Isolation and Identification of Nematode-Infecting Microsporidia
Hala Tamim El Jarkass, Hala Tamim El Jarkass, Aaron W. Reinke, Aaron W. Reinke
Abstract
Nematodes are naturally infected by the fungal-related pathogen microsporidia. These ubiquitous eukaryotic parasites are poorly understood, despite infecting most types of animals. Identifying novel species of microsporidia and studying them in an animal model can expedite our understanding of their infection biology and evolution. Nematodes present an excellent avenue for pursuing such work, as they are abundant in the environment and many species are easily culturable in the laboratory. The protocols presented here describe how to isolate bacterivorous nematodes from rotting substrates, screen them for microsporidia infection, and molecularly identify the nematode and microsporidia species. Additionally, we detail how to remove environmental contaminants and generate a spore preparation of microsporidia from infected samples. We also discuss potential pitfalls and provide suggestions on how to mitigate them. These protocols allow for the identification of novel microsporidia species, which can serve as an excellent starting point for genomic analysis, determination of host specificity, and infection characterization. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Gathering samples
Support Protocol 1 : Generating 10× and 40× Escherichia coli OP50 and seeding NGM plates
Basic Protocol 2 : Microsporidia screening, testing for Caenorhabditis elegans susceptibility, and sample freezing
Basic Protocol 3 : DNA extraction, PCR amplification, and sequencing to identify nematode and microsporidia species
Basic Protocol 4 : Removal of contaminating microbes and preparation of microsporidia spores
Support Protocol 2 : Bleach-synchronizing nematodes
INTRODUCTION
Microsporidia are ubiquitous obligate intracellular parasites within the fungal kingdom. They infect a large diversity of animal hosts, including humans, in which they can cause disease and death (Han & Weiss, 2017; Jarkass & Reinke, 2020). These parasites have highly reduced genomes, making them exceptionally reliant on their host for resources (Corradi, 2015; Nakjang et al., 2013; Wadi & Reinke, 2020). However, despite their widespread nature, much remains unknown about their infection biology. This, coupled with their host reliance, has made the development of effective treatment strategies challenging. Recently, Caenorhabditis elegans and other free-living terrestrial nematodes were found to harbor microsporidia infection in the wild. Although microsporidia only grow inside host cells, infections in nematodes are almost always horizontally transferred, and large amounts of spores are generated. This allows the infection to be propagated, and spores can be frozen for preservation. To date, C. elegans has been used to uncover various aspects of microsporidia infection and the host response to infection (Balla et al., 2015, 2016; Estes et al., 2011; Luallen et al., 2016; Reddy et al., 2017, 2019; Szumowski et al., 2014, 2016; Tamim El Jarkass et al., 2022; Troemel et al., 2008; Willis et al., 2021).
Sampling of wild nematodes has revealed microsporidia species with the ability to infect various tissues, including the intestine, muscle, and epidermis (Luallen et al., 2016; Wadi et al., 2023; Zhang et al., 2016). These sampling efforts have also revealed microsporidia species with varying host specificity patterns, ranging from specialists that infect only one or two closely related species to generalists that can infect many species of nematodes, including those from different genera. These studies highlight the diversity of microsporidia within the natural habitats of nematodes. Identifying novel species of microsporidia allows for the characterization and understanding of genome evolution, infection dynamics, and host specificity. The most common method for identifying microsporidia infecting bacterivorous nematodes has been light microscopy, with which multiple stages of the parasite life cycle can be identified (Luallen et al., 2016; Troemel et al., 2008; Zhang et al., 2016). This technique relies on mounting a few animals onto a glass slide and looking for microsporidia meronts or spores. The advantages of this approach are that nematodes do not need to be fixed, both proliferative and spore stages can be observed, and fluorescent microscopy is not needed. However, identifying signs of infection can be challenging for beginners, and microsporidia from low-infection populations may be missed. An alternative approach is to use the fluorescent dye Direct Yellow 96 (DY96), which stains the chitin-containing microsporidia spores (Wadi et al., 2023).
Here, we provide multiple protocols to identify, characterize, and propagate microsporidia infection in wild bacterivorous nematodes (Fig. 1). In Basic Protocol 1, we discuss how to identify environmental nematode sources and the steps involved in propagating these nematodes in the laboratory. Support Protocol 1 describes the steps required for generating Escherichia coli OP50, the standardized nematode laboratory food source. Once nematodes are identified, Basic Protocol 2 describes the steps involved in screening these samples for microsporidia infection using the fluorescent dye DY96. Basic Protocol 3 provides a framework to identify the host and pathogen species. Antibiotic cleanup of samples as well as generating a spore preparation is covered in Basic Protocol 4. In Support Protocol 2, we discuss how to generate a bleach-synchronized population of nematodes, which is critical for the steps outlined in Basic Protocol 4.
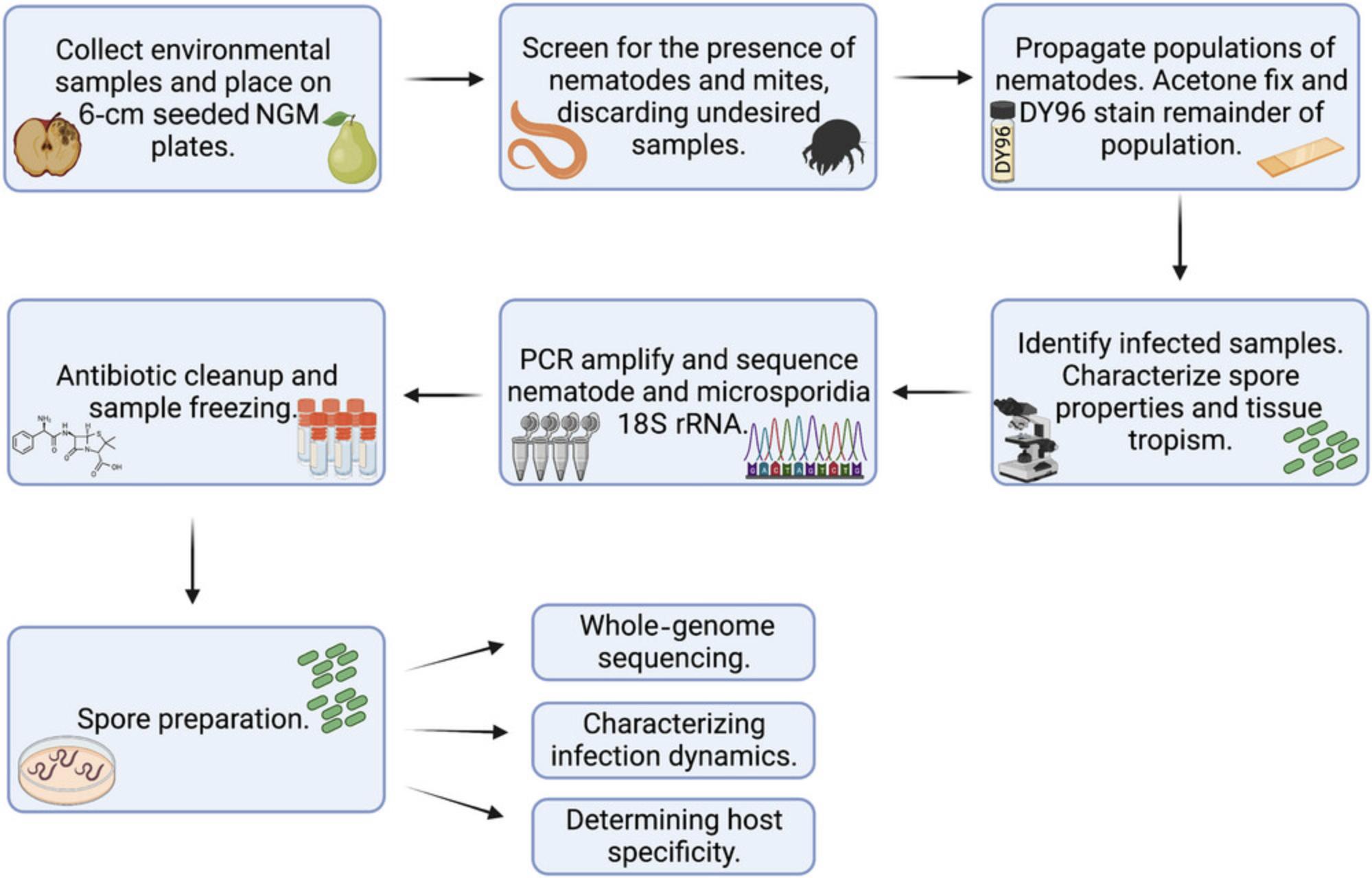
STRATEGIC PLANNING
Sampling (Basic Protocol 1) should be prioritized during a time of year that will yield the desired nematodes. This will depend on the geographic location; in Toronto, Ontario, nematodes are most abundant in September through November.
Prior to beginning Basic Protocol 1, ensure that E. coli OP50 and 6- and 10-cm seeded nematode growth medium (NGM) plates are prepared (see Support Protocol 1).
Order the oligonucleotides required for PCR amplification (Basic Protocol 3) ahead of time.
Acquire the ERT54 IPR reporter strain for Basic Protocol 2 from the Caenorhabditis Genetics Center (CGC; https://cgc.umn.edu/strain/ERT54).
NOTE : Follow all the appropriate safety guidelines and regulations for the use and handling of pathogenic microorganisms. Nematode-infecting microsporidia are considered Biosafety Level-1 (BSL-1) organisms, as they have never been observed to infect any other animals besides nematodes (Murareanu et al., 2021). Environmental bacteria or fungi that are inadvertently isolated along with nematodes may be considered BSL-1 or BSL-2 organisms depending on the organization where this work is performed.
NOTE : Follow all relevant safety procedures for field work involved in isolating nematodes. Ensure that if any permits or permissions to sample nematodes are needed, these are obtained before isolation occurs.
Basic Protocol 1: GATHERING SAMPLES
In this protocol, we describe the steps involved in identifying and isolating wild samples. The protocol begins with instructions to identify natural substrates in the environment that have a high likelihood of containing nematodes. We describe how to sample these substrates and how to handle them in the laboratory. We also describe how to scan samples for the presence of nematodes and how to discard those containing undesired animals such as mites. The samples that are screened in the laboratory may reveal no, few, or many nematodes and will certainly contain environmental microbes such as bacteria and fungi. Although the goal of this protocol is to identify nematodes that are infected with microsporidia, the nematode strains themselves can be useful to the research community. In particular, the Caenorhabditis species C. elegans , C. briggsae , and C. tropicalis can be sent to CaeNDR, the Caenorhabditis Natural Diversity Resource (https://caendr.org/) (Crombie et al., 2024).
Materials
-
6-cm NGM plates seeded with 10× E. coli OP50 (see Support Protocol 1)
-
Likely substrates for nematodes
-
Sealable sandwich bags (Ziploc® or VWR, 89005-284)
-
Marker
-
Gloves
-
Parafilm
-
Plastic storage container with lid
-
Dissecting microscope with transmitted light (Zeiss Stemi 305 or equivalent)
-
Plastic beaker
-
Microwave
-
Biosafety bin liner
CAUTION : Mites can move between stacks of plates in a box. In doing so, nematode embryos attached to their legs can get transferred between plates. This results in the contamination of nematode stocks. Therefore, we do not recommend storing environmental samples in the same incubator as sterile samples until they have been screened. Double-wrapping plates in parafilm may help limit the transfer of mites between stacks of plates.
1.Prepare the desired number of 6-cm NGM plates seeded with 10× E. coli OP50 (see Support Protocol 1).
