger | 1-20 μM | Merck: ferrostatin, SML0583 |
| Liproxstatin | Lipid ROS scavenger | 50-300 nM | Merck: liproxstatin-1, SML1414 |
| Deferoxamine | Iron chelator | 10-100 μM | Merck: deferoxamine mesylate salt, D9533 |
- Phase-contrast microscope
- Hemacytometer (optional)
- High-resolution camera (optional)
1.Incubate cells for 6-18 hr in tissue culture medium in the presence of the cytotoxic agent under investigation, alone as well as in the presence of zVAD-FMK (10 μM) or another poly-caspase inhibitor, necrostatin (10 μM), ferrostatin-1 (10 μM), or other inhibitors (see Table 2).
2.Under each condition, count live, apoptotic, and necrotic cells (as illustrated in Fig. 7) among 3 × 100 cells from at least three separate fields.
3.Calculate the percentage of apoptotic or necrotic cells in each condition as follows:
4.Use a high-resolution camera to image cells.
REAGENTS AND SOLUTIONS
Annexin V binding buffer
- 10 mM HEPES-NaOH, pH 7.4
- 150 mM NaCl
- 5 mM KCl
- 1 mM MgCl2
- 1.8 mM CaCl2
- Adjust to pH 7.4 with concentrated NaOH
- Store up to 6 months at 4°C
Annexin V-FITC staining solution, 1 μg/ml
Add 10 μg annexin V-FITC to 10 ml annexin V binding buffer (see recipe). Prepare fresh.
Eosin Y staining solution, 1%
- Add 1 g eosin Y to 100 ml water. Store for 12 months at room temperature.
FITC-conjugated anti-BrdU staining solution
Dilute an appropriate amount of the FITC-conjugated anti-BrdU (according to the manufacturer's instruction) in PBS (pH 7.2) containing 0.15% Triton X-100 and 1% BSA. Prepare fresh. Protect from light.
Hoechst 33342 staining solution, 2 μg/ml
Add 20 μl of 1 mg/ml Hoechst 33342 stock solution (see recipe) to 10 ml PBS (pH 7.4). Store up to several months at 4°C in foil-wrapped tubes.
Hoechst 33342 stock solution, 1 mg/ml
Dissolve 10 mg Hoechst 33342 (bisbenzimide H 33342 trihydrochloride) in 10 ml water. Store up to several months in a foil-wrapped tube at 4°C.
Methylene blue staining solution, 1%
Add 1 g of methylene blue to 100 ml water. Store for 12 months at room temperature.
Paraformaldehyde, 3.5%
Dissolve 3.5 g paraformaldehyde in 100 ml PBS (pH 7.4) in a foil-wrapped bottle. Heat the paraformaldehyde to 70°C on a stirring hot plate in a fume hood. Store up to a month at 4°C or aliquot into foil-wrapped tubes and freeze for long-term storage.
Phosphate-citrate wash buffer
- 200 mM Na2HPO4
- 100 mM citric acid
- Adjust to pH 7.4
- Store up to 6 months at 4°C
Propidium iodide (PI) staining solution, 50 μg/ml, containing 100 μg/ml RNase A (DNase-free)
Add 500 μl of 1 mg/ml PI stock solution (see recipe) and 1 mg RNase A to 10 ml PBS (pH 7.4). Prepare fresh.
Propidium iodide (PI) stock solution, 1 mg/ml
Dissolve 10 mg PI in 10 ml water. Store up to several months in a foil-wrapped tube at 4°C.
TdT equilibration buffer
- 25 mM Tris
- 0.2 M potassium cacodylate
- 1 mM CoCl2
- 1 mM dithiothreitol (DTT)
- 0.025% bovine serum albumin (BSA)
- Adjust pH to 6.6
- Store aliquots up to several months at –20°C
TdT reaction solution
- 25 mM Tris
- 0.2 M potassium cacodylate
- 1 mM CoCl2
- 1 mM DTT
- 0.025% bovine serum albumin (BSA)
- 80 μM BrdUTP
- 0.5 U/μl TdT enzyme
- Adjust pH to 6.6
- Prepare just before use on ice, protected from light
The enzyme should be added at the last minute just before use.
Triton X-100, 0.15% (v/v)
Dissolve 150 μl Triton X-100 in 100 ml PBS (pH 7.4). Store for up to several months at room temperature.
COMMENTARY
Background Information
Apoptosis is a mode of programmed cell death that is critical in multicellular organisms for the disposal of superfluous cells during development and also for the elimination of transformed, infected, and injured cells (Jacobson et al., 1997; Kerr et al., 1972). One of the main underlying forces driving apoptosis appears to be the avoidance of inflammation by ensuring that dying cells are recognized and engulfed before they can leak their intracellular contents into the extracellular space (Martin, 2016). Release of intracellular contents can provoke inflammation due to the liberation of pro-inflammatory molecules, called damage-associated molecular patterns (DAMPs), but this is avoided in apoptosis as cells are removed by phagocytes after the appearance of ‘eat me’ signals such as phosphatidylserine on the surface apoptotic cells.
Apoptosis is executed at the molecular level by a dedicated set of proteases called caspases (Lüthi et al., 2008; Slee et al., 1999). Activation of members of this family of cysteine proteases results in the proteolysis of hundreds of caspase substrates, leading to the controlled demolition of the cellular architecture (Martin et al., 2012; Taylor et al., 2008). Apoptosis is characterized by a series of striking morphological and biochemical alterations. During apoptosis, cells become rounded and detach from their neighbors. This is associated with extensive plasma membrane blebbing, which frequently leads to the release of small vesicular fragments of the cell, called apoptotic bodies (Fig. 3). Apoptosis is also accompanied by loss of plasma membrane lipid asymmetry, organelle fragmentation, chromatin condensation, DNA hydrolysis, and nuclear fragmentation (Jacobson et al., 1997; Taylor et al., 2008). As noted above, the ordered dismantling of apoptotic cells facilitates their removal by professional phagocytes and ensures minimal disturbance and damage to neighboring cells. Many of the above features of apoptosis, such as cell detachment, membrane blebbing, nuclear condensation, and fragmentation, can be observed under the microscope (Fig. 9).
