Exploring Curated Conformational Ensembles of Intrinsically Disordered Proteins in the Protein Ensemble Database
Federica Quaglia, Federica Quaglia, András Hatos, András Hatos, Damiano Piovesan, Damiano Piovesan, Silvio C. E. Tosatto, Silvio C. E. Tosatto, Tamas Lazar, Tamas Lazar, Peter Tompa, Peter Tompa
conformational ensembles
database
intrinsically disordered proteins
literature curation
PED
structural modeling
Abstract
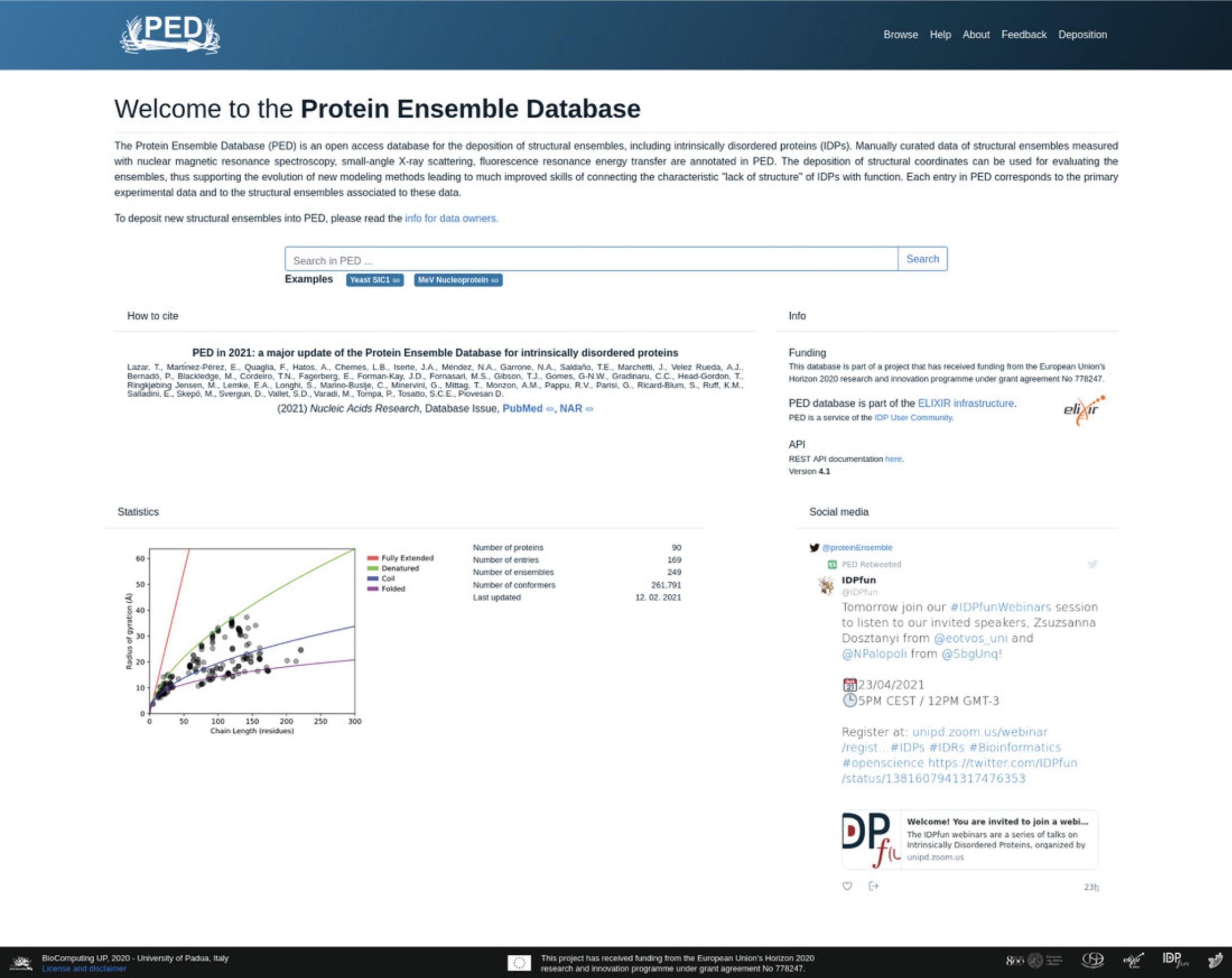
The Protein Ensemble Database (PED; https://proteinensemble.org/) is the major repository of conformational ensembles of intrinsically disordered proteins (IDPs). Conformational ensembles of IDPs are primarily provided by their authors or occasionally collected from literature, and are subsequently deposited in PED along with the corresponding structured, manually curated metadata. The modeling of conformational ensembles usually relies on experimental data from small-angle X-ray scattering (SAXS), fluorescence resonance energy transfer (FRET), NMR spectroscopy, and molecular dynamics (MD) simulations, or a combination of these techniques. The growing number of scientific studies based on these data, along with the astounding and swift progress in the field of protein intrinsic disorder, has required a significant update and upgrade of PED, first published in 2014. To this end, the database was entirely renewed in 2020 and now has a dedicated team of biocurators providing manually curated descriptions of the methods and conditions applied to generate the conformational ensembles and for checking consistency of the data.