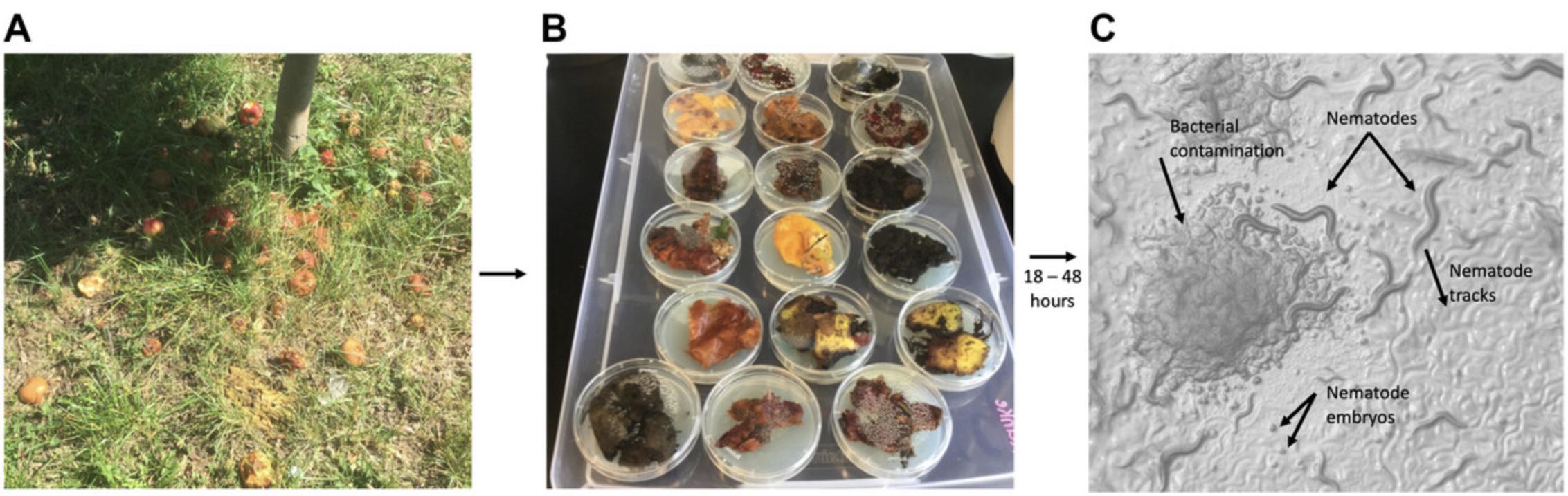
2.Identify areas with likely substrates for nematodes, which are pieces of fruit in an advanced stage of rotting that should be malleable and not dry (Fig. 2A).
3.Invert a sealable sandwich bag and collect a sample that is ∼6 cm in diameter. Using the free hand, re-invert and seal the bag. Make note of the substrate, geographical location, and coordinates of the sample. Give each sample a unique identifier (e.g., code or number) and write this identifier on the bag using a marker. Repeat until desired number of samples are obtained, keeping samples in separate bags.
4.While wearing gloves, remove the lid from a 6-cm seeded NGM plate (see step 1) and place the sample on the plate. Spread the sample out across the plate, replace plate lid, and wrap with parafilm. Store right side up in a plastic storage container with a lid at room temperature for 18 to 48 hr (Fig. 2B).
5.Generate a spreadsheet containing information on the sample substrate and location (see Table S1 in the Supporting Information for an example). As more information becomes available about these samples, update accordingly.
6.Remove as much of the substrate as possible and wipe any condensation off the lid. Try to remove any insects or other invertebrates, if large and visible. Wrap the plate in parafilm and incubate at room temperature until screening the plates for nematodes (see step 7).
7.Screen the parafilmed plates using a dissecting microscope with transmitted light and look for the presence of nematodes.
8.Characterize nematodes by making note of their appearance and the presence/absence of embryos and/or males. Keep an eye out for mites and set these samples aside (Figs. 2C and 3A).
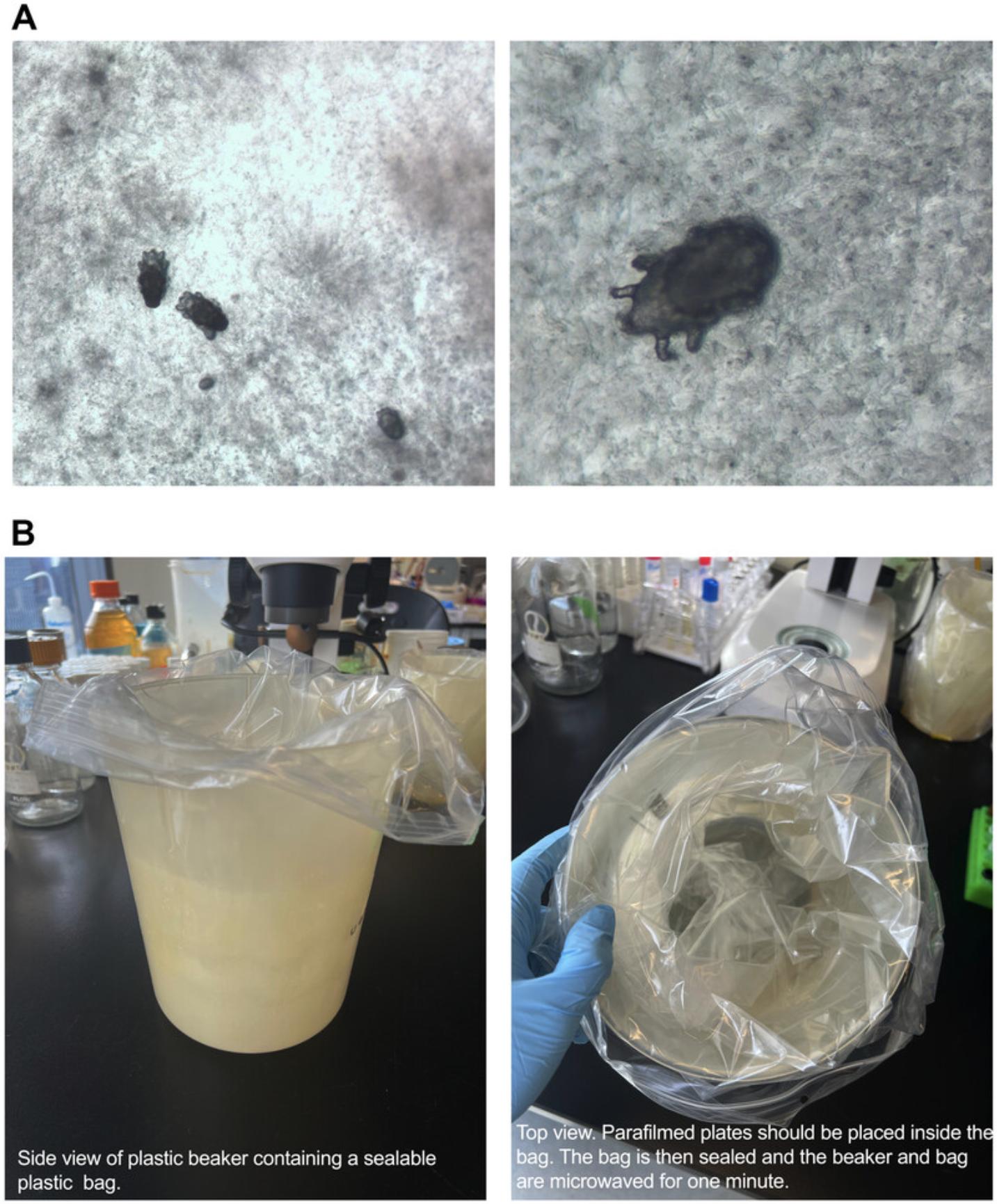
9.Place a sealable plastic bag inside a plastic beaker (Fig. 3B). Place all the parafilm-wrapped plates containing mites into the plastic bag and seal it (Fig. 3B). Microwave the beaker for 1 min, until the agar has liquified. Toss the sealed bag into a biosafety bin liner and immediately discard the liner as biohazardous waste.
Support Protocol 1: GENERATING 10× AND 40× Escherichia coli OP50 AND SEEDING NGM PLATES
This protocol describes the steps involved in generating 10× and 40× E. coli OP50 stocks. The 10× OP50 is used for seeding NGM plates, whereas 40× E. coli OP50 is used in spore preparations to provide a large amount of food in a small volume. We also describe how to seed and store NGM plates.
Materials
-
E. coli OP50-1 (available from the CGC)
-
10-cm LB agar plate (see recipe)
-
Sterile 100-ml bottle containing 100 ml sterile LB (see recipe)
-
Two sterile 2-L flasks containing 1 L sterile LB (see recipe)
-
6% (v/v) bleach (sodium hypochlorite; Fisher, SS290-1)
-
6-cm unseeded NGM plates (see recipe)
-
10-cm unseeded NGM plates (see recipe; optional)
-
37°C incubator
-
21°C and 37°C shaking incubator (EppendorfTM InnovaTM S44i shaker or equivalent)
-
500-ml conical tubes, sterile (Fisher Scientific, C431123)
-
Scale
-
Serological pipets
-
500-ml centrifuge tube cushions (Corning, 431124)
-
Centrifuge (Beckman Coulter Allegra X-14R or equivalent)
-
50-ml conical tubes, sterile (FroggaBio, TB50-500)
-
Dissecting microscope with transmitted light (Zeiss Stemi 305 or equivalent)
-
Metal spreader, ethanol (95%, Fisherbrand, HC11001GL) and flame sterilized
-
Plastic storage container with lid
NOTE : All steps in this protocol should be carried out near a flame (i.e., using a Bunsen burner) and on an ethanol-sterilized bench [sterilized with 70% (v/v) ethanol (diluted from 95% ethanol, Fisherbrand, HC11001GL)] to prevent any contamination.
CAUTION : Ethanol is flammable.
Generating E. coli OP50 cultures
1.Streak out E. coli OP50-1 onto a 10-cm LB agar plate and incubate at 37°C overnight.
2.Working near a flame, pick a single E. coli OP50-1 colony into a sterile 100-ml bottle containing 100 ml sterile LB.
3.Cap the bottle and shake to ensure even distribution throughout the medium.
4.Pour 50 ml LB from step 3 into each of two sterile 2-L flasks containing 1 L sterile LB.
5.Place inoculated flasks into 37°C shaking incubator at 180 rpm for 18 hr.
Generating 10× E. coli OP50
6.Retrieve flasks and pour the contents into 500-ml conical tubes.
7.Using a scale, ensure that conical tubes are the same weight. Use a serological pipet to transfer volume from one conical tube to the other if required to balance out the weights.
8.Spin conical tubes on 50-ml centrifuge tube cushions for 10 min at 3500 × g.
9.Pour 45 ml supernatant from the 500-ml conical tubes into 50-ml conical tubes, keeping track of the sample origins.
10.Pour the remaining 450 ml supernatant into the corresponding culture flask from step 6.
11.Pour the 45 ml from step 9 back into the corresponding 500-ml conical tube containing the cell pellet.
12.Use a serological pipet to resuspend the bacteria or shake the conical tubes by placement in the shaking incubator at 21°C and 180 rpm for ∼20 min.
13.Transfer the resuspended 50-ml culture into the corresponding 50-ml conical tube used in step 9.
14.Label this aliquot with “10× OP50”, the date, and the initials of the person who prepared it.
15.Store aliquots at 4°C.
16.Decontaminate the 2-L flasks containing bacterial supernatants (see step 10) by adding 6% bleach to a final concentration of 1% for ≥30 min and then discard.
Testing for contamination
17.Transfer 400 μl of each 10× OP50 aliquot from step 15 onto an individual 6-cm unseeded NGM plate.
18.Incubate plates at 37°C for 16 hr or room temperature for 48 hr.
19.Screen the plates using a dissecting microscope with transmitted light.
20.Discard any contaminated aliquots and store the clean aliquots for ≤1 month at 4°C.
Generating a 40× E. coli aliquot
21.Spin down a 50-ml 10× OP50 aliquot from step 15 for 5 min at 7197 × g.
22.Remove the supernatant until 12.5 ml remains and resuspend the cell pellet.
23.Store ≤1 month at 4°C.
Seeding NGM plates
24.Seed NGM plates by pipetting 200 μl (for 6-cm plates) or 500 μl (for 10-cm plates) 10× E. coli OP50 onto the center of each plate.
25.For 10-cm plates, spread the culture using an ethanol- and flame-sterilized metal spreader.
26.Incubate the plates at room temperature for ≥24 hr to promote E. coli growth.
27.Place seeded NGM plates in a plastic storage container with a lid, and store ≤1 month at 4°C.