The proteases belonging to the caspase family normally reside as inactive precursors in healthy cells. Apoptotic caspases are subdivided into two groups: the initiator caspases (caspase-2, -8, -9, and -10), which are activated as part of oligomeric complexes in response to particular pro-apoptotic stimuli, and the effector caspases (caspase-3, -6, and -7), which become activated after specific cleavage by the initiator caspases. The effector caspases are responsible for the controlled demolition of cellular structures (reviewed in Stennicke & Salvesen, 1998).
During apoptosis, a profound rearrangement of the cytoskeletal network occurs under the control of activated caspases. Several microtubule and microfilament proteins and their associated proteins are caspase substrates, and their proteolysis results in the disruption of the cytoskeletal network. These substrates include proteins such as actin, gelsolin, myosin, tubulin, and fodrin (Chen et al., 1996; Gerner et al., 2000; Kothakota et al., 1997; Martin, O'Brien et al., 1995). Although the caspase-mediated cleavage of these cytoskeletal components probably promotes the cell rounding and shrinking seen during apoptosis, it is important to note that the link between proteolysis of the above substrates and any functional outcome during apoptosis has been inferred rather than conclusively demonstrated (reviewed in Taylor et al., 2008).
Proteolytic cleavage of the Rho-associated kinase ROCK-I is one exception to this. ROCK kinases are key regulators of actin cytoskeleton organization and are responsible for cell motility (Maekawa et al., 1999). Caspase-mediated proteolysis of ROCK-I removes its C-terminal auto-inhibitory domain, generating a constitutively active form of ROCK-I. Active ROCK-I phosphorylates myosin light chains, increasing the contraction of the actin cytoskeleton and leading to plasma membrane blebbing (Coleman et al., 2001; Sebbagh et al., 2001). Interestingly, ROCK-mediated membrane blebbing seems to be dispensable for efficient phagocyte clearance, but not for plasma membrane remodeling (Shiratsuchi et al., 2002).
Recognition and removal of apoptotic cells by phagocytes is provoked by changes to the plasma membrane of apoptotic cells that occur early in apoptosis, before membrane permeabilization. These changes trigger the recognition and engulfment of early apoptotic cells (Fadok et al., 1992), minimizing the release of intracellular contents (especially DAMPs) that can initiate inflammatory responses upon liberation into the extracellular space (Martin, 2016). One of the key triggers of macrophage recognition is the externalization of phosphatidylserine (PS) on the outer leaflet of the plasma membrane, which occurs before loss of plasma membrane integrity and uptake of vital dyes (Martin, Reutelingsperger et al., 1995). PS is normally maintained on the cytoplasmic leaflet of the plasma membrane in healthy cells by P-type ATPases, an ATP-dependent family of flippases (reviewed by Leventis & Grinstein, 2010). During apoptosis, PS exposure is caspase-dependent and can be inhibited by poly-caspase inhibitors such as zVAD-FMK (Martin et al., 1996). This suggests that one or more proteins that are involved in maintaining the asymmetry of the plasma membrane may be cleaved by caspases. Cleavage of the Xk-related protein 8 (Xkr8) by caspases has been shown to promote the externalization of PS during apoptosis (Suzuki et al., 2013). Thus, PS externalization during apoptosis promotes the clearance of apoptotic cells, preventing membrane rupture, release of cytoplasmic contents, damage to neighboring cells, and the initiation of inflammation.
Another highly noticeable characteristic of apoptosis that occurs during the early phase of death is the fragmentation of the nucleus. Disintegration of the nuclear lamina and generation of contractile forces by the actin cytoskeleton seem to be the key events leading to nuclear fragmentation. Coating the nuclear side of the nuclear envelope, the nuclear lamina is an intermediate filament network essentially composed of two types of lamins (A and B). The lamina forms a structural scaffold for the nucleus. It provides structural integrity to the nuclear envelope, connecting the chromatin (on the nuclear side) and the actin cytoskeleton (on the cytosolic side) to the nuclear envelope (reviewed by Burke & Stewart, 2013). During apoptosis, the lamins are targeted by caspase-dependent proteolysis (Rao et al., 1996). However, weakening the lamina integrity does not cause the nucleus to passively fall apart. To ensure fragmentation, the nucleus must be physically torn apart. The forces required to achieve this are again delivered by the contraction of the actin bundle that is in part generated by ROCK-I-mediated phosphorylation of the myosin light chains (Croft et al., 2005). Interestingly, inhibition of microtubule contraction, in addition to preventing apoptotic bodies formation, also blocks the relocation of nuclear fragments to bleb regions, suggesting that cytoskeletal rearrangements are critical for efficient nuclear breakdown (Moss et al., 2006).