Here, we present a detailed description on how to explore PED with its protein pages and experimental pages, and how to interpret entries of conformational ensembles. We describe how to efficiently search conformational ensembles deposited in PED by means of its web interface and API. We demonstrate how to make sense of the PED protein page and its associated experimental entry pages with reference to the yeast Sic1 use case. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
[Correction added on May 17, 2022, after first online publication: CRUI-CARE funding statement has been added.]
Basic Protocol 1 : Performing a search in PED
Support Protocol 1 : Programmatic access with the PED API
Basic Protocol 2 : Interpreting the protein page and the experimental entry page—the Sic1 use case
Support Protocol 2 : Downloading options
Support Protocol 3 : Understanding the validation report—the Sic1 use case
Basic Protocol 3 : Submitting new conformational ensembles to PED
Basic Protocol 4 : Providing feedback in PED
INTRODUCTION
Intrinsically disordered proteins (IDPs) and disordered regions (IDRs) represent segments of a polypeptide chain that lack a stable three-dimensional tertiary fold (Dunker et al., 2001; Tompa, 2002; Wright & Dyson, 1999). The diverse roles of these regions stem from the dynamic nature of their structure, e.g., IDRs can function by molecular recognition, or display sites for post-translational modification, flexible linkers, or entropic bristles (Dunker, Brown, David Lawson, Iakoucheva, & Obradović, 2002; Lee et al., 2014; Piovesan et al., 2017). These experimentally verified IDRs and their corresponding functions, as well as their interaction partners and structural transitions, are catalogued in the manually curated DisProt database (Hatos et al., 2019). As a consequence of their inherently dynamic structure, instead of a single snapshot, this group of proteins is characterized by a representative set of conformers reflecting internal heterogeneity, called the conformational ensemble (Fisher, Huang, & Stultz, 2010; Jensen, Ruigrok, & Blackledge, 2013; Tompa, 2011).
Experimental data to characterize conformational ensembles are primarily derived from nuclear magnetic resonance (NMR) spectroscopy measurements that provide residue-specific information in the form of structural restraints (proximity, dynamics and secondary structure) and small-angle X-ray scattering (SAXS) curves characteristic of the shape and compactness of the proteins (Bernadó & Svergun, 2012; Jensen et al., 2013; Marsh & Forman-Kay, 2012; Schramm et al., 2019). These data can be readily accessed in dedicated primary databases such as the BioMagResDB (BMRB; Romero et al., 2020) and SASBDB (Kikhney, Borges, Molodenskiy, Jeffries, & Svergun, 2020). Förster resonance energy transfer (FRET) experiments can also provide data on the global compactness and dynamics of the protein in the form of FRET efficiencies between donor and acceptor fluorescent tags grafted onto the polypeptide chain (Fuertes et al., 2017; Nath et al., 2012; Schramm et al., 2019).