Basic Protocol 2: MICROSPORIDIA SCREENING, TESTING FOR Caenorhabditis elegans SUSCEPTIBILITY, AND SAMPLE FREEZING
Once nematodes are identified (Basic Protocol 1), they can be screened for the presence of microsporidia. Here, we describe how to maintain samples to ensure they are continually being propagated. We then discuss how to identify microsporidia infection, detailing what to expect and what characteristics to document. We also provide a strategy to test if the identified microsporidia species can infect C. elegans. Lastly, we provide instructions on how to freeze down infected samples of interest for long-term storage. For incubation of nematodes, a range of temperatures can be used, from 15° to 25°C. The recommended temperature of 21°C is provided as a guideline.
Materials
-
Candidate plates (see Basic Protocol 1, step 7)
-
95% (v/v) ethanol (Fisherbrand, HC11001GL)
-
6- and 10-cm seeded NGM plates (see Support Protocol 1)
-
M9 + 0.1% (v/v) Tween-20 (see recipe)
-
Acetone
-
DY96 solution (see recipe)
-
EverBrite Mounting Medium (Biotium, 23002), 4°C
-
ERT54 L1 animals (C. elegans strain ERT54 jyIs8 [pals-5p ::GFP + myo-2p ::mCherry] X, CGC; maintain at 21°C)
-
Freezing medium (see recipe)
-
Dissecting microscope with transmitted light (Zeiss Stemi 305 or equivalent)
-
Metal spatula
-
Bunsen burner
-
Microcentrifuge tubes
-
Tube rack
-
Microcentrifuge (Eppendorf S424 or equivalent)
-
Liquid waste container
-
Tinfoil
-
Sample rotator (VWR, 82007-202)
-
Glass microscope slides
-
Coverslips (thickness #1.5)
-
Fluorescence microscope with GFP and DAPI filters (Zeiss Axio Imager M2 or equivalent)
-
21°C incubator (15°C and 25°C optional) (Thermo Scientific Forma environmental chamber or equivalent)
-
Cryotubes, sterile (Thermo Fisher, 366656)
-
Fluorescence dissecting microscope with GFP and RFP filters (Zeiss Axiozoom V16 or equivalent)
-
Styrofoam tube rack
CAUTION : Store acetone in a flame-proof safety cabinet.
Identifying candidate plates, fixation, and staining
1.Screen candidate plates daily under a dissecting microscope with transmitted light and identify those with lots of large adult animals that have mostly consumed the E. coli OP50.
2.Sterilize a metal spatula by dipping it in 95% ethanol and carefully placing into the flame of a Bunsen burner. Once the spatula cools down, chunk ¼ of the plate onto a new 6-cm seeded NGM plate. To do this, use the spatula to cut through the agar, separating the chunk from the rest of the plate. Then, push the spatula under the chunk, lifting it out of the plate. Finally, put the chunk over the recipient plate and invert the spatula so that the chunk is now face down on the new plate. Wash off the animals on the remaining ¾ of the plate with 700 μl M9 + 0.1% Tween-20 into a microcentrifuge tube.
3.Place the microcentrifuge tubes in a tube rack on the bench to promote gravity settling. Once a nematode pellet is clearly visible, remove the supernatant but avoid disrupting the pellet.
4.Wash the samples with 700 μl M9 + 0.1% Tween-20, invert to mix, and gravity settle. Repeat this step as many times as necessary until the sample supernatant is clear. Then, add 700 μl acetone to the pellet.
5.Centrifuge the samples for 30 s at 6010 × g.
6.Remove the supernatant and discard in an appropriate liquid waste container.
7.Add 500 μl DY96 solution, wrap the tubes in tinfoil, and rotate the samples for 30 min in the dark.
8.Centrifuge samples for 30 s at 6010 × g and discard supernatant.
9.Add 10 to 20 μl cold EverBrite Mounting Medium and pipet up and down gently to evenly mix.
10.Load 10 μl sample onto a glass microscope slide and place a coverslip on top.
11.Image the samples using a fluorescence microscope with GFP and DAPI filters.
Characterizing infected nematode samples
12.Screen samples using a 10× or 20× objective and the GFP channel to identify microsporidia.
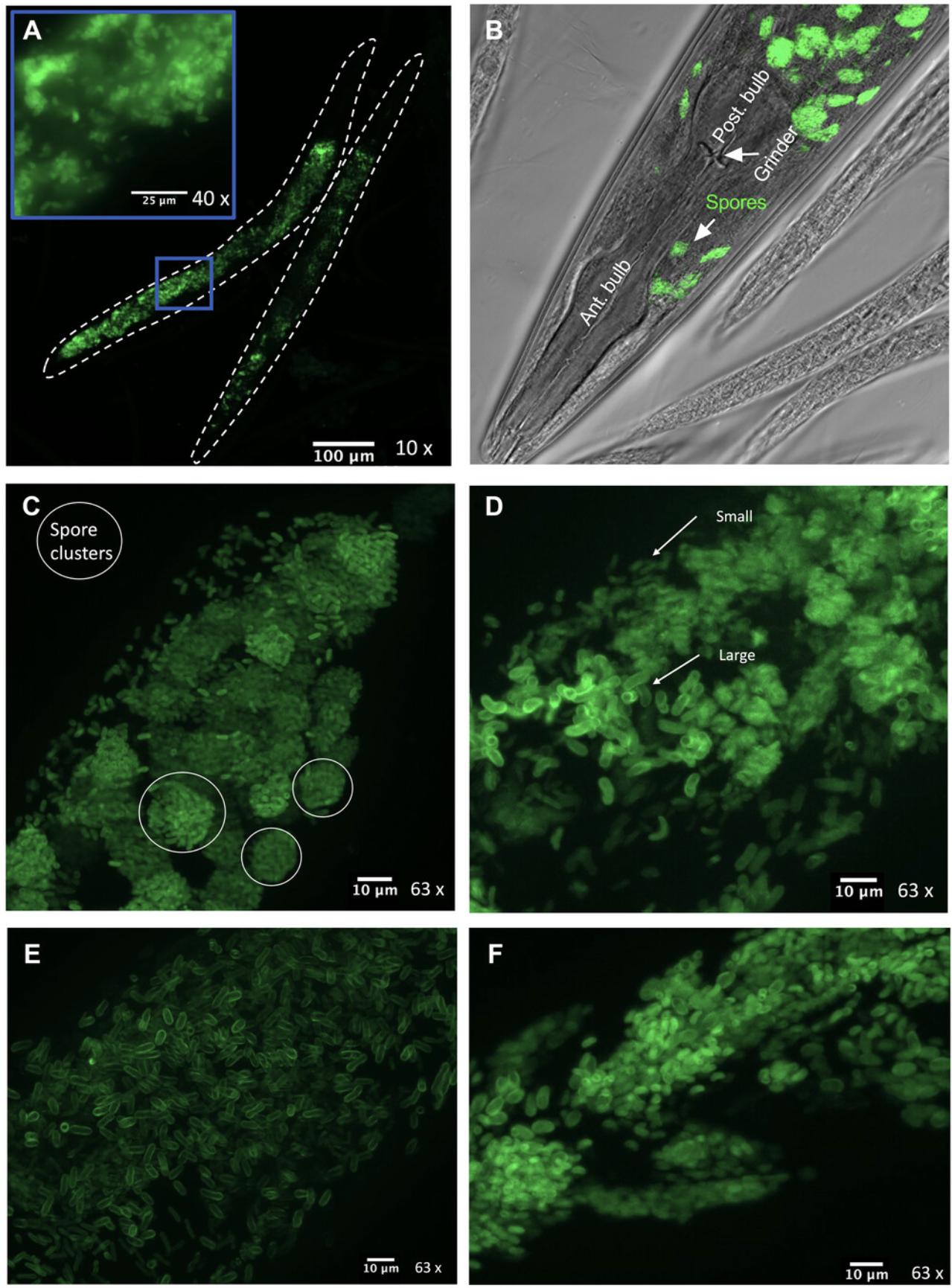
13.Identify the fraction of adults that contain spores by counting about 100 to 200 adults. Count adult worms that contain spores throughout the body and not just in the intestinal lumen (Figs. 4 and 5).
14.Characterize the spore morphology by determining the size of the spores present. To do this, take Z-stack images of individual infected worms using a 40× or 63× objective. Determine the plane at the top of the worm where spores are in focus (this will be the starting height) and determine the plane at the bottom of the worm where there are no spores in focus (this is the ending height). Determine the exposure time that allows the brightest signal possible without any saturation at any of the Z-planes. After capturing the Z-stack, create a maximum intensity projection. Using this image, measure the length and width of at least 50 spores from at least two infected animals and determine the mean length and width. Only measure spores that are not touching other spores and that appear to be parallel to the plane of the image.
15.Characterize the infection properties by assessing the tissues infected and if the spores form large clusters.
16.If the infection is not very prevalent in the population, chunk a densely populated plate and propagate at different temperatures (e.g., 15°C and 25°C) to promote spread, followed by determination of whether any improvements have occurred (see steps 2 to 15).
Determining if a microsporidia species can infect C. elegans
17.Chunk about 50 large animals from the infected plates generated in step 2 after about 5 to 6 days and add about 20 ERT54 L1 animals to a 10-cm seeded NGM plate. Incubate this plate at 21°C or room temperature.
18.After ∼1 day, determine what percentage of mCherry-positive animals also display GFP fluorescence using a fluorescent dissecting microscope with GFP and RFP filters. After 4 days, fix and stain with DY96 (as in steps 4 to 10) and determine what proportion of mCherry-positive adult animals have microsporidia spores in them.
Freezing of samples
19.To freeze down animals that contain microsporidia infection, first chunk the infected plate onto a 10-cm seeded NGM plate as in step 2.Monitor the plate for a few days. Once lots of L1 larvae with little E. coli OP50 are visible on the plate, wash the animals into four sterile cryotubes using 5 ml freezing medium.
20.If C. elegans (ERT54) can be infected with microsporidia, then freeze these animals as well. Freeze down all nematode samples regardless of infection status if they will be sent to nematode repositories. Place the tubes in a Styrofoam tube rack and place at −80°C.
21.Thaw the fourth vial ≥24 hr post-freezing to ensure nematodes are successfully recovered.
Basic Protocol 3: DNA EXTRACTION, PCR AMPLIFICATION, AND SEQUENCING TO IDENTIFY NEMATODE AND MICROSPORIDIA SPECIES
In this protocol, we discuss how to extract DNA from infected samples (Basic Protocol 2) and provide the PCR parameters and the primer sequences used for amplification. As the nematodes are infected, lysis and DNA extraction of these animals result in the release of host and pathogen DNA. PCR amplification involves the use of either Caenorhabditis -specific or nematode-specific PCR primers that amplify the 18S rRNA of the host. For microsporidia, there are multiple primer pairs that can be used, with amplification of the 18S rRNA being the most common.
Materials
- Oligonucleotides (IDT; see Table 1; store at −20°C)
- Single-worm lysis buffer (see recipe)
- Infected samples (see Basic Protocol 2, step 1)
- PCR reaction components (see Table 2)
- DNA ladder (1 Kb Plus DNA Ladder, NEB, N0550L; store at 4°C)
- 6× loading dye (NEB, B7025S)
- Monarch PCR and DNA Cleanup Kit (NEB, T1030S)
- Monarch DNA Gel Extraction Kit (NEB, T1020S)
| Target | Oligonucleotide label | Sequence (5’-3’) | Annealing temperature (°C) | Product size (kb) |
|---|---|---|---|---|
| Nematode 18S | G18S4a (Forward) | GCTCAAGTAAAAGATTAAGCCATGC | 68a | ∼1 |
| DF18S-8 (Reverse) | GTTTACGGTCAGAACTASGGCGG | |||
| Caenorhabditis 18S | 5.8S-1 (Forward) | CTGCGTTACTTACCACGAATTGCARAC | 68a | ∼2 |
| KK28S-4 (Reverse) | GCGGTATTTGCTACTACCAYYAMGATCTGC | |||
| Microsporidia beta tubulin | Bn1F (Forward) | ACAAACAGGNCARTGYGGNAAYCA | 64 | ∼1.2 |
| Bn1R (Reverse) | TGCTTCAGTRAAYTCCATYTCRTCCAT | |||
| Microsporidia SSU rDNAb (Zhang et al., 2016; this study) | V1F (Forward) | CACCAGGTTGATTCTGCCTGAC | 60a | ∼1.3 |
| 1492r (Reverse) | GGTTACCTTGTTACGACTT | |||
| 18SR1492 (Reverse) | GGAAACCTTGTTACGACTT | |||
| NEM_R1 (Reverse) | CGGARACCTTGTTACGACTTTTATCC | |||
| VPE_R1 (Reverse) | CACCGACCGCRACCTTGTTACG | |||
| Microsporidia SSU rDNA (Zhang et al., 2016) | SPF (Forward) | GATACGAGGAATTGGGGTTTG | 61 | ∼1 |
| SPR (Reverse) | GGGTACTGGAAATTCCGTGTT |
- a
Indicates experimentally determined optimal annealing temperatures. All other annealing temperatures are recommended based on primer melting temperature (Tm) via the NEB Tm calculator and Phusion (https://tmcalculator.neb.com/#!/main).