Before the fragmentation of the nucleus, the chromatin is subject to profound rearrangements. Genomic DNA cleavage at internucleosomal sites was one of the first biochemical markers of apoptosis to be characterized (Williams et al., 1974). In apoptosis, DNA is hydrolyzed at ∼180-bp intervals by a nuclease called caspase-activated DNase (CAD; Enari et al., 1998). During apoptosis, CAD is released from its inhibitor, ICAD, and activated as a result of caspase-mediated cleavage of ICAD (Liu et al., 1997; Sakahira et al., 1998). CAD–/– cells fail to undergo oligonucleosomal DNA fragmentation, although cell death otherwise proceeds (Samejima et al., 2001). Caspases are also required for the chromatin condensation that usually accompanies DNA fragmentation of apoptotic cells. Chromatin condensation observed within apoptotic nuclei seems to involve the phosphorylation of histone H2B, a core component of nucleosomes, by a caspase-activated kinase known as mammalian sterile 20-like kinase 1 (Mst1; Cheung et al., 2003; Ura et al., 2001).
Passive necrosis is typically initiated by high concentrations of toxins, cytotoxic drugs, reactive oxygen species, extremes of temperature, physical injury, and other severe insults (Wyllie et al., 1980). This mode of death is not under specific molecular control but appears to be the direct result of severe plasma membrane damage leading to permeabilization of the plasma membrane. During necrosis, influx of water and efflux of intracellular potassium and other small molecules are swiftly followed by high-amplitude cell swelling, leading to rupture of the plasma membrane and release of soluble intracellular contents into the extracellular space (Wyllie et al., 1980). Because of the extreme nature of the conditions that provoke passive necrosis, this mode of cell death is usually observed under severe pathological conditions and affects large numbers of cells simultaneously. Consequently, necrosis typically occurs very rapidly (within 2-3 hr) after exposure to a necrosis-inducing stimulus and affects numerous cells in tandem. This contrasts sharply with apoptosis, in which apoptotic cells usually initiate this process asynchronously and over a prolonged period of time (8-20 hr). Unlike apoptosis, necrosis is a highly inflammatory mode of cell death due to the release of pro-inflammatory molecules, called damage-associated molecular patterns (DAMPs), which include most IL-1 family members (reviewed in Martin, 2016; Martin et al., 2022). Although passive necrosis is not under molecular control, recent data suggest that the final phase of passive necrosis may be modulated in some circumstances by the pore-forming protein ninjurin1 that is activated as a consequence of cell swelling (Kayagaki et al., 2021).
Necroptosis is a mode of programmed necrosis that is typically seen in response to engagement of members of the death receptor subset of the TNFR family (TNFR1, CD95/Fas, TRAIL-R1/2) where caspase-8 activation is inhibited, due to either pharmacological caspase inhibitors (e.g., zVAD-FMK, emricisan), genetic inactivation of caspase-8 or the caspase-8-activating protein FADD, or the inhibition of caspases by virus-encoded caspase inhibitors (e.g., Crm-A). During necroptosis, activation of the FADD-binding kinase RIPK1 leads to the formation of a complex with RIPK3 (called the necrosome), followed by RIPK3-mediated phosphorylation and activation of the pore-forming protein MLKL (Vanden Berghe et al., 2014). Activation of MLKL leads to its oligomerization and the formation of MLKL pores, followed by plasma membrane rupture (reviewed in Kearney & Martin, 2017). It is currently unclear how commonly necroptosis is induced under physiological conditions, but this is an area of active investigation. Necroptosis can usually be inhibited by means of small molecules (e.g., necrostatin-1, see Table 2) that block the kinase activity of RIPK1 and the formation of the necrosome (RIPK1/RIPK3) complex (Degterev et al., 2005, 2008).
Ferroptosis is a mode of programmed necrosis that is initiated via the iron-dependent accumulation of lipid hydroperoxides that promote plasma membrane rupture. This is normally prevented by the hydroperoxide-detoxifying enzyme GPX4, and ferroptosis can be induced through GPX4 inactivation or glutathione depletion (Dixon et al., 2012; Friedmann Angeli et al., 2014). The accumulation of lipid hydroperoxides can occur by two major mechanisms: autoxidation due to iron-catalyzed spontaneous peroxyl radical generation, or oxidation due to enzyme-catalyzed (non-heme) iron-dependent lipoxygenases (Jiang et al., 2021; Stockwell et al., 2017). It is currently unclear whether ferroptosis is a bona fide cell death mechanism or simply a detoxification pathway that results in necrosis when overwhelmed.
Critical Parameters and Troubleshooting
Controls
Appropriate controls are required for the successful analysis and interpretation of any biological assay. Negative controls, consisting of the cells of interest left untreated and processed similarly to the samples to be tested, allow several experimental parameters to be controlled. These include determination of the background cell death level, estimation of the autofluorescence background of the sample, and verification that the experimental procedure did not generate any false-positive result. It is also essential to be able to discriminate between a negative result (absence of apoptosis or necrosis) and an ineffective detection of apoptosis due to a technical issue. To this end, positive controls, consisting of the cells under investigation treated with a stimulus known to induce apoptosis in the cells under investigation (see Table 1), should be processed in parallel with the tested samples. It can also be useful to consider using a positive control for necrosis, in which case an appropriate dose of hydrogen peroxide (typically 200 to 300 mM) or alternatively one freeze/thaw cycle should be used to treat the cells. Such positive controls are also helpful for acquiring experience in recognizing the morphological features of apoptotic and necrotic cells. Finally, positive and negative controls are necessary to properly adjust flow cytometer settings.
The expression of exogenous proteins is frequently used to analyze the function of a specific gene or to dissect a particular signaling pathway. With the exception of some transformed cell lines, transfection efficiency often becomes a limiting factor for the analysis of transfected cells. Cotransfection of the plasmid of interest along with a reporter plasmid encoding a green fluorescent protein (GFP) or a red fluorescent protein (mCherry or DsRed) easily allows the identification of the transfected cells when performing single-cell analysis using a fluorescence microscope or flow cytometer. When using a fluorescence microscope, the investigator can then simply restrict the analysis to the green or red cells. When cell death analysis is carried out by flow cytometry using a fluorescent reporter gene, a gate selecting the green or red cells can be created to restrict the analysis to only the transfected population.