Reweighting approaches for ensemble calculation usually rely on the generation of a pool of thousands of conformers based on random coil generation tools [e.g., Flexible-Meccano (Ozenne et al., 2012), TraDES (Feldman & Hogue, 2002)] or molecular dynamics (MD) simulations (Chan-Yao-Chong et al., 2019; Rauscher et al., 2015; Robustelli, Piana, & Shaw, 2018), followed by the refinement of the pools by the addition of experimental data to select sub-ensembles that better fit the data (Köfinger et al., 2019; Krzeminski, Marsh, Neale, Choy, & Forman-Kay, 2013; Rangan et al., 2018; Varadi et al., 2014). In contrast, restraining approaches introduce experimentally derived constraints/restraints and only generate conformations that comply with the measurement data (Cavalli, Camilloni, & Vendruscolo, 2013; Rangan et al., 2018; Varadi et al., 2014). The resulting final ensembles are a set of conformers with ideally atomic coordinates that sufficiently reflect the measurements. However, multiple sets of conformational ensembles can fit the data equally well, due to the limitations of the set of experimental constraints, causing it to be an inherently underdetermined problem (Lazar et al., 2020). However, as the number of available ensembles covering a given protein region accumulates, the alternative ensembles might shed light on this problem. The Protein Ensemble Database (abbreviated now as PED, initially pe-DB) was launched to create a deposition database making these conformational ensembles available to the public in a structured and well-organized manner (Varadi et al., 2014).
PED is now a service of the Italian node of ELIXIR, the European infrastructure for biological data, and a central resource of the ELIXIR IDP user community (Davey et al., 2019). First published in 2014 and updated in 2016 (Varadi & Tompa, 2015; Varadi et al., 2014), PED was revamped in 2020 and extensively expanded to include six times more data than the previous version (Lazar et al., 2021). A re-designed graphical interface allows an improved user experience. Two types of pages can be found in PED, the Protein entry pages that group together experiments at the protein level, and the Experimental entry pages, revolving around the experiments and corresponding to the submitted entries. The high quality of the PED experiment-centric entries is now ensured both by manual curation from a dedicated team of biocurators and an automatic pipeline for validation.
In this article we describe in detail how to perform a search in PED (Basic Protocol 1), visualize and interpret protein-centric and experiment-centric entries in PED (Basic Protocol 2), submit new conformational ensembles (Basic Protocol 3), and provide feedback (Basic Protocol 4). We also describe programmatic access with the PED REST API (Support Protocol 1), the downloading options in PED (Support Protocol 2), and how to understand the conformational ensemble validation report (Support Protocol 3).
For abbreviations used in the text, see List of Abbreviations at the end of this article.
Basic Protocol 1: PERFORMING A SEARCH IN PED
To search the Protein Ensemble Database (PED), please visit the webpage at https://proteinensemble.org/. PED is an open-access database freely available without registration. In this protocol, we describe the different options for searching the database, browsing the ensembles, and retrieving relevant information.
Necessary Resources
Hardware
- While PED works best on laptop or desktop computers, it is also accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
No input data are required
Performing a text search
1.Open a web browser and connect to the Main page of the Protein Ensemble Database at https://proteinensemble.org/.
2.Searches in PED can be performed in two ways:
- Users can perform a search using the “Search” box on the top-middle of the PED home page. After typing the query text in the box, hit the “Search” button (Fig.1).
Users can look for a specific protein, e.g., Sic1 from yeast, by submitting the protein name, e.g.,Sic1or its corresponding UniProtKB (UniProt Consortium,2019) accession,P38634. This will redirect them to a list of associated PED entries.
Users might also be interested in looking for a specific publication. To do so, please provide a valid identifier of the publication, i.e., the corresponding PubMed (White,2020) identifier (PMID) or DOI.