- b
The forward primer V1F can be used in combination with any of the subsequent reverse primers in this table. NEM_R1 was designed to potentially amplify additional Nematocida species not captured by the previous two primers. VPE_R1 was designed to potentially amplify species from the genera Vittaforma, Pancytospora, and Enterospectra.
| Component | Volume (μl)a |
|---|---|
| Phusion HF buffer (NEB, M0530L) | 10 |
| 10 mM dNTP mix (NEB, N0447L; store at −20°C) | 1 |
| 10 μM Forward primer (see Basic Protocol 3, step 1; diluted from 100 μM stock) | 2.5 |
| 10 μM Forward primer (see Basic Protocol 3, step 1; diluted from 100 μM stock) | 2.5 |
| DNA (from lysate generated in Basic Protocol 3, step 4) | 2 |
| Nuclease-free H2O (Invitrogen, LS10977015) | 31.5 |
| Phusion DNA polymerase (NEB, M0530L; store at −20°C) | 0.5 |
- a
These volumes are per the NEB Phusion kit instructions. Other DNA polymerases can be used, but the exact volume of each component may need to be modified per the manufacturer's instructions. Scale up the volumes according to the number of reactions being set up.
-
PCR strip tubes, sterile
-
Worm pick, sterile
-
Thermocycler (Bio-Rad C1000 Touch thermal cycler or equivalent)
-
Gel imaging apparatus (Bio-Rad ChemiDoc imaging system or equivalent)
-
Razor blade or scalpel
-
Blue light box (Blue Light Transilluminator, VWR, 76151-834, or equivalent)
-
Spectrophotometer (Thermo Scientific NanoDrop 2000c or equivalent)
-
Additional reagents and equipment for preparing a 1% (w/v) agarose (see recipe) gel, for agarose gel electrophoresis (see Current Protocols article: Voytas, 2000), and for Sanger sequencing
NOTE : Experiments involving PCR require extremely careful technique to prevent contamination.
DNA extraction
1.Acquire oligonucleotides listed in Table 1 for PCR amplification. For nematodes, start with the Nematode 18S primers, and if the species is within Caenorhabditis , use the Caenorhabditis 18S to determine the species identity. For microsporidia, start with V1F paired with either 18SR1492r or 1492r. If these pairs do not provide sufficient amplification, try the other primer pairs.
2.Pipet 10 μl single-worm lysis buffer into a sterile PCR strip tube for each sample.
3.Pick about 20 large adult animals from an infected sample into the same PCR tube containing the lysis buffer using a sterile worm pick.
4.Place the strip tubes into a thermocycler and run the following worm lysis program:
- 65°C for 60 min
- 95°C for 15 min.
5.Prepare a PCR master mix by multiplying the amounts of PCR components in Table 2 by the number of samples + 1.
6.Place the PCR reactions in a thermocycler using the following parameters:
| Initial step: | 30 s | 98°C |
| 34 cycles: | 10 s | 98°C |
| 30 s | X°C | |
| 1 min | 72°C | |
| Final steps: | 10 min | 72°C |
| indefinitely | 4°C. |
Assessing PCR amplicons via DNA gel electrophoresis
7.Prepare a 1% agarose gel.
8.Load 10 μl DNA ladder onto gel. Mix 1 μl of 6× loading dye with 5 μl of each sample and then load the entire volume. Run the gel at 80 V for 50 min.
9.Image the gel using a gel imaging apparatus. Assess the success of the PCR reaction by the presence of bands of the correct size, as outlined in step 1.
10.Purify the PCR reaction using the Monarch PCR and DNA Cleanup Kit or perform a gel extraction by running the entire remaining volume (45 μl) on a 1% agarose gel as before (see steps 7 to 9). Excise the band of the correct size using a razor blade or scalpel and image with a blue light box following the Monarch DNA Gel Extraction Kit instructions.
11.Assess the quality and concentration of the purified amplicon using a spectrophotometer.
12.Prepare and submit samples for Sanger sequencing by following the instructions of the facility that will perform the sequencing.
13.Trim any regions of poor sequencing quality. Align the sequences obtained from the forward and reverse primers using software such as ApE (https://www.frontiersin.org/articles/10.3389/fbinf.2022.818619/full) and fill in any sequencing gaps to generate a single sequence.
14.Use BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to reveal the closest known sequence match. Keep track of this information in the spreadsheet generated in Basic Protocol 1.
Basic Protocol 4: REMOVAL OF CONTAMINATING MICROBES AND PREPARATION OF MICROSPORIDIA SPORES
Once infected samples of interest are identified (Basic Protocol 3), they can be prioritized for the removal of contaminating bacteria using antibiotics. Although bleaching is an excellent way to destroy contaminating bacteria, bleach will also destroy the microsporidia present within the nematodes. In contrast, antibiotics are a selective way to remove contaminating bacteria. Once the samples are cleared of bacterial contamination, any residual fungal contamination can be dealt with by chunking the contaminated fraction of the plate away if the fungus is filamentous (e.g., mold). Once a clean population of nematodes containing the microsporidia of interest is obtained, these samples can then be used to generate a spore preparation. This preparation involves using the nematodes as a factory to produce large quantities of dormant microsporidia spores. These spores are then aliquoted and stored frozen for use in subsequent experiments.
Materials
-
10-cm seeded NGM plates (see Support Protocol 2)
-
70% (v/v) ethanol (diluted from 95% ethanol, Fisherbrand, HC11001GL)
-
M9 (see recipe), sterile
-
2.0-mm zirconia beads, sterile (BioSpec, 11079124ZX)
-
6-cm seeded and unseeded antibiotic NGM plates (see recipe)
-
10× and 40× E. coli OP50 (see Support Protocol 1)
-
Spore-counting solution (see recipe)
-
2-ml microcentrifuge tubes, sterile
-
Bead disruptor (Scientific Industries Digital Disrupter Genie or equivalent)
-
21°C and 25°C (and another appropriate temperature; optional) incubator (Thermo Scientific Forma environmental chamber or equivalent)
-
Laminar flow cabinet
-
Parafilm
-
Sealable plastic box
-
Metal spatula
-
Plastic boxes with lids
-
15-ml conical tubes, sterile (Thermo, 430053)
-
Centrifuge (Eppendorf 5430R or equivalent)
-
Dissecting microscope with transmitted light (Zeiss Stemi 305 or equivalent)
-
50-ml conical tubes (FroggaBio, TB50-500), sterile
-
5-μm syringe filters, sterile (Fisher Scientific, SLSV025LS)
-
3-ml syringes, sterile (Fisher Scientific, 14-955-457)
-
CELL-VU™ disposable counting chamber (Millennium Sciences, DRM600), including slide and coverslip
-
Fluorescence microscope with GFP filter (Zeiss Axio Imager M2 or equivalent)
-
Additional reagents and equipment for chunking (see Basic Protocol 2, step 2), bleach synchronization (see Support Protocol 2), and freezing down sample aliquots (see Basic Protocol 2, steps 19 to 21)
CAUTION : Ethanol is highly flammable. When spraying the bench, ensure that the Bunsen burner is not lit.
NOTE : It is crucial that proper sterile technique is utilized and that filter tips are used at every step in this protocol. Any contaminated plates must immediately be discarded to prevent spread of contamination.
Antibiotic cleanup
About 4 to 5 days before cleanup
1.Chunk the environmental sample for cleanup onto a 10-cm seeded NGM plate (see Basic Protocol 2, step 2) and grow until densely populated.
Cleanup day
2.Sterilize the benchtop and micropipets using 70% ethanol. Assess the contaminants present on the plate, making note of the properties and colors of bacterial colonies.
3.Using 1 ml sterile M9, wash animals off the plate and into a sterile 2-ml microcentrifuge tube. Add ∼100 μl sterile 2.0-mm zirconia beads.
4.Place tubes in a bead disruptor and vortex at 3000 rpm for 5 min.
5.Let the beads settle and transfer supernatant to a clean tube. Store the supernatant for ≥1 hr at −80°C.
6.Place the entire supernatant on a 6-cm unseeded antibiotic NGM plate. Incubate the plate for 24 to 48 hr at 21°C.
7.Bleach-synchronize a population of animals and hatch overnight (see Support Protocol 2).
8.Add 400 µl of 10× E. coli OP50 and 1000 synchronized L1 animals to the plate from step 6.Dry the plate either on a bench or in a laminar flow cabinet.
9.Parafilm and incubate the plate for ∼4 days at the appropriate temperature in a sealed plastic box.
10.Chunk the animals onto a new 6-cm seeded antibiotic NGM plate (see Basic Protocol 2, step 2) and incubate for several days.
11.Chunk the animals onto a 10-cm seeded NGM plate (no antibiotics) (see Basic Protocol 2, step 2) and incubate for several days. Using the remaining animals on the plate, determine the percentage of the adult population containing microsporidia spores, as described in Basic Protocol 2, steps 12 and 13.
12.Assess the degree of contamination present on the plate. If the sample is deemed to be clean, proceed to using it to generate a spore preparation. If not, use the plate generated in step 10 and repeat steps 2 to 11 until no bacterial contamination is detected.
13.Freeze down several clean aliquots of the sample, as described in Basic Protocol 2, steps 19 to 21.
Spore preparation
Stage 1: Setting up source plates
14.Spray the bench with 70% ethanol to sterilize.
15.Thaw an aliquot of an infected population (see step 13) or chunk a densely populated contaminant-free plate (see step 12) evenly onto three 10-cm seeded NGM plates.
16.Dry the plates on a bench or in a laminar flow cabinet and incubate at 21°C until the population has crashed out.
Stage 1, continued: 18 to 24 hr before using source plates
17.Bleach-synchronize a population of animals (see Support Protocol 2) to be used in step 19.Hatch overnight.
18.For each of the three source plates, prepare 24 “harvesting plates” (10-cm seeded NGM plates, 72 plates total).
Stage 2: Expanding the infected population
19.The next day, add 2640 L1 animals from step 17 to 6600 μl of 40× E. coli OP50 for every 24 harvesting plates.
20.Pipet 250 μl of the nematode– E. coli mixture onto each of the 72 plates from step 18.
21.Chunk each of the three source plates into 24 sectors (pie slices) using a metal spatula dipped in ethanol and flamed as in Basic Protocol 2, step 2.Place 1/24 of a sector upside down on top of the nematode– E. coli mix.
22.Store the plates right side up in plastic boxes with a lid. Incubate at 21°C for about 4 to 7 days.
23.Screen the plates every 2 days for contamination, discarding any plates that display dark bacterial colonies or fungal mold.
24.Monitor the population density by looking at the number and size of animals on the plates.
Stage 3: Harvesting animals
25.Pool the animals from about 5 to 6 plates together into a single sterile 15-ml conical tube by washing animals off plates using 2 to 3 ml sterile M9 per plate.
26.Pellet worms by centrifuging 15-ml conical tubes for 30 s at 1400 × g.
27.Remove the supernatant and wash the worms with 10 ml sterile M9.
28.Repeat steps 26 and 27 until the supernatants are clear.
29.Centrifuge tubes for 30 s at 7197 × g and remove the supernatant down to 1 ml.
30.Transfer the 1 ml into a sterile 2-ml microcentrifuge tube. Transfer 50 μl into a separate microcentrifuge tube for contamination checks (see steps 32 and 33).