Cell concentration
Although fluorescence microscopy analysis generally allows a qualitative analysis of the fluorescence of a cell population, flow cytometry is a powerful technique to accurately measure the fluorescence intensity emitted at the single-cell level. Precision is thus required to obtain reproducible staining with minimal intersample variations. This is particularly crucial when using propidium iodide (PI) to measure cellular DNA content. Because PI binds to DNA in a stoichiometric manner, DNA staining intensity may vary depending on the availability of DNA-binding sites and thus is sensitive to cell number. If a very high concentration of cells is used for the sub-G1 assay, PI availability might become limiting. Consequently, intersample variation in cell numbers could affect sample-to-sample staining intensity and prevent the investigator from comparing the samples with each other. The concentrations of the fluorescent probes used in this unit are usually optimal at cell densities of up to 106 cells. Being able to measure the cell concentration and appropriate volumes of the dyes with precision is essential to ensure consistency and reproducibility.
Fixation
A wide variety of different fixation methods are available and routinely used for microscopy and flow cytometry. Fixatives have different properties and achieve different effects on cellular constituents. Alcohol-based fixation procedures will dehydrate cells, remove membrane lipids, and frequently alter protein structures. Cross-linking fixatives (such as paraformaldehyde) are less aggressive and are thus usually preferred in the case of immunostaining protocols (especially of membrane-bound proteins). Another major difference between alcohol-based fixatives and cross-linking fixatives is their ability to permeabilize cells. In contrast to ethanol or methanol fixing solutions, paraformaldehyde is much more protective of membrane integrity. As a consequence, a permeabilization step (generally using 0.15% Triton X-100) is required before staining with Hoechst 33342 or before immunostaining.
For cell cycle analysis, ethanol is preferred over any cross-linking fixatives for several reasons. First, paraformaldehyde cross-links the chromatin and stabilizes chromatin structure, thus reducing the accessibility of DNA to PI. As a consequence, the accurate measurement of DNA content based on stoichiometric binding of PI to DNA is compromised and the resolution of the various peaks is reduced. Second, DNA fragments generated during apoptosis are trapped within the cross-linked chromatin and are not efficiently extracted during the wash steps. The use of paraformaldehyde thus compromises the generation of the sub-G1 peak.
Although fixation is a useful procedure for long-term storage of the sample and for immunostaining, it is also, unfortunately, incompatible with some protocols. This is the case when PI is used as a vital dye in the annexin V/PI assay as cells should never be fixed before or after PI staining. If fixation is performed after incubation with PI, the dye will diffuse out of dead cells.
Conversely, if fixation is performed before PI staining, all cells, live and dead, become permeable to PI. Similarly, it is not recommended to fix cells before performing the annexin V test. The fixation-permeabilization step would allow the annexin V to diffuse into the cells and to stain live, apoptotic, and necrotic cells in a similar way. However, if required, cells can still be fixed after staining with annexin V, in which case paraformaldehyde made with a calcium-containing buffer should be used.
Experimental endpoint
The choice of the endpoint for an experiment is a crucial parameter when analyzing cell death. Apoptosis tends to take 8-20 hr to occur, whereas passive necrosis, necroptosis, or ferroptosis tend to occur rapidly, within 2-3 hr of application of the stimulus. As apoptosis is proceeding, morphological and biochemical alterations progressively appear and accumulate over a number of hours, and individual cells typically manifest these morphological alterations independently. In contrast, where passive or programmed necrosis is occurring, most cells in the culture will typically exhibit similar morphological alterations (e.g., swelling, ballooning, plasma membrane rupture) in tandem. In vitro , advanced stages of apoptosis are characterized by plasma membrane permeabilization, which can lead to a necrotic-like morphology. This late apoptotic stage is called secondary necrosis, a stage that cells normally don't reach in vivo as phagocyte-mediated clearance is normally very efficient. Cells that have undergone secondary necrosis present with necrotic characteristics such as permeability to vital dyes but also retain some apoptotic features such as nuclear condensation and fragmentation. Thus, it can become difficult to discriminate between apoptosis and necrosis when apoptosis is in the very late stages.
Usually, morphological and biochemical features of apoptosis (e.g., blebbing, nuclear fragmentation, DNA condensation, DNA fragmentation, caspase activation, PS externalization) become detectable within 6 to 8 hr after treatment, depending on the pro-apoptotic stimulus used, the stimulus dose, and the cell line. It is highly recommended to perform time course experiments to successfully measure and characterize cell death.
Fluorescence analysis
One of the most basic precautions that has to be taken regarding the preparation of fluorescently labeled samples is to protect the fluorescent probe and stained samples from light. Prolonged exposure of a fluorescent probe to direct light or to laser light will dramatically reduce the fluorescence emitted by the dye. It is also highly advisable not to overexpose the sample to the laser light to prevent photobleaching, a process that could irreversibly inhibit the fluorescent property of the dye. In the case of the preparation of fixed samples for fluorescence or confocal microscopy analysis, it is thus essential to use a mounting medium containing an antifade reagent to reduce the incidence of photobleaching.