-
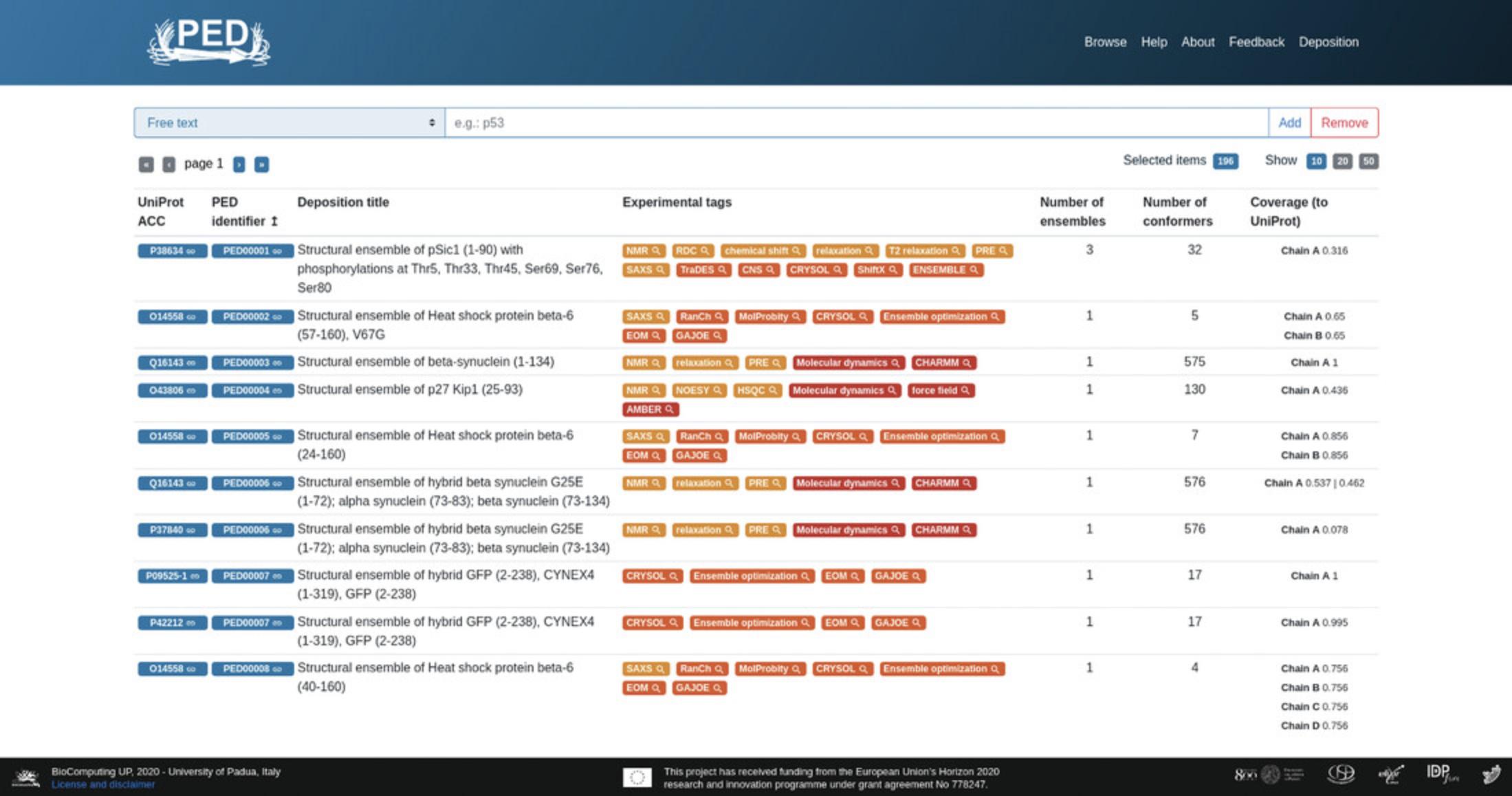
Alternatively, users can perform an advanced search by connecting to the Browse page of the Protein Ensemble Database athttps://proteinensemble.org/browse/. Navigation to the Browse page is also possible from the Main page by clicking the corresponding button in the navigation panel, i.e., the first button on the top-right, titled “Browse” (Fig.1). A search can be performed here using the search box by typing the query text, or a combination of queries, e.g., “UniProt ACC” and “experimental tag.” This search field is auto-updating, so that there is no need to click a button to register each change.
3.Select a term from the drop-down menu on the Browse page. The option selected from the drop-down list will limit the content being searched. Users can perform the following functions:
-
It is possible to perform a free text search in the database by selecting the “free text” term from the drop-down menu.
-
For a specific experiment-centric page, select the “Entry identifier” term and look for a specific PED identifier, e.g., PED00001.
-
For all the experiment-centric pages, i.e., entries of conformational ensembles, associated to a specific protein, select the “UniProt ACC” term, e.g., P38634. This will retrieve all the experiment-centric entries associated with yeast pSic1.
-
Select the “Experimental tag” term to look for a specific experimental tag, e.g., “NMR” or “EOM.” This will retrieve all the entries associated with the tag(s). Three categories of tags are available—depending on the experimental setup used to generate the conformational ensemble—and are highlighted in the Browse table in yellow for the tags related to “measurement methods,” in orange for the tags describing “ensemble generation methods,” and in red for the tags associated to “molecular dynamics” simulations.