31.Freeze tubes at −80°C for ≥24 hr.
Stage 4: Contamination checks
32.Thaw tubes containing 50-μl samples. Pipet 10 μl undiluted sample and 10 μl of a 1:50 dilution (in sterile M9) onto separate 6-cm unseeded antibiotic NGM plates. Incubate each dilution at 21°C and 25°C for 5 days.
33.Assess the plates under a dissecting microscope with transmitted light, looking for any colonies that do not look like OP50.
Stage 5: Break up worms and filter lysate
34.Thaw uncontaminated 1-ml aliquots from step 31 and add ∼500 μl sterile 2.0-mm zirconia beads. Vortex in a bead disruptor at 3000 rpm for 5 min.
35.Transfer the supernatants into a sterile 50-ml conical tube. Wash the beads twice more with 1 ml sterile M9 and add supernatants to the same tube.
36.Filter the lysates through a sterile 5-μm syringe filter attached to a sterile 3-ml syringe.
37.Aliquot the filtered lysate into desired volumes in microcentrifuge tubes and store at −80°C. Reserve some unfrozen lysate for step 38.
Stage 6: Measure spore concentration
38.Determine the concentration of spores by combining 10 μl spore prep lysate (see step 37) and 90 μl spore-counting solution in a new microcentrifuge tube. Mix by pipetting to ensure spores are in solution. Add 4 μl stained spores onto a CELL-VU™ slide by putting the drop on the inner edge of the circle. Place the coverslip so that the edge just covers the drop of spores and then carefully slide the coverslip so that the circle is fully covered.
39.Using a fluorescence microscope with a 20× objective, count the total number of green spores in 10 grids. Divide the number of spores counted by 200 to determine the concentration of the spore preparation lysate in millions of spores per μl.
Stage 7: Quality control for contamination
40.Thaw one aliquot of the spore prep from step 37 to test for contamination as described in steps 32 and 33.
Support Protocol 2: BLEACH-SYNCHRONIZING NEMATODES
This protocol describes the use of a sodium hypochlorite and sodium hydroxide mixture (bleach solution) to isolate embryos from adult nematodes. In doing so, the population becomes synchronized as the embryos hatch into M9 in the absence of food. Thus, all nematodes are unable to continue development until placed on a food source.
Materials
-
Densely populated but unstarved nematodes
-
M9 (see recipe), sterile
-
6-cm seeded NGM plates
-
1.7-ml microcentrifuge tubes, sterile
-
Timer
-
Microcentrifuge (Eppendorf S424 or equivalent)
-
Dissecting microscope with transmitted light (Zeiss Stemi 305 or equivalent)
-
15-ml conical tubes, sterile (Fisher, 430053)
-
Rotisserie rotator (Thermo Scientific, 415110)
-
21°C incubator (Thermo Scientific Forma environmental chamber or equivalent)
-
Centrifuge (Eppendorf 5702 or equivalent)
-
Additional reagents and equipment for preparing bleach solution (see recipe)
1.Prepare the required volume of bleach solution.
2.Wash densely populated but unstarved nematodes off plate and into a sterile 1.7-ml microcentrifuge tube with 1 ml sterile M9.
3.Allow nematodes to gravity settle, generating a worm pellet.
4.Remove supernatant.
5.Add 1 ml bleach solution from step 1 to the nematode pellet and set a timer for 1 min.
6.Centrifuge samples for 30 s at 845 × _ g_.
7.Remove supernatant and add another 1 ml bleach solution.
8.Observe the samples under a dissecting microscope with transmitted light, looking for populations where ∼80% of the carcasses have dissolved and released embryos into the environment.
9.Centrifuge samples for 30 s at 845 × g.
10.Remove supernatant and replace with 1 ml sterile M9.
11.Repeat steps 9 and 10 two more times.
12.Add 1 ml sterile M9 to the worm pellet and transfer to a sterile 15-ml conical tube containing 4 ml sterile M9.
13.Place tube on a rotisserie rotator overnight at 21°C.
14.Observe the tube under the dissecting microscope.
15.Spin down the tube for 30 s at 1400 × g.
16.Concentrate the worm pellet down to 1 ml.
17.Resuspend the pellet and estimate the concentration of nematodes present by aliquoting 5 μl onto a 6-cm seeded NGM plate and counting the number of thrashing L1s.
REAGENTS AND SOLUTIONS
Agarose, 1%
- 0.5 g ultra-pure agarose (Invitrogen, LS16500500)
- 50 ml 1× TAE (diluted from 50× TAE; see recipe)
- Microwave for 1 min
- Add 2.5 μl 10,000× SYBR® Safe DNA Gel Stain (Invitrogen, S33102)
- Pour mixture into gel cast with comb and let solidify
- Wrap in plastic wrap followed by tinfoil
- Store ≤1 week at 4°C
Volumes should be adjusted to the size and dimensions of the gel-casting equipment used.
Antibiotic NGM plates
- 50 mg/ml carbenicillin
- 25 mg/ml kanamycin
- 12.5 mg/ml tetracycline
- 37.5 mg/ml chloramphenicol (resuspended in 95% ethanol)
- 100 mg/ml cefotaxime
- 50 mg/ml gentamycin
- For generating unseeded plates, create antibiotic mixture by adding 12.5 μl of each antibiotic solution except for cefotaxime and gentamycin, for which 25 μl should be added, to 300 μl M9 (see recipe)
- For generating seeded plates, centrifuge 1 ml 10× E. coli OP50 (see Support Protocol 1) for 1 min at 7197 × g , remove all but 300 μl of supernatant, resuspend pellet, and add bacteria to 100 μl antibiotic mixture
- Add the 400 μl solution to the top of an 6-cm unseeded NGM plate and gently rock plate to spread liquid evenly
- Dry in laminar flow cabinet or on bench
- Store ≤1 week at 4°C
Bleach solution
- 125 μl sodium hypochlorite (6% bleach; Fisher, SS290-1; store at 4°C in the dark)
- 200 μl 1 N NaOH (Fisher, SS266-1; store at 4°C)
- 675 μl MilliQ H2O
- Prepare fresh immediately before use, store for maximum of 1 hr at room temperature
Each sample will require 2 ml bleach solution, so scale up as required.
Calcium chloride (CaCl2), 1 M
- 219 g CaCl2 (Fisher Scientific, C78500)
- 1 L MilliQ H2O
- Aliquot 90 ml into 100-ml bottles
- Autoclave for 30 min
- Store ≤2 years at room temperature
Cholesterol, 5 mg/ml
- 250 mg cholesterol (Sigma-Aldrich, C3045-5G; store at −20°C)
- 50 ml 95% (v/v) ethanol (Fisherbrand, HC11001GL)
- Wrap in tinfoil
- Store ≤1 year at room temperature
Direct Yellow 96 (DY96) solution
- 5 ml 10× phosphate-buffered saline (PBS; BioBasic, UPD8117)
- 0.5 ml 10% (v/v) Tween-20 (see recipe)
- 0.5 ml 10% (w/v) SDS (see recipe)
- 200 μl 5 mg/ml DY96 stock solution (see recipe)
- 43.8 ml MilliQ H2O
- Mix by inverting tube
- Store ≤2 years at room temperature in the dark
The solution will be a pale yellow color.
If desired, the tube can be wrapped in tinfoil to minimize light exposure.
Direct Yellow 96 (DY96) stock solution, 5 mg/ml
- 50 mg DY96 (1PlusChem, 1P00EDJN)
- 10 ml MilliQ H2O
- Mix by inverting tube
- Wrap tube in tinfoil and store in the dark
- Store ≤2 years at room temperature in the dark
The solution will be a bright yellow color.
Freezing medium
- Mix equal volumes of S-basal and freezing solutions (see recipes)
- Store ≤1 year at 4°C
Freezing solution
- 5.85 g NaCl (BioShop Canada, SOD002.1)
- 1 g K2 HPO4 (potassium phosphate dibasic; BioShop Canada, PPD303.1)
- 6 g KH2PO4 (potassium phosphate monobasic; BioShop Canada, PPM302.1)
- 300 ml glycerol (BioShop Canada, Gly002.1)
- 700 ml MilliQ H2O
- Aliquot
- Autoclave for 30 min
- Add 300 µl 1 M MgSO4 (see recipe)
- Store ≤2 years at room temperature
LB agar plates
- 10 g NaCl (BioShop Canada, SOD002.1)
- 10 g tryptone (BioShop Canada, TRP402.1)
- 5 g yeast extract (Fisher Scientific, BP1422-2)
- 15 g agar (Fisher Scientific, BP1423-2)
- 1 L MilliQ H2O
- Autoclave for 30 min
- Cool to 55°C before pouring into 10-cm plates
- Store ≤1 month at 4°C
Agar will solidify if cooled for too long.
Luria broth (LB)
- 10 g NaCl (BioShop Canada, SOD002.1)
- 10 g tryptone (BioShop Canada, TRP402.1)
- 5 g yeast extract (Fisher Scientific, BP1422-2)
- 975 ml MilliQ H2O
- Combine in 2-L flask or aliquot into desired volumes for smaller cultures
- Autoclave for 30 min
- Store ≤6 months at room temperature
Always ensure that LB is sterile before use (sterile LB is clear and not cloudy).
Wait for the LB to cool down prior to inoculation with bacteria.
M9
- 5.8 g Na2HPO4 (disodium phosphate; BioShop Canada, SPD307.1)
- 3.0 g KH2PO4 (potassium phosphate monobasic; BioShop Canada, PPM302.1)
- 0.5 g NaCl (BioShop Canada, SOD002.1)
- 1.0 g NH4Cl (Fisher Chemical, A661-500)
- 998 ml MilliQ H2O
- Aliquot into 50-ml bottles
- Autoclave for 30 min
- Store ≤2 years at room temperature
M9 + 0.1% Tween-20
- 49.5 ml M9 (see recipe)
- 0.5 ml 10% (v/v) Tween-20 (see recipe)
- Store ≤2 years at room temperature
Magnesium sulfate (MgSO4), 1 M
- 246.5 g MgSO4 (Fisher Scientific, M63-500)
- 1 L MilliQ H2O
- Aliquot 90 ml into 100-ml bottles
- Autoclave for 30 min
- Store ≤2 years at room temperature
Nematode growth medium (NGM) plates
- 20 g agar (Fisher Scientific, BP1423-2)
- 3 g NaCl (BioShop Canada, SOD002.1)
- 2.5 g Bacto Peptone (Thermo Fisher Scientific, DF0118-07-2)
- 980 ml MilliQ H2O
- Autoclave for 30 min
- 25 ml 1 M KPi (see recipe)
- 1 ml 1 M MgSO4 (see recipe)
- 1 ml 1 M CaCl2 (see recipe)
- 1 ml 5 mg/ml cholesterol (see recipe)
- Pour 12 ml medium in 6-cm plate or 30 ml medium in 10-cm plate
- Once solidified, place upside down and store ≤6 months at 4°C in tightly sealed container
Potassium chloride (KCl), 1 M
- 74.55 g KCl (Sigma, P3911)
- 1 L MilliQ H2O
- Autoclave 20 min
- Store ≤2 years at room temperature
Potassium phosphate (KPi), 1 M
- 30.1 g K2HPO4 (potassium phosphate dibasic; BioShop Canada, PPD303.1)
- 118.1 g KH2PO4 (potassium phosphate monobasic; BioShop Canada, PPM302.1)
- 1 L MilliQ H2O
- Aliquot
- Autoclave 30 min
- Store ≤2 years at room temperature
S-basal solution
- 5.85 g NaCl (BioShop Canada, SOD002.1)
- 1 g K2HPO4 (potassium phosphate dibasic; BioShop Canada, PPD303.1)
- 6 g KH2PO4 (potassium phosphate monobasic; BioShop Canada, PPM302.1)
- 1 ml 5 mg/ml cholesterol (see recipe)
- 995 ml MilliQ H2O
- Aliquot
- Autoclave for 30 min
- Store ≤2 years at room temperature
Single-worm lysis buffer
- 2 ml 1 M Tris HCl (Fisher Scientific, J22638-AP)
- 1 ml 1 M KCl (see recipe)
- 50 μl 1 M MgCl2 (Thermo Fisher, J61014.AK)
- 90 μl 100% Tween-20 (Sigma-Aldrich, P7945)
- 60 μl 20 mg/ml proteinase K (NEB, P8107S)
- 90 μl NP40 substitute (Sigma, 492018)
- 16.71 ml nuclease-free H2O (Invitrogen, 10977015)
- Aliquot into 1-ml microcentrifuge tubes
- Store ≤2 years at −20°C
- Thaw aliquot before use and store ≤6 months at 4°C
Sodium dodecyl sulfate (SDS), 10%
- 50 g SDS (Sigma, L3771)
- 400 ml MilliQ H2O
- Stir until dissolved
- MilliQ H2O to 500 ml
- Aliquot
- Store ≤2 years at room temperature
Do not autoclave.