Taking single fluorescence intensity measurements is extremely easy using a flow cytometer. However, accurate fluorescence recording is slightly more complex, and some fluorescence properties must be taken into account when analyzing multicolor samples. Under excitation with an appropriate fluorescent laser, fluorochromes emit photons at different wavelengths, forming an emission spectrum. Although a specific maximal emission wavelength can be determined for each fluorescent molecule, emission spectra frequently cover a large range of wavelengths. This is notably the case for green-fluorescence-emitting probes such as FITC, Alexa 488, or fluorescein, which have emission spectra that are prolonged toward the red wavelengths. As a consequence, green and red fluorochromes often present with an emission spectra overlap in the red wavelength range, thus complicating the accurate measurement of individual fluorescence intensity with the red channel of the flow cytometer. To overcome this potential source of artefact, the flow cytometer can be set up to subtract a fraction of the green signal from the red signal detected by the red channel, a procedure called color compensation. The preparation of appropriate controls is indispensable to properly adjust the compensation parameters. These controls include unstained cells and green-positive cells (i.e., apoptotic cells stained with annexin V-FITC or FLICA, depending on the protocol).
Annexin V-FITC/PI staining
The annexin V assay is relatively straightforward to perform. The main critical parameter with this protocol is the Ca2+ concentration (1 to 3 mM) in the annexin V binding buffer: too low a concentration will prevent the binding of annexin V to phosphatidylserine, whereas too high a concentration can promote binding to other phospholipids. Attention must also be paid to the incubation duration with PI, as prolonged exposure leads to PI uptake by viable cells and generates false-positive results.
Sub-G1 analysis
As previously mentioned, the use of ethanol as a fixative is crucial for the stoichiometric staining of DNA with PI and for the extraction of DNA fragments generated during apoptosis. However, it is important to note that the use of 100% ethanol is not recommended as it will favor the formation of cell clumps, which affect the measurement of the DNA content and the generation of the sub-G1 peak. If such cell aggregates are present in the sample, they need to be removed from the analysis as detailed in Basic Protocol 3.
Other parameters are likely to influence the resolution of the sub-G1 peak. These include the efficiency of the washing step following fixation and the use of RNase A. As PI can bind RNA as well as DNA, it is important to treat the sample with RNase A so as to accurately measure the DNA content only.
Finally, it is important to note that in the case of necrosis (but not secondary necrosis), DNA is not fragmented, and thus a sub-G1 peak will not be generated.
Anticipated Results
Basic Protocol 1: Analysis of cell morphology by phase-contrast microscopy
Under a phase-contrast microscope, apoptotic cells are readily recognizable by a progressive loss in cell volume, cell detachment, the formation of membrane blebs, and the loss of the regular shape characteristic of live cells (Figs. 1 and 3). At later apoptotic stages, the morphology of apoptotic cells changes as they undergo secondary necrosis. Very similar to necrotic cells (Figs. 4 and 5), these cells are swollen and present with a condensed nucleus and a ghost appearance.
Alternate Protocol 1: Analysis of cell morphology with eosin-methylene blue
The eosin-methylene blue staining is universal, but the staining pattern can vary from cell to cell. The cytoplasm should be stained in blue and nuclei in pink. The nuclei of healthy cells normally display a regular staining, whereas chromatin condensation and nuclear fragmentation should easily be observed within apoptotic cells (Fig. 8). This protocol allows the identification of necrotic cells that usually exhibit as ruptured or “ghost” cells releasing their intracellular content as a result of plasma membrane rupture. The nuclei of necrotic cells are frequently relatively intact and slightly swollen.
Alternate Protocol 2: Analysis of nuclear morphology with Hoechst 33342
Hoechst 33342 is a cell-permeable dye that stains the nucleus of any cell in blue. The nucleus of live cells is normally relatively homogeneously stained with Hoechst 33342.
Conversely, nuclei of early apoptotic cells display condensed chromatin easily discernable from noncondensed chromatin by a brighter Hoechst 33342 staining pattern (Fig. 9). Hoechst 33342 also reveals the nuclear fragmentation associated with apoptosis. In fact, apoptotic cells frequently exhibit several nuclear fragments densely stained with Hoechst 33342 (Fig. 9).
Basic Protocol 2: Measurement of plasma membrane alteration with annexin V-FITC and PI
Although basal annexin V staining can vary from cell to cell, live cells are normally annexin V- and PI-negative and are distributed in the bottom left quadrant of the flow cytometer dot plot. Early apoptotic cells with externalized phosphatidylserine, but non-permeabilized plasma membranes, are annexin V-positive and PI-negative. These cells are thus shifted to the bottom right quadrant of the dot plot. When apoptotic cells progress toward secondary necrosis, their plasma membrane becomes permeant and cells progressively get stained with PI thus shifting in the top right quadrant of the dot plot (Fig. 10). Necrotic cells, which are characterized by very early rupture of their plasma membrane, move directly to the top right corner (i.e., double positive) of the dot plot, similarly to secondary necrotic cells.
Basic Protocol 3: Measurement of DNA fragmentation with propidium iodide
In this assay, PI is used to measure the DNA content of cells. The histogram displaying the PI staining intensity of healthy cells presents two peaks corresponding to cells in the G0/G1 and in the G2/M phases of the cell cycle. The latter has twice as much DNA (i.e., 4 N chromosomes) as the G0/G1 population. In between these peaks, cells in S phase are characterized by a heterogeneous DNA content. Due to DNA fragments formed during apoptosis being extracted during the staining, apoptotic cells display a reduction in their DNA content and form a new cell population to the left of the G0/G1 peak (Fig. 11).
Alternate Protocol 3: Analysis of DNA fragmentation with the TUNEL assay
As the TUNEL procedure stains fragmented DNA, apoptotic cells can be stained using Alternate Protocol 3. Live cells are thus FITC-negative and distributed on the left hand side of the FL1 histogram. Cells undergoing apoptosis and DNA fragmentation become TUNEL-positive and display an increased green fluorescence intensity. They are shifted on the right hand side of the histogram. Similar to the sub-G1 population, late apoptotic but not necrotic cells are detected as TUNEL-positive.