-
Find conformational ensembles from a specific author by selecting the “Data owner” name and looking for the author's name. A list of the entries from that author will appear.
-
Find all the entries curated by a specific biocurator by selecting the “Biocurator name” and specifying the biocurator's name.
-
Find a specific protein. By selecting the “Protein name” term and looking for “Sic1” it is possible to retrieve all the entries deposited in PED related to that protein, i.e., PED00001, PED00023, PED00159, PED00160, and PED00161.
-
A specific publication. In this case it is possible to select the “Publication title” option and enter the title of the publication into the search box or to search for the identifier of a publication—PubMed identifier or DOI—by selecting the “Publication ID” term.
-
Look for PED entries cross-referenced to entries in DisProt, e.g., DP00631, or to raw experimental data deposited in SASBDB or BMRB, e.g., “16659” is a BMRB cross-reference, by selecting the “Cross-reference (PDB, BMRB, SASBDB)” term.
4.Users can customize the table columns shown on the Browse page to visualize more/fewer details of the entries. By default, all columns are shown, i.e., “UniProt ACC,” “PED identifier,” “PED title,” “Coverage (to UniProt),” “Number of ensembles,” and “Number of conformers.”
Searching for PED entries associated with a specific protein: The p53 use case
This section demonstrates how to look for all the entries—protein-centric and experiment-centric—associated with a specific use case, the p53 protein.
5.Open a web browser and connect to PED at https://proteinensemble.org/, then select the “Browse” button from the top bar. Alternatively, directly access the Browse page of the Protein Ensemble Database at https://proteinensemble.org/browse/ and select the type of search next to the search box.
6.Entries associated with p53 can be retrieved by searching for the “Protein name” term and entering p53 in the Search box. This will immediately retrieve all the entries related to p53 in PED. The displayed results now include PED identifiers associated with more than one UniProt ACC, since p53 is part of multiple protein complexes with other proteins, e.g., for PED00037 (https://proteinensemble.org/PED00037), the experimental entry titled “Solution Structure of p53-TAD::Cbp-TAZ1 fusion protein” is related to both the CREBBP (https://proteinensemble.org/P45481) and p53 (https://proteinensemble.org/P04637) protein pages.
7.Users can then choose if they want to explore the protein-centric or the experiment-centric pages.
-
To access the protein-centric page of p53, click on the “P04637” button in the first column of the result table. This will redirect the user to the protein entry, where it is possible to visualize all the conformational ensembles associated with the protein.
-
To access the experimental entry “Solution Structure of p53-TAD::Cbp-TAZ1 fusion protein,” click on the PED00037 button in the second column of the results table of the Browse page to be redirected to the page dedicated to this conformational ensemble.
Users can also directly search for the UniProt accession number of the p53 protein, i.e., P04637.
8.Open a web browser, connect to PED at https://proteinensemble.org/, then select the “Browse” button from the top bar.
9.Alternatively, directly access the Browse page of the Protein Ensemble Database at https://proteinensemble.org/browse/ and select the type of search next to the search box.
10.Select the “UniProt ACC” term from the list of the drop-down menu. Type the given UniProt accession, e.g., P04637 for the p53 protein, into the search box to find the experimental entries containing protein region(s) of the specified protein (e.g., P04637) (Fig. 2).
11.Users can then choose if they want to explore the protein-centric page associated to the p53 protein or the experiment-centric pages.
-
To access the protein-centric page of p53, click on the “P04637” button (https://proteinensemble.org/PED00037) in the first column of the result table. This will redirect the user to the protein entry, where it is possible to visualize all the conformational ensembles associated with the protein.
-
To access a specific experimental entry, “Structural ensemble of the complex between Tfb1 (2-115) and the activation domain of p53 (20-73),” click on “PED00087” (https://proteinensemble.org/PED00087) in the second column of the results table of the Browse page to be redirected to the page dedicated to this conformational ensemble.