Spore-counting solution
- 800 μl 50% (v/v) glycerol
- 90 μl M9 (see recipe) or 10× PBS (BioBasic, UPD8117)
- 10 μl 5 mg/ml DY96 stock solution (see recipe)
- Store ≤1 year at room temperature in the dark
TAE, 50×
- 242 g Tris base (BioShop Canada, TRS001.1)
- 100 ml 0.5 M EDTA (Thermo Fisher, J62786.AP)
- 57.1 ml glacial acetic acid (Sigma, 695092)
- MilliQ H2O to 1 L
- Store ≤2 years at room temperature
- Dilute to 1× in MilliQ H2O if needed
Do not autoclave.
Tween-20, 10%
- 5 ml 100% Tween-20 (Sigma-Aldrich, P7945)
- 45 ml MilliQ H2O
- Store ≤2 years at room temperature
COMMENTARY
Background Information
Microsporidia are a major threat to many commercially and ecologically important animals (Stentiford et al., 2013, 2016). They are prevalent in the environment and can cause disease in the immunocompromised (Han & Weiss, 2017; Ruan et al., 2021). Microsporidia are currently known to infect about 1500 animal species, though this is likely a large underestimation of the true diversity of microsporidia (Larsen et al., 2017; Murareanu et al., 2021). Despite their widespread nature, many aspects of their infection biology remain enigmatic. Given their extreme host dependance, studying microsporidia within a whole-animal model is advantageous and can reveal host-specific aspects of infection. Approximately 15 years ago, microsporidia were found to naturally infect C. elegans (Tecle & Troemel, 2022; Troemel et al., 2008). The discovery of the nematode-infecting species N. parisii provided an excellent opportunity to study this genetically intractable parasite in a genetically tractable host. Additionally, studying microsporidia infection in nematodes offers several advantages. First, nematodes can be readily sampled from the environment and screened for the presence of microsporidia. In fact, such efforts have already identified at least 15 species of nematode-infecting microsporidia, many of which can also infect C. elegans (Luallen et al., 2016; Reinke et al., 2017; Wadi et al., 2023; Zhang et al., 2016). Second, many species of nematodes can be easily cultured in the laboratory under the same conditions. Third, many nematodes have short generation times and can be cultured at high densities, allowing for infection of large populations of animals. Lastly, and particularly for C. elegans , the many gene editing and manipulation tools available can circumvent the lack of those in microsporidia (Reinke & Troemel, 2015). As multiple species of microsporidia infect C. elegans , this has become a powerful system for understanding evolution of transcriptional responses and host resistance mechanisms and for identifying microsporidia inhibitors with broad specificity (Mok et al., 2023; Murareanu et al., 2022; Wan et al., 2022).
There are thousands of known free-living terrestrial species of nematodes, though this is likely an underestimation (Schratzberger et al., 2019; Smythe et al., 2019). Many species of terrestrial free-living nematodes are often found in microbially rich environments, including decomposing fruits and stems, as discussed in Basic Protocol 1 (Barriere & Felix, 2014). The geographical location of sampling will dictate the nematode species isolated, as will the time of year. For example, C. elegans are found in temperate regions primarily in the fall (Frézal & Félix, 2015; Schulenburg & Félix, 2017). From our own sampling efforts in southern Ontario, C. elegans does not appear to be present, but we have had success identifying other Caenorhabditis species, as well as those from the genera Oscheius , Pristionchus , and Panagrellus. Microsporidia can infect many different animal tissues, but the intestine or muscle/epidermis is the primary site of infection in nematodes (Murareanu et al., 2021).
Species of the genera Nematocida , Enterospectra , and Pancytospora have been found to infect various Caenorhabditis and related nematodes. The spores of many Nematocida species have been characterized and can be divided into two main groups. Those that form two classes of spore sizes are characteristic of the Parisii group, whereas those forming a single spore size and large spore clusters are more likely to be part of the Displodere group. Species in the Parisii group have so far only been found to infect the intestine, whereas some species in the Displodere group have been observed to infect the muscle and epidermis. There are currently respectively seven and eight members of the two groups (Wadi et al., 2023; Zhang et al., 2016). Nematocida spores range from 1.3 to 2.4 μm in length and 0.5 to 1.0 μm in width. Pancytospora species are typically longer, up to 4 μm in length (Zhang et al., 2016). Therefore, characterizing spore size and tissue tropism can provide insight into the identity of the species.
Performing infection characterization in a genetically tractable host is desirable, and many species infect C. elegans in the lab, even if they were originally isolated from a different nematode species. We provide a strategy to assess if C. elegans is susceptible to infections from isolated microsporidia species by utilizing a transgenic strain carrying an infection reporter, as described in Basic Protocol 2. An immune response termed the intracellular pathogen response (IPR) is upregulated in C. elegans upon microsporidia infection. One of the most highly upregulated IPR genes is pals-5. The use of the P pals5::GFP reporter, coupled with DY96 staining, can be used to determine if a species can infect C. elegans (Willis et al., 2022). This enables the potential study of novel microsporidia species within a genetically tractable host.
Although several primer pairs exist for amplifying molecular barcode genes in microsporidia, amplification of nematode-infecting microsporidia is not always successful (Zhang et al., 2016). Although the small subunit 18S rRNA is the most commonly used phylogenetic marker in microsporidia, several other options exist (Bojko et al., 2022). Additionally, as genomic sequencing technology such as Nanopore DNA sequencing becomes cheaper and more available, multigene species identification that is independent of PCR will become possible (Taxt et al., 2020). The molecular characterization of nematode-infecting microsporidia is explored thoroughly in Basic Protocol 3.
The generation of a large quantity of dormant spores can enable multiple different areas of investigation and is explored in Basic Protocol 4. These spores can serve as a template for whole-genome sequencing, allowing for genomic comparison between multiple different species and assessment of microsporidian evolution. They could be used to characterize infection dynamics compared to other nematode-infecting species as well as the host response to infection (Bakowski et al., 2014; Wan et al., 2022). Lastly, as microsporidia demonstrate great host specificity, the availability of multiple different species of microsporidia that infect related nematodes can help uncover the host-specific cues that drive host specificity (Wadi et al., 2023; Willis & Reinke, 2022; Zhang et al., 2016). Given the number of species of nematode-infecting microsporidia identified to date, we have likely just scratched the surface (see Table S2 in the Supporting Information for properties of reported nematode-infecting microsporidia species). Expanding the known diversity of these species will allow us to gain insight into novel aspects of microsporidian evolution and infection mechanisms.
Critical Parameters
Basic Protocol 1
The sampling process may not yield any nematodes. The key parameters are the time of year, temperature, location, type of substrate, and level of substrate decay. In our own experience, from sampling in and around Toronto, Canada, up to ∼50% of samples collected between September and November contain nematodes. Once nematodes are identified, it is expected that the population will continue to proliferate. However, some nematode populations may fail to do so. It is important to consider the temperature and conditions from which the sample was obtained and assess if these differ drastically from the laboratory environment. It is worth considering if the nematodes are a male-female species or hermaphrodites. In the unfortunate event that the population obtained from the substrate was exclusively male or female, the population will likely not be able to be propagated. Careful observation and characterization of the population should be done as soon as nematodes are detected.
To increase the chance of detecting microsporidia in a population, allow the nematodes to grow for at least 5 to 6 days after substrate removal. As DY96 dye stains only spores, the microsporidia need to reach this stage to be observed. Although the time to sporulation will depend on the species, we have observed that most species will form spores within 96 hr after infection. Harvesting samples too soon may result in missing potential microsporidia infection. Additionally, as we propagate samples by chunking the plates, there is a higher chance that dormant spores are also present on the agar if animals had sufficient time to grow and sporulate. This is especially critical if an epidermal infection is present, where spores are shed only when old animals rupture and release new spores. This is different from intestinal-infecting species, where spores are continually shed into the environment. Always remember to propagate a sample prior to washing it off for fixation. Once the sample is fixed, the nematodes and microsporidia are not viable and cannot be recovered.
Support Protocol 1
Working in a sterile environment to avoid contaminating the E. coli OP50 stock is critical. This also applies when pouring and seeding NGM plates. Avoid using seeded NGM plates that are older than 1 month. Always inspect plates to ensure that the agar is not peeling off the sides.
Basic Protocol 2
When screening DY96-stained animals for microsporidia, be aware that many things may fluoresce in the green channel. Use the flowchart in Figure 4A as a guide. When assessing the fraction of infected animals, do so in at least 100 animals and make sure that only the adult animals are being quantified. Younger animals will not have been around for ≥72 hr, meaning that microsporidia will not have had sufficient time to sporulate. Thus, they will appear uninfected but may harbor the intracellular stages that cannot be stained with DY96. In contrast, smaller animals full of spores may indicate heavily infected animals that succumbed to the infection, as microsporidia infection can impact host fitness in a dose-dependent manner (Willis et al., 2021). If only a small fraction of worms are infected, consider how to modify the propagation approach to ensure that the population is not bottlenecked when chunking. If only 10% of the population is infected, consider leaving animals on the plates for longer than 5 to 6 days and adding additional E. coli to the plates instead. Alternatively, chunk half the plate to propagate the population, rather than a quarter, and continually supplement the plate with additional E. coli. If the infectivity is still low, consider incubating the plates at different temperatures. One other possibility for low infectivity is that the infection is not in the native nematode host. It is possible that this microsporidia species propagates better in other nematode species. If the species can infect C. elegans , this is a convenient host in which to grow and characterize the microsporidia species.
Basic Protocol 3
When generating worm lysates, it is important to pick about 15 large, old animals into the lysis buffer. This increases the chances of picking infected animals, as they will have been around the longest. If animals are stuck on the sides of the PCR tube, this will not result in successful DNA extraction. If a sample was found to be only mildly infected, this may influence the ability to amplify microsporidia-specific sequences.
Basic Protocol 4
When performing antibiotic cleanup, it is critical that no additional contamination is acquired. Having a clean sample will ensure that during the spore preparation, only pure microsporidia spores are isolated. Keeping a sterile work environment and using proper sterile technique should help limit this. When harvesting animals for spore preparation, it is critical that plates are densely populated with large animals. This ensures maximum spore yield.
Support Protocol 2
The bleach solution should always be prepared fresh. Densely populated plates with large, unstarved animals that have many embryos should be used.