Basic Protocol 4: Measurement of caspase activation with FLICA probes
Although expressed by healthy cells, caspases only become activated in response to pro-apoptotic stimuli. A FLICA/PI staining, healthy cells are negative for FLICA and PI staining. Early apoptotic cells have activated caspases and an intact plasma membrane. They are thus detected as a FLICA-positive and PI-negative population. As apoptosis progresses and plasma membrane integrity is lost, apoptotic cells become FLICA- and PI-positive. This protocol is very useful to discriminate late apoptosis from necrosis, necroptosis, and ferroptosis, as necrotic cells take up PI but have inactive caspases, are FLICA-negative, and are PI-positive, thus forming a very distinctive population on the FLICA versus PI dot plot.
Basic Protocol 5: Discrimination between apoptosis, necrosis, necroptosis, and ferroptosis
In this assay, specific inhibitors of apoptosis, necroptosis, or ferroptosis (Table 2) are used to determine the mode of cell death elicited by a treatment. Typically, poly-caspase inhibitors will significantly delay the kinetics of apoptosis but will not block this mode of cell death unless cells are undergoing apoptosis in a death-receptor-dependent (i.e., TNFR, FasR, TRAIL-R) manner. Caspase inhibitors can also alter the phenotype of apoptosis to resemble necrosis when cells do eventually die in the presence of such inhibitors. Furthermore, caspase inhibitors in combination with certain stimuli (e.g., TNF, FasL, TRAIL) can result in cells undergoing necroptosis under some circumstances (typically in cells that express RIPK3), and this usually accelerates the kinetics of cell death as compared with exposure to such stimuli in the absence of caspase inhibitors. If this is suspected, treatment with the cell death stimulus, in the presence or absence of poly-caspase inhibitors plus or minus necrostatin, can be used to determine whether this is occurring. Cells undergoing ferroptosis are typically rescued through treatment with the stimulus in combination with the radical-trapping antioxidants ferrostatin-1 or liproxstatin-1, which slow the accumulation of lipoperoxidases that are associated with this mode of cell death.
Time Considerations
Analysis of cell morphology by phase-contrast microscopy (Basic Protocol 1) is extremely rapid, as it does not require any preparation or processing of the sample. The duration of the analysis at the microscope varies with the experience level of the investigator and the number of samples that need to be analyzed.
Analysis of cell morphology with eosin-methylene blue staining by microscopy (Alternate Protocol 1) can take up to 1 hr for staining including the mounting of the slides. The amount of time required to analyze the stained slides then depends on the number of slides and should take <2 min per sample. Preparation of cytospins (Support Protocol), which is a prerequisite for Alternate Protocol 1, can usually be achieved within 30 min, but 1 hr might be needed depending on the number of cytospins to be prepared.
Analysis of nuclear morphology with Hoechst 33342 staining of fixed cells by fluorescence microscopy (Alternate Protocol 2) requires ∼45 min from the fixation step to the mounting step. The actual time required to count the cells at the microscope may vary between 1 and 2 min per sample.
Annexin V and PI staining to measure plasma membrane alterations (Basic Protocol 2) can be achieved in <30 min. Then, 1 to 2 min are required for the measurement of each sample by flow cytometry, and an extra 10 to 15 min might be spent to set up the flow cytometer before acquisition. The time for the final analysis may vary from 5 to 15 min depending on the number of samples.
To measure DNA fragmentation, PI staining for sub-G1 analysis (Basic Protocol 3) can take between 3.5 and 4 hr. The same amount of time should be spent at the flow cytometer as for the other analyses.
Analysis of DNA fragmentation with the TUNEL assay by flow cytometry (Alternate Protocol 3) can take between 3.5 and 4 hr for TUNEL staining. The same amount of time should be spent at the flow cytometer as for the other analysis.
FLICA/PI staining to measure caspase activation (Basic Protocol 4) can be achieved in 1.5 to 2 hr. The same amount of time should be spent at the flow cytometer as for the other analysis.
Acknowledgments
The Martin laboratory is supported by an ERC Advanced grant (101020534, DESTRESS) as well as an Irish Research Council Advanced Laureate Award (IRCLA/2019/133). The Hollville laboratory is supported by a Royal Society Research grant (RG16955).
Open access funding provided by IReL.
Author Contributions
Aoife Costigan : Writing—review and editing. Emilie Hollville : Funding acquisition; writing—original draft; writing—review and editing. Seamus J. Martin : Conceptualization; funding acquisition; supervision; writing—original draft.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Brumatti, G., Sheridan, C., & Martin, S. J. (2008). Expression and purification of recombinant annexin V for the detection of membrane alterations on apoptotic cells. Methods , 44, 235–240. https://doi.org/10.1016/j.ymeth.2007.11.010
- Burke, B., & Stewart, C. L. (2013). The nuclear lamins: Flexibility in function. Nature Reviews Molecular Cell Biology , 14, 13–24. https://doi.org/10.1038/nrm3488
- Chen, Z., Naito, M., Mashima, T., & Tsuruo, T. (1996). Activation of actin-cleavable interleukin 1beta-converting enzyme (ICE) family protease CPP-32 during chemotherapeutic agent-induced apoptosis in ovarian carcinoma cells. Cancer Research , 56, 5224–5229.