Exploring specific experimental setup using the controlled vocabulary of PED
To explore whether a specific experimental setup has been used by someone to model ensembles, users can employ the searchable experimental tags of the controlled vocabulary created and maintained by the PED biocurator team. For instance, one might be interested in a setup where both NMR and SAXS data were used to refine an MD-based pool of conformations generated by an AMBER force field.
12.Open a web browser and connect to PED at https://proteinensemble.org/, then select the “Browse” button from the top bar. Alternatively, directly access the Browse page of the Protein Ensemble Database at https://proteinensemble.org/browse/ and select the type of search next to the search box.
13.To search terms from the PED controlled vocabulary (https://proteinensemble.org/about#cv; Fig. 3A), select “Experimental tag” from the drop-down menu.
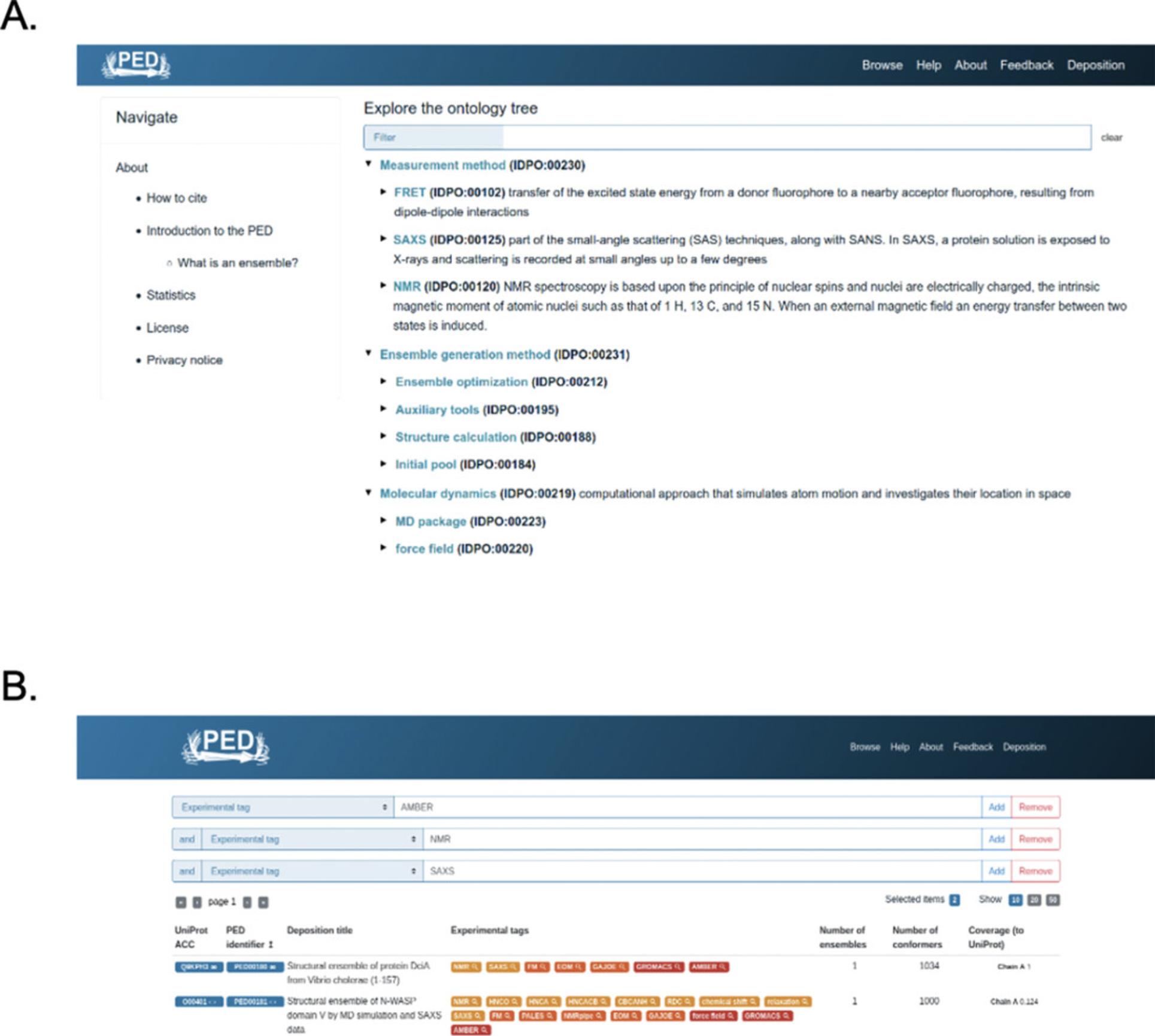
14.Type the ’AMBER’ keyword into the search box to see the entries that contain the term in their ensemble determination methodology.