Troubleshooting
Commonly encountered issues and their potential causes and solutions are listed in Table 3.
| Problem | Possible cause | Solution |
|---|---|---|
| Limited/no nematode proliferation | Toxic contaminant or improper growth conditions | Try to pick off as many worms as possible onto a new 6-cm seeded plate |
| Lack of a mate | This is unfortunately a dead end | |
| Animals keep starving | Lack of food | Use a higher concentration of OP50 and constantly monitor plate |
| Mites are present in most of the sampled plates | Leaving the samples for too long in a box, stacked on top of one another, without screening for mites. These mites have now likely spread between plates. | It is best to discard the plates rather than trying to save the samples. Microwave samples and dispose of in appropriate biosafety waste. |
| Under the fluorescence microscope, none of the animals is large | Samples were fixed too early, or the population has starved and only larvae remain | Ensure that samples are washed off only when plates are densely populated with large adult animals |
| Fewer than 100 animals are present on the glass slide | Not enough animals were washed off the plate | Ensure that as much of the population as possible is washed off the plate. Multiple washes may be necessary. |
| Animals were lost during washes due to improper settling/centrifugation | This is most likely to happen during the first step, which is gravity settling. Ensure that when removing the supernatant, that the pellet is not disrupted. If the sample has been disrupted, place the sample back into the tube and let it settle again. | |
| Not enough sample on the slide | Load more sample on a separate slide | |
| Thawed vials of a once-infected sample are no longer infected | Sample was not heavily infected to begin with and may have been bottlenecked | Try to recover the other frozen aliquots. If the issue persists, try freezing another set if the plate is still being propagated. |
| Frozen aliquot was not successfully recovered | Not enough L1 animals present during freezing | L1 animals are the most resilient to freezing. Ensuring that the plate being harvested for freezing contains many freshly hatched L1 animals and is either nearly or freshly starved is best. |
| Microsporidia PCR products not visible on DNA gel | Lysate does not contain enough microsporidia DNA | Assess if sample is not very infected, and if so, attempt to boost infectivity and try again |
| Improper cycling parameters | Consider gradient PCR to identify the optimal annealing temperature. Consider if the extension time is sufficient based on the polymerase used. | |
| Species cannot be amplified with current primers | Some species of microsporidia are not efficiently amplified with the current primer pairs. If PCR is not possible with any primer pairs, consider whole-genome sequencing. | |
| Bacterial contaminants are still growing despite multiple rounds of antibiotic cleanup | Worms have not been fully broken up or worms are still alive after the bead extraction | Consider a longer bead extraction or longer freezing period |
| Bacteria may have some amount of antibiotic resistance | Incubate extracted spores for several more days before adding bleached worms | |
| Low spore yield from spore preparation | Animals were harvested too early | Only wash nematodes off plates when there are many large, sick animals. Washing before this point may result in animals that contain intracellular stages that have not yet sporulated, resulting in low yield. |
| Spore preparation infectivity differs between aliquots | Spores were not continually resuspended while aliquoting | Ensure that spores are continually mixed either by using a pipet or by gently shaking the conical tube to minimize this issue. |
| All L1 animals are dead/few to no L1 animals recovered after bleach synchronization | Sample is overbleached | Avoid exposing animals to the bleach solution for too long. Aim for ∼20% visible animal carcasses before stopping the reaction. Nematode species being bleached may not carry as many embryos internally, resulting in low yield. Some species that do not lay embryos, such as Panagrellus redivivus, are also difficult to bleach. |
Understanding Results
When collecting substrates from the environment (Basic Protocol 1), those containing nematodes will be easy to distinguish, as nematodes will proliferate on the NGM agar and populate the plate. Nematode tracks in the E. coli OP50 lawn or embryos on the plate can also indicate the presence of nematodes. To monitor how long nematodes have been on plates, labeling each with the date is strongly encouraged. In parallel to this, keeping a log of sample history is very useful and can help avoid confusion when dealing with multiple samples. Table S1 in the Supporting Information provides an example of the type of information to collect on the samples.
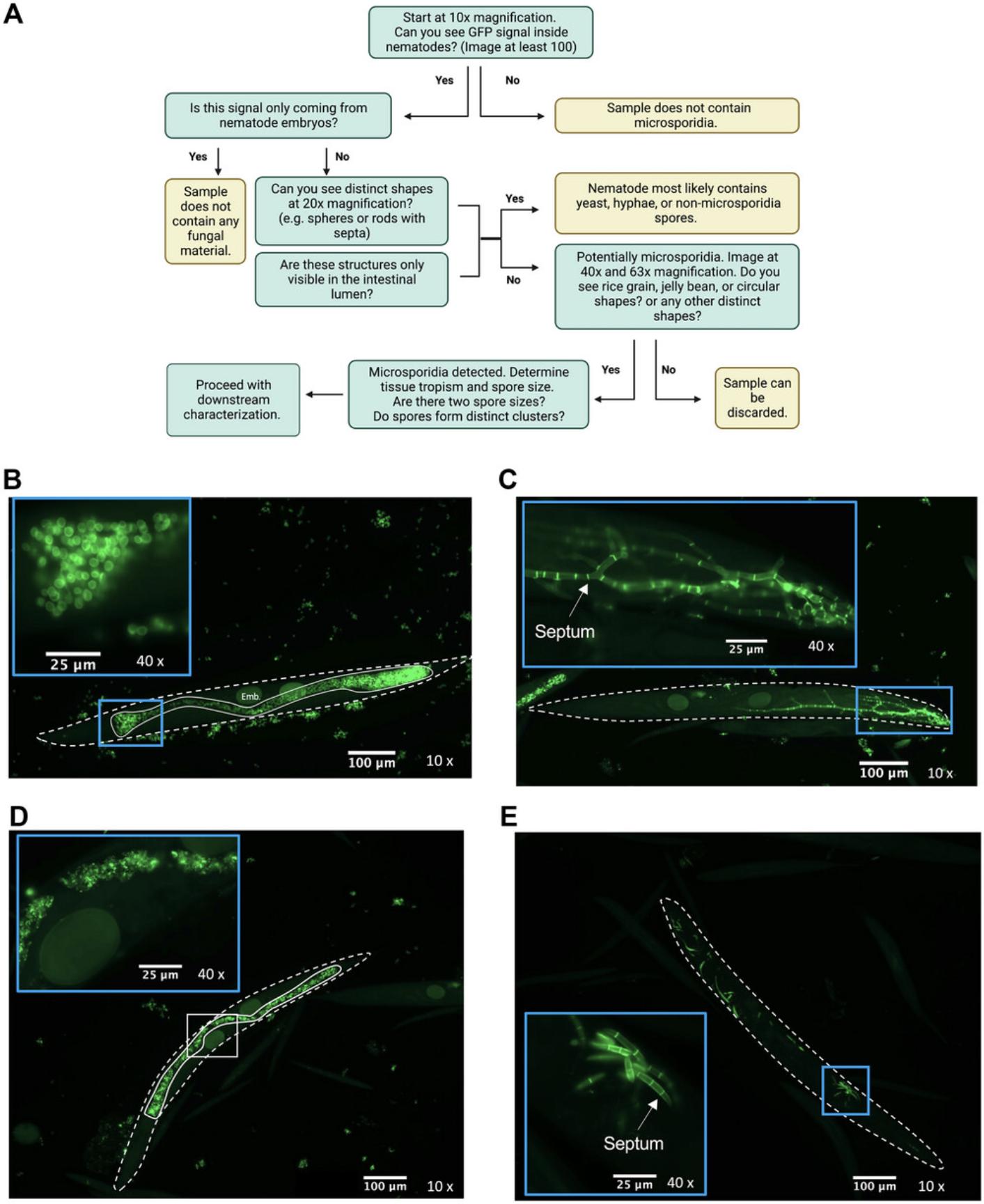
The most critical part of this set of protocols is correctly identifying microsporidia infection in nematodes (Basic Protocol 2). Figure 4A provides a flowchart to help assess if any of the samples contain microsporidia. This flowchart assumes that the previous steps (sample propagation) have been performed as outlined. We also provide examples of non-microsporidia fungal material that is frequently observed in environmental samples. We often see yeast cells that are circular in structure and accumulate in the intestinal lumen (Fig. 4B). Fungal hyphae that contain cell walls or septa between segments can also be seen growing inside of animals (Fig. 4C). This is not seen in microsporidia. We also observe GFP material accumulating in the intestinal lumen, with no defined structure or shape, that looks like speckles (Fig. 4D). The identity of this material is unknown but may represent digested fungal material in the intestinal lumen. Fungal spores that contain septa can also be seen inside the nematode intestine. However, these are quite large and are easily visible even at 10× magnification (Fig. 4E). Large amounts of chitinous structures that outline the intestinal lumen are not microsporidia. Microsporidia spores will often be present both in the intestinal lumen and within the cells of the nematode. Epidermal-infecting species must first be ingested but should not be seen exclusively within the intestinal lumen and should also be visible in the target tissue. In the rare event that microsporidia spores are seen exclusively in the intestinal lumen, this indicates that this species may not be able to infect the current nematode host.
Figure 5 provides examples of what microsporidia look like and how to identify if parasite cells display spore clusters (Fig. 5C) and assess if there are two classes of spore sizes (Fig. 5D). We also provide examples of how microsporidia spores may appear (Figs. 5E and 5F). Some spores look like rice grains (Fig. 5E) or have a short and stubby morphology (Fig. 5F). This is not an exhaustive list of what may be seen but should serve as a good starting point to help understand what to look for. Microsporidia morphology is generally not visible at 10× or 20× magnification and appears as a cluster of fluorescent material that is not restricted to the intestinal lumen. The example provided in Figure 5A depicts an intestinal-infecting species. Keep in mind that there may be microsporidia species that grow in the muscle or epidermis. These can be detected by assessing if spores can be observed anterior to the pharynx of the nematode (Fig. 5B).
PCR amplification of microsporidia SSU RNA (Basic Protocol 3) has had variable success in our experience. The presence of a clear band at ∼1300 bp with V1F and any of its accompanying reverse primers is a good indication of success. However, the lack of a PCR amplicon coupled with the lack of a positive control makes it difficult to troubleshoot this part of the protocol. Using different primer pairs can be useful in ensuring that at least one of them amplifies a microsporidia sequence. The primer pair yielding the strongest signal on a DNA gel with the least amount of nonspecific amplification is prioritized for Sanger sequencing. It is not uncommon to have difficulty amplifying microsporidian SSU RNA. Part of the problem lies in the large amount of diversity present in microsporidian genomes, making the design of a completely universal primer challenging. Whole-genome sequencing can be considered for species identification if amplification proves unsuccessful.
For antibiotic cleanup of samples (Basic Protocol 4), the lack of visible foreign microbial growth on the plate will indicate success. These samples can now be prioritized for spore preparations. A successful spore prep will yield a large quantity of contaminant-free spores that can be used for controlled infection experiments.
Time Considerations
The entire set of protocols can be completed in about 2.5 to 4 months. Initial sample collection and screening (Basic Protocols 1 and 2) take ∼3 weeks. Molecular characterization of nematode and microsporidia species (Basic Protocol 3) can take up to 1 month if troubleshooting of PCR parameters is required. The most time-consuming part will be the antibiotic cleanup (anywhere from 2 weeks to 1.5 months) and the spore preparation (∼1 month) in Basic Protocol 4.
Acknowledgments
We thank Winnie Zhao, Carolyn Chen, and Jonathan Tersigni for providing helpful comments on the manuscript. We thank Lisa van Sluijs for providing some of the infected nematode samples. This work was supported by a Canadian Institutes of Health Research grant no. 400784 (to A.W.R.), and H.T.E.J was supported by a University of Toronto Open Fellowship and an Ontario Graduate Scholarship.
Author Contributions
Hala Tamim El Jarkass : Conceptualization; data curation; investigation; methodology; visualization; writing—original draft; writing—review and editing. Aaron W. Reinke : Conceptualization; Funding acquisition; Methodology; Supervision; Writing—review and editing.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Open Research
Data Availability Statement
All the data that support the findings of this study are included within this article.