- Cheung, W. L., Ajiro, K., Samejima, K., Kloc, M., Cheung, P., Mizzen, C. A., Beeser, A., Etkin, L. D., Chernoff, J., Earnshaw, W. C., & Allis, C. D. (2003). Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell , 113, 507–517. https://doi.org/10.1016/S0092-8674(03)00355-6
- Coleman, M. L., Sahai, E. A., Yeo, M., Bosch, M., Dewar, A., & Olson, M. F. (2001). Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nature Cell Biology , 3, 339–345. https://doi.org/10.1038/35070009
- Croft, D. R., Coleman, M. L., Li, S., Robertson, D., Sullivan, T., Stewart, C. L., & Olson, M. F. (2005). Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration. Journal of Cell Biology , 168, 245–255. https://doi.org/10.1083/jcb.200409049
- Degterev, A., Hitomi, J., Germscheid, M., Ch'en, I. L., Korkina, O., Teng, X., Abbott, D., Cuny, G. D., Yuan, C., Wagner, G., Hedrick, S. M., Gerber, S. A., Lugovskoy, A., & Yuan, J. (2008). Identification of RIP1 kinase as a specific cellular target of necrostatins. Nature Chemical Biology , 4(5), 313–321. https://doi.org/10.1038/nchembio.83
- Degterev, A., Huang, Z., Boyce, M., Li, Y., Jagtap, P., Mizushima, N., Cuny, G. D., Mitchison, T. J., Moskowitz, M. A., & Yuan, J. (2005). Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nature Chemical Biology , 1(2), 112–119. https://doi.org/10.1038/nchembio711
- Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., Patel, D. N., Bauer, A. J., Cantley, A. M., Yang, W. S., Morrison, B. 3rd., & Stockwell, B. R. (2012). Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell , 149(5), 1060–1072. https://doi.org/10.1016/j.cell.2012.03.042
- Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A., & Nagata, S. (1998). A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature , 391, 43–50. https://doi.org/10.1038/34112
- Fadok, V. A., Voelker, D. R., Campbell, P. A., Cohen, J. J., Bratton, D. L., & Henson, P. M. (1992). Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. Journal of Immunology , 148, 2207–2216.
- Friedmann Angeli, P. J., Schneider, M., Proneth, B., Tyurina, Y. Y., Tyurin, V. A., Hammond, V. J., Herbach, N., Aichler, M., Walch, A., Eggenhofer, E., Basavarajappa, D., Rådmark, O., Kobayashi, S., Seibt, T., Beck, H., Neff, F., Esposito, I., Wanke, R., … Conrad, M. (2014). Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nature Cell Biology , 16(12), 1180–1191. https://doi.org/10.1038/ncb3064
- Gerner, C., Frohwein, U., Gotzmann, J., Bayer, E., Gelbmann, D., Bursch, W., & Schulte-Hermann, R. (2000). The Fas-induced apoptosis analyzed by high throughput proteome analysis. Journal of Biological Chemistry , 275, 39018–39026. https://doi.org/10.1074/jbc.M006495200
- Jacobson, M. D., Weil, M., & Raff, M. C. (1997). Programmed cell death in animal development. Cell , 88, 347–354. https://doi.org/10.1016/S0092-8674(00)81873-5
- Jiang, X., Stockwell, B. R., & Conrad, M. (2021). Ferroptosis: Mechanisms, biology and role in disease. Nature Reviews. Molecular Cell Biology , 22(4), 266–282. https://doi.org/10.1038/s41580-020-00324-8
- Kayagaki, N., Kornfeld, O. S., Lee, B. L., Stowe, I. B., O'Rourke, K., Li, Q., Sandoval, W., Yan, D., Kang, J., Xu, M., Zhang, J., Lee, W. P., McKenzie, B. S., Ulas, G., Payandeh, J., Roose-Girma, M., Modrusan, Z., Reja, R., Sagolla, M., … Dixit, V. M. (2021). NINJ1 mediates plasma membrane rupture during lytic cell death. Nature , 591(7848), 131–136. https://doi.org/10.1038/s41586-021-03218-7
- Kearney, C. J., & Martin, S. J. (2017). An inflammatory perspective on necroptosis. Molecular Cell , 65(6), 965–973. https://doi.org/10.1016/j.molcel.2017.02.024
- Kerr, J. F., Wyllie, A. H., & Currie, A. R. (1972). Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. British Journal of Cancer , 24, 239–275. https://doi.org/10.1038/bjc.1972.33
- Kothakota, S., Azuma, T., Reinhard, C., Klippel, A., Tang, J., Chu, K., McGarry, T. J., Kirschner, M. W., Koths, K., Kwiatkowski, D. J., & Williams, L. T. (1997). Caspase-3-generated fragment of gelsolin: Effector of morphological change in apoptosis. Science , 278, 294–298. https://doi.org/10.1126/science.278.5336.294
- Leventis, P. A., & Grinstein, S. (2010). The distribution and function of phosphatidylserine in cellular membranes. Annual Review of Biophysics , 39, 407–427. https://doi.org/10.1146/annurev.biophys.093008.131234
- Liu, X., Zou, H., Slaughter, C., & Wang, X. (1997). DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell , 89, 175–184. https://doi.org/10.1016/S0092-8674(00)80197-X
- Logue, S. E., Elgendy, M., & Martin, S. J. (2009). Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nature Protocols , 4, 1383–1395. https://doi.org/10.1038/nprot.2009.143
- Lüthi, A. U., Cullen, S. P., & Martin, S. J. (2008). Two-dimensional gel-based analysis of the demolition phase of apoptosis. Methods in Enzymology , 442, 343–354. https://doi.org/10.1016/S0076-6879(08)01417-1
- Maekawa, M., Ishizaki, T., Boku, S., Watanabe, N., Fujita, A., Iwamatsu, A., Obinata, T., Ohashi, K., Mizuno, K., & Narumiya, S. (1999). Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science , 285, 895–898. https://doi.org/10.1126/science.285.5429.895