15.You can add more tags to your search by clicking on the “Add” button on the right. The new search field that appears will be set to the default value “Free text.”
16.Change the type of the second search field to “Experimental tag” similarly as before and enter the second keyword ’NMR.’
17.Click again on the “Add” button on the right.
18.Change the field from “Free text” to “Experimental tag” and type your third keyword SAXS (Fig. 3B). As of today, there are two ensembles that fulfill the above selected criteria—PED00180 and PED00181—generated by the same authors with almost identical methodology (Chan-Yao-Chong et al., 2019, 2020).
Support Protocol 1: PROGRAMMATIC ACCESS WITH THE PED API
PED has a RESTful API for the programmatic access to retrieve entries and corresponding ensemble models, or to perform a customized search in the database. The Help page of PED lists all API endpoints with short descriptions and examples (https://proteinensemble.org/help#api). These endpoints are available from the https://proteinensemble.org/api/{endpoint_name} URL. Below, we exemplify three procedures, with explanations, using a Python script as a client: (i) how to retrieve metadata of entries; (ii) how to download ensembles; and (iii) how to search for the available ensembles associated with a protein accession number (UniProt accession) and with both an accession number and a given method.
Necessary Resources
Hardware
- While PED works best on laptop or desktop computers, it is also accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
No input data are required
How to retrieve the metadata of entries
Metadata of experimental entries can be retrieved by using their corresponding PED identifiers. The following syntax must be used to retrieve the metadata of a single entity from PED: https://proteinensemble.org/api/{identifier}, where the “identifier” must be a valid PED ID, for example PED00001. The query is customizable with version and release. Here we provide two python3 code snippets to access the metadata of an entry in JSON format (Code Snippet 1).
Code Snippet 1 :
-
#!/usr/bin/env python3
-
import requests
-
ped_id = "PED00001"
-
url = "https://proteinensemble.org/api/" + ped_id
-
resp_json = requests.get(url).json()
-
print(resp_json)
The JSON output format is easy to parse in Python and in other programming languages using popular JSON parser modules. Code Snippet 2 provides an example to extract information from the JSON outputs of Code Snippet 1. In the following lines of code, the title of the entry and the ensemble identifiers are printed. In addition, all ensemble identifiers are stored in the list variable ensembles_ids.
Code Snippet 2 :
-
#!/usr/bin/env python3
-
import requests
-
ped_id = "PED00001"
-
url = "https://proteinensemble.org/api/" + ped_id
-
resp_json = requests.get(url).json()
-
print(resp_json["title"])
-
ensembles_ids = []
-
for ensemble in resp_json["ensembles"]:
- ensembles_ids.append(ensemble["ensemble_id"])
-
print(ensembles_ids)
Code Snippet 2 can access a given entry (here PED00001) and print the corresponding ensembles identifiers.
How to download ensembles
In this step, we demonstrate how the PDB models of ensembles can be downloaded. For this, in the following code (Code Snippet 3), we make use of the information obtained in Code Snippet 2, specifically the list variable ensembles_ids. Ensemble identifiers stored in the variable are inserted into the URL https://proteinensemble.org/api/get_ensembles as a parameter.
Code Snippet 3 :
-
#!/usr/bin/env python3
-
import requests
-
get list of ensemble ids
-
ped_id = "PED00001"
-
url = "https://proteinensemble.org/api/" + ped_id
-
resp_json = requests.get(url).json()
-
print(resp_json["title"])
-
ensembles_ids = []
-
for curr_ensemble in resp_json["ensembles"]:
- ensembles_ids.append(curr_ensemble["ensemble_id"])
-
get direct link to the downloadable file (url as string)
-
parameters = {
- `ensemble_id': ensembles_ids
-
}
-
download_link = requests.get(url, params=parameters).text
-
print(download_link)
-
download ensembles
-
resp_file = requests.get(download_link.replace(`"', "))
-
with open(
PED00001_pdbs.tar',wb') as f:- f.write(resp_file.content)
The response of the endpoint https://proteinensemble.org/api/get_ensembles is a dedicated link stored in the download_link variable that can be used to download the selected ensemble(s). For security reasons, the link remains active for only 30 min. In the end, Code Snippet 3 downloads the selected ensemble into a .tar file.