Supporting Information
| Filename | Description |
|---|---|
| cpz11035-sup-0001-tableS1.xlsx11.3 KB | Table S1. An example spreadsheet containing relevant information for sample tracking. |
| cpz11035-sup-0002-tableS2.xlsx24.1 KB | Table S2. Characteristics of microsporidia that infect free-living terrestrial nematodes. Table is modified from Murareanu et al. (2021). |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Bakowski, M. A., Desjardins, C. A., Smelkinson, M. G., Dunbar, T. A., Lopez-Moyado, I. F., Rifkin, S. A., Cuomo, C. A., & Troemel, E. R. (2014). Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLOS Pathogens , 10(6), e1004200. https://doi.org/10.1371/journal.ppat.1004200
- Balla, K. M., Andersen, E. C., Kruglyak, L., & Troemel, E. R. (2015). A wild C. elegans strain has enhanced epithelial immunity to a natural microsporidian parasite. PLOS Pathogens , 11(2), e1004583. https://doi.org/10.1371/journal.ppat.1004583
- Balla, K. M., Luallen, R. J., Bakowski, M. A., & Troemel, E. R. (2016). Cell-to-cell spread of microsporidia causes Caenorhabditis elegans organs to form syncytia. Nature Microbiology , 1(11), 16144. https://doi.org/10.1038/nmicrobiol.2016.144
- Barriere, A., & Felix, M.-A. (2014). Isolation of C. elegans and related nematodes. WormBook: The Online Review of C. Elegans Biology , 1–19. https://doi.org/10.1895/wormbook.1.115.2
- Bojko, J., Reinke, A. W., Stentiford, G. D., Williams, B., Rogers, M. S. J., & Bass, D. (2022). Microsporidia: A new taxonomic, evolutionary, and ecological synthesis. Trends in Parasitology , 38(8), 642–659. https://doi.org/10.1016/j.pt.2022.05.007
- Corradi, N. (2015). Microsporidia: Eukaryotic intracellular parasites shaped by gene loss and horizontal gene transfers. Annual Review of Microbiology , 69(1), 167–183. https://doi.org/10.1146/annurev-micro-091014-104136
- Crombie, T. A., McKeown, R., Moya, N. D., Evans, K. S., Widmayer, S. J., LaGrassa, V., Roman, N., Tursunova, O., Zhang, G., Gibson, S. B., Buchanan, C. M., Roberto, N. M., Vieira, R., Tanny, R. E., & Andersen, E. C. (2024). CaeNDR, the Caenorhabditis Natural Diversity Resource. Nucleic Acids Research , 52(D1), D850–D858. https://doi.org/10.1093/nar/gkad887
- Crombie, T. A., Tanny, R. E., Buchanan, C. M., Roberto, N. M., & Andersen, E. C. (2022). A highly scalable approach to perform ecological surveys of selfing Caenorhabditis nematodes. JoVE (Journal of Visualized Experiments) , 181, e63486. https://doi.org/10.3791/63486
- de Albuquerque, N. R. M., & Haag, K. L. (2022). Using average nucleotide identity (ANI) to evaluate microsporidia species boundaries based on their genetic relatedness. Journal of Eukaryotic Microbiology , 70(2), e12944. https://doi.org/10.1111/jeu.12944
- Estes, K. A., Szumowski, S. C., & Troemel, E. R. (2011). Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLOS Pathogens , 7(9), e1002227. https://doi.org/10.1371/journal.ppat.1002227
- Frézal, L., & Félix, M.-A. (2015). C. elegans outside the Petri dish. eLife , 4, e05849. https://doi.org/10.7554/eLife.05849
- Gimond, C., Poullet, N., & Braendle, C. (2022). Isolating Caenorhabditis elegans from the natural habitat. In G. Haspel & A. C. Hart (Eds.), C. elegans: Methods and Applications (pp. 283–292). Springer. https://doi.org/10.1007/978-1-0716-2181-3_15
- Han, B., & Weiss, L. M. (2017). Microsporidia: Obligate intracellular pathogens within the fungal kingdom. Microbiology Spectrum , 5(2). https://doi.org/10.1128/microbiolspec.FUNK-0018-2016
- Jarkass, H. T. E., & Reinke, A. W. (2020). The ins and outs of host-microsporidia interactions during invasion, proliferation and exit. Cellular Microbiology , 22(11), e13247. https://doi.org/10.1111/cmi.13247
- Larsen, B. B., Miller, E. C., Rhodes, M. K., & Wiens, J. J. (2017). Inordinate fondness multiplied and redistributed: The number of species on Earth and the new pie of life. The Quarterly Review of Biology , 92(3), 229–265. https://doi.org/10.1086/693564
- Luallen, R. J., Reinke, A. W., Tong, L., Botts, M. R., Félix, M.-A., & Troemel, E. R. (2016). Discovery of a natural microsporidian pathogen with a broad tissue tropism in Caenorhabditis elegans. PLOS Pathogens , 12(6), e1005724. https://doi.org/10.1371/journal.ppat.1005724
- Mok, C., Xiao, M. A., Wan, Y. C., Zhao, W., Ahmed, S. M., Luallen, R. J., & Reinke, A. W. (2023). High-throughput phenotyping of infection by diverse microsporidia species reveals a wild C. elegans strain with opposing resistance and susceptibility traits. PLOS Pathogens , 19(3), e1011225. https://doi.org/10.1371/journal.ppat.1011225
- Murareanu, B. M., Antao, N. V., Zhao, W., Dubuffet, A., El Alaoui, H., Knox, J., Ekiert, D. C., Bhabha, G., Roy, P. J., & Reinke, A. W. (2022). High-throughput small molecule screen identifies inhibitors of microsporidia invasion and proliferation in C. elegans. Nature Communications , 13(1), 1. https://doi.org/10.1038/s41467-022-33400-y
- Murareanu, B. M., Sukhdeo, R., Qu, R., Jiang, J., Reinke, A. W., & Weiss, L. M. (2021). Generation of a microsporidia species attribute database and analysis of the extensive ecological and phenotypic diversity of microsporidia. mBio , 12(3), e01490–21. doi: 10.1128/mBio.01490-21
- Nakjang, S., Williams, T. A., Heinz, E., Watson, A. K., Foster, P. G., Sendra, K. M., Heaps, S. E., Hirt, R. P., & Martin Embley, T. (2013). Reduction and expansion in microsporidian genome evolution: New insights from comparative genomics. Genome Biology and Evolution , 5(12), 2285–2303. https://doi.org/10.1093/gbe/evt184
- Reddy, K. C., Dror, T., Sowa, J. N., Panek, J., Chen, K., Lim, E. S., Wang, D., & Troemel, E. R. (2017). An intracellular pathogen response pathway promotes proteostasis in C. elegans. Current Biology , 27(22), 3544–3553.e5. https://doi.org/10.1016/j.cub.2017.10.009
- Reddy, K. C., Dror, T., Underwood, R. S., Osman, G. A., Elder, C. R., Desjardins, C. A., Cuomo, C. A., Barkoulas, M., & Troemel, E. R. (2019). Antagonistic paralogs control a switch between growth and pathogen resistance in C. elegans. PLOS Pathogens , 15(1), e1007528. https://doi.org/10.1371/journal.ppat.1007528
- Reinke, A. W., Balla, K. M., Bennett, E. J., & Troemel, E. R. (2017). Identification of microsporidia host-exposed proteins reveals a repertoire of rapidly evolving proteins. Nature Communications , 8, 14023. https://doi.org/10.1038/ncomms14023
- Reinke, A. W., & Troemel, E. R. (2015). The development of genetic modification techniques in intracellular parasites and potential applications to microsporidia. PLOS Pathogens , 11(12), e1005283. https://doi.org/10.1371/journal.ppat.1005283
- Ruan, Y., Xu, X., He, Q., Li, L., Guo, J., Bao, J., Pan, G., Li, T., & Zhou, Z. (2021). The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasites & Vectors, 14(1), 186. https://doi.org/10.1186/s13071-021-04700-x
- Schratzberger, M., Holterman, M., van Oevelen, D., & Helder, J. (2019). A worm's world: Ecological flexibility pays off for free-living nematodes in sediments and soils. BioScience , 69(11), 867–876. https://doi.org/10.1093/biosci/biz086
- Schulenburg, H., & Félix, M.-A. (2017). The natural biotic environment of Caenorhabditis elegans. Genetics , 206(1), 55–86. https://doi.org/10.1534/genetics.116.195511
- Smythe, A. B., Holovachov, O., & Kocot, K. M. (2019). Improved phylogenomic sampling of free-living nematodes enhances resolution of higher-level nematode phylogeny. BMC Evolutionary Biology , 19(1), 121. https://doi.org/10.1186/s12862-019-1444-x
- Stentiford, G. D., Becnel, J. J., Weiss, L. M., Keeling, P. J., Didier, E. S., Williams, B. A. P., Bjornson, S., Kent, M. L., Freeman, M. A., Brown, M. J. F., Troemel, E. R., Roesel, K., Sokolova, Y., Snowden, K. F., & Solter, L. (2016). Microsporidia – emergent pathogens in the global food chain. Trends in Parasitology , 32(4), 336–348. https://doi.org/10.1016/j.pt.2015.12.004
- Stentiford, G. D., Feist, S. W., Stone, D. M., Bateman, K. S., & Dunn, A. M. (2013). Microsporidia: Diverse, dynamic, and emergent pathogens in aquatic systems. Trends in Parasitology , 29(11), 567–578. https://doi.org/10.1016/j.pt.2013.08.005
- Szumowski, S. C., Botts, M. R., Popovich, J. J., Smelkinson, M. G., & Troemel, E. R. (2014). The small GTPase RAB-11 directs polarized exocytosis of the intracellular pathogen N. parisii for fecal-oral transmission from C. elegans. Proceedings of the National Academy of Sciences , 111(22), 8215–8220. https://doi.org/10.1073/pnas.1400696111
- Szumowski, S. C., Estes, K. A., Popovich, J. J., Botts, M. R., Sek, G., & Troemel, E. R. (2016). Small GTPases promote actin coat formation on microsporidian pathogens traversing the apical membrane of Caenorhabditis elegans intestinal cells: Actin coats on microsporidia-containing vesicles. Cellular Microbiology , 18(1), 30–45. https://doi.org/10.1111/cmi.12481
- Tamim El Jarkass, H., Mok, C., Schertzberg, M. R., Fraser, A. G., Troemel, E. R., & Reinke, A. W. (2022). An intestinally secreted host factor promotes microsporidia invasion of C. elegans. eLife , 11, e72458. https://doi.org/10.7554/eLife.72458
- Taxt, A. M., Avershina, E., Frye, S. A., Naseer, U., & Ahmad, R. (2020). Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Scientific Reports , 10(1), 1. https://doi.org/10.1038/s41598-020-64616-x
- Tecle, E., & Troemel, E. R. (2022). Insights from C. elegans into microsporidia biology and host-pathogen relationships. Experientia Supplementum (2012) , 114, 115–136. https://doi.org/10.1007/978-3-030-93306-7_5
- Troemel, E. R., Félix, M.-A., Whiteman, N. K., Barrière, A., & Ausubel, F. M. (2008). Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLOS Biology , 6(12), e309. https://doi.org/10.1371/journal.pbio.0060309
- Voytas, D. (2000). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51(1), 2.5A.1–2.5A.9. https://doi.org/10.1002/0471142727.mb0205as51
- Wadi, L., Jarkass, H. T. E., Tran, T. D., Islah, N., Luallen, R. J., & Reinke, A. W. (2023). Genomic and phenotypic evolution of nematode-infecting microsporidia. PLOS Pathogens , 19(7), e1011510. https://doi.org/10.1371/journal.ppat.1011510
- Wadi, L., & Reinke, A. W. (2020). Evolution of microsporidia: An extremely successful group of eukaryotic intracellular parasites. PLOS Pathogens , 16(2), e1008276. https://doi.org/10.1371/journal.ppat.1008276
- Wan, Y. C., Troemel, E. R., & Reinke, A. W. (2022). Conservation of Nematocida microsporidia gene expression and host response in Caenorhabditis nematodes. PLOS ONE , 17(12), e0279103. https://doi.org/10.1371/journal.pone.0279103
- Willis, A. R., Jarkass, H. T. E., & Reinke, A. W. (2022). Studying inherited immunity in a Caenorhabditis elegans model of microsporidia infection. JoVE (Journal of Visualized Experiments) , 182, e63636. https://doi.org/10.3791/63636
- Willis, A. R., & Reinke, A. W. (2022). Factors that determine microsporidia infection and host specificity. Experientia Supplementum (2012) , 114, 91–114. https://doi.org/10.1007/978-3-030-93306-7_4
- Willis, A. R., Zhao, W., Sukhdeo, R., Wadi, L., Tamim El Jarkass, H., Claycomb, J. M., & Reinke, A. W. (2021). A parental transcriptional response to microsporidia infection induces inherited immunity in offspring. Science Advances , 7(19), eabf3114. doi: 10.1126/sciadv.abf3114
- Zhang, G., Sachse, M., Prevost, M.-C., Luallen, R. J., Troemel, E. R., & Félix, M.-A. (2016). A large collection of novel nematode-infecting microsporidia and their diverse interactions with Caenorhabditis elegans and other related nematodes. PLOS Pathogens , 12(12), e1006093. https://doi.org/10.1371/journal.ppat.1006093