- Martin, S. J. (2016). Cell death and inflammation: The case for IL-1 family cytokines as the canonical DAMPs of the immune system. The FEBS Journal , 283(14), 2599–2615. https://doi.org/10.1111/febs.13775
- Martin, S. J., Frezza, V., Davidovich, P., Najda, Z., & Clancy, D. M. (2022). IL-1 family cytokines serve as 'activity recognition receptors' for aberrant protease activity indicative of danger. Cytokine , 157, 155935. https://doi.org/10.1016/j.cyto.2022.155935
- Martin, S. J., Henry, C. M., & Cullen, S. P. (2012). A perspective on mammalian caspases as positive and negative regulators of inflammation. Molecular Cell , 46, 387–397. https://doi.org/10.1016/j.molcel.2012.04.026
- Martin, S. J., Finucane, D. M., Amarante-Mendes, G. P., O'Brien, G. A., & Green, D. R. (1996). Phosphatidylserine externalisation during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity. Journal of Biological Chemistry , 271, 28753–28756. https://doi.org/10.1074/jbc.271.46.28753
- Martin, S. J., O'Brien, G. A., Nishioka, W. K., McGahon, A. J., Mahboubi, A., Saido, T. C., & Green, D. R. (1995). Proteolysis of fodrin (non-erythroid spectrin) during apoptosis. Journal of Biological Chemistry , 270, 6425–6428. https://doi.org/10.1074/jbc.270.12.6425
- Martin, S. J., Reutelingsperger, C. P., McGahon, A. J., Rader, J. A., van Schie, R. C., LaFace, D. M., & Green, D. R. (1995). Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: Inhibition by overexpression of Bcl-2 and Abl. Journal of Experimental Medicine , 182, 1545–1556. https://doi.org/10.1084/jem.182.5.1545
- Moss, D. K., Betin, V. M., Malesinski, S. D., & Lane, J. D. (2006). A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. Journal of Cell Science , 119, 2362–2374. https://doi.org/10.1242/jcs.02959
- Rao, L., Perez, D., & White, E. (1996). Lamin proteolysis facilitates nuclear events during apoptosis. Journal of Cell Biology , 135, 1441–1455. https://doi.org/10.1083/jcb.135.6.1441
- Sakahira, H., Enari, M., & Nagata, S. (1998). Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature , 391, 96–99. https://doi.org/10.1038/34214
- Samejima, K., Tone, S., & Earnshaw, W. C. (2001). CAD/DFF40 nuclease is dispensable for high molecular weight DNA cleavage and stage I chromatin condensation in apoptosis. Journal of Biological Chemistry , 276, 45427–45432. https://doi.org/10.1074/jbc.M108844200
- Sebbagh, M., Renvoizé, C., Hamelin, J., Riché, N., Bertoglio, J., & Bréard, J. (2001). Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nature Cell Biology , 3, 346–352. https://doi.org/10.1038/35070019
- Shiratsuchi, A., Mori, T., & Nakanishi, Y. (2002). Independence of plasma membrane blebbing from other biochemical and biological characteristics of apoptotic cells. Journal of Biochemistry , 132, 381–386. https://doi.org/10.1093/oxfordjournals.jbchem.a003233
- Slee, E. A., Harte, M. T., Kluck, R. M., Wolf, B. B., Casiano, C. A., Newmeyer, D. D., Wang, H. G., Reed, J. C., Nicholson, D. W., Alnemri, E. S., Green, D. R., & Martin, S. J. (1999). Ordering the cytochrome c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. Journal of Cell Biology , 144, 281–292. https://doi.org/10.1083/jcb.144.2.281
- Stennicke, H. R., & Salvesen, G. S. (1998). Properties of the caspases. Biochimica et Biophysica Acta , 1387, 17–31. https://doi.org/10.1016/S0167-4838(98)00133-2
- Stockwell, B. R., Friedmann Angeli, J. P., Bayir, H., Bush, A. I., Conrad, M., Dixon, S. J., Fulda, S., Gascón, S., Hatzios, S. K., Kagan, V. E., Noel, K., Jiang, X., Linkermann, A., Murphy, M. E., Overholtzer, M., Oyagi, A., Pagnussat, G. C., Park, J., Ran, Q., … Zhang, D. D. (2017). Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell , 171(2), 273–285. https://doi.org/10.1016/j.cell.2017.09.021
- Suzuki, J., Denning, D. P., Imanishi, E., Horvitz, H. R., & Nagata, S. (2013). Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science , 341, 403–406. https://doi.org/10.1126/science.1236758
- Taylor, R. C., Cullen, S. P., & Martin, S. J. (2008). Apoptosis: Controlled demolition at the cellular level. Nature Reviews Molecular Cell Biology , 9, 231–241. https://doi.org/10.1038/nrm2312
- Ura, S., Masuyama, N., Graves, J. D., & Gotoh, Y. (2001). Caspase cleavage of MST1 promotes nuclear translocation and chromatin condensation. Proceedings of the National Academy of Sciences of the United States of America , 98, 10148–10153. https://doi.org/10.1073/pnas.181161698
- Williams, J. R., Little, J. B., & Shipley, W. U. (1974). Association of mammalian cell death with a specific endonucleolytic degradation of DNA. Nature , 252, 754–755. https://doi.org/10.1038/252754a0
- Vanden Berghe, T., Linkermann, A., Jouan-Lanhouet, S., Walczak, H., & Vandenabeele, P. (2014). Regulated necrosis: The expanding network of non-apoptotic cell death pathways. Nature Reviews. Molecular Cell Biology , 15(2), 135–147. https://doi.org/10.1038/nrm3737
- Wyllie, A. H., Kerr, J. F., & Currie, A. R. (1980). Cell death: The significance of apoptosis. International Review of Cytology , 68, 251–306. https://doi.org/10.1016/s0074-7696(08)62312-8