How to search for the available ensembles associated with a protein accession number (UniProt Acc) or both a protein accession number and a given method
In PED there is a built-in, customizable search engine that can also be accessed programmatically via the URL https://proteinensemble.org/api/browse. In the query, accession numbers (here UniProt accession, “acc”), controlled vocabulary terms, and free text can be given. In Code Snippet 4, we exemplify a very simple search for a UniProt accession (P04637 for human p53). In Code Snippet 5, we extend this to experimental PED entries corresponding to the UniProt accession P38634 (for yeast Sic1) solved by the combination of NMR and SAXS.
Code Snippet 4 :
-
#!/usr/bin/env python3
-
import requests
-
uniprot_acc = "P04637"
-
url = "https://proteinensemble.org/api/browse?acc=" + uniprot_acc
-
resp_json = requests.get(url).json()
-
print(resp_json)
Code Snippet 5 :
-
#!/usr/bin/env python3
-
import requests
-
parameters = {
acc' :P38634',term' : [NMR', `SAXS']
-
}
-
resp_json = requests.get(url, params = parameters).json()
-
print(resp_json)
The resulting JSON output provides a list of objects representing different PED entries, each including the UniProt accession (uniprot_acc), the number of conformers (number_of_conformations), the number of ensemble replicas (number_of_ensembles), and the PED entry identifiers (entry_id).
Basic Protocol 2: INTERPRETING THE PROTEIN PAGE AND THE EXPERIMENTAL ENTRY PAGE—THE SIC1 USE CASE
Here we present a use case, the Sic1 protein of yeast (https://proteinensemble.org/P38634), to describe how to interpret protein-centric and experiment-centric entries in PED. The Sic1 entry—also shown in the home page examples of PED—was first annotated in 2014 and has been extensively reviewed and updated in 2020 (Lazar et al., 2021). It has also been significantly expanded by adding new conformational ensembles.
Sic1—a cyclin-dependent kinase (CDK) inhibitor—can interact with Cdc4, and the interaction requires the phosphorylation of the N-terminal tail of Sic1.Phosphorylation of the N-terminal domain of Sic1 is indeed sufficient—and required—to enable its targeting to Cdc4.However, although a significant amount of transient secondary and tertiary structure can be found in Sic1, neither binding nor phosphorylation can induce its complete folding. Both experiment-centric entries of the unphosphorylated N-terminal targeting domain (1-90) of Sic1 and of its multi-site phosphorylated state (pSic1) are available in PED (Gomes et al., 2020; Mittag et al., 2010).
PED experiment-centric entries are annotated by an expert team of biocurators who aim at capturing the experimental setup used to generate the conformational ensemble, the model organism, and cross-references not only to related entries in other databases, e.g., DisProt (Hatos et al., 2019) and IntAct (Orchard, Ammari, Aranda, & Breuza, 2014), but also to raw experimental data deposited in BMRB (Romero et al., 2020) and/or SASBDB (Kikhney et al., 2020) when available. Moreover, PED biocurators carefully analyze the PDB files of each submitted ensemble to ensure that they satisfy the required standards of the database and define the ensemble construction, annotating each chain of the deposited PDB file, i.e., name and description, along with the corresponding UniProt ACCs, the boundaries of each protein in the ensemble, and tag sequences, if any.
Necessary Resources
Hardware
- While PED works best on laptop or desktop computers, it is also accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
No input data are required
Exploring a protein-centric page of PED
Navigating to the page
1.Navigate to the protein page. Protein pages use the UniProt accession number as identifier; therefore, their URLs are compiled as https://proteinensemble.org/ + UniProt ACC. Protein pages can be accessed by two ways:
-
Look up the UniProt ACC for the protein of interest by copy-pasting it in the above formula, e.g., the UniProt ACC for the Sic1 protein of yeast is P38634 and will result in the URLhttps://proteinensemble.org/P38634.
-
Access a protein page from the Browse menu option by clicking on a UniProt ACC of a search hit (or from the Home page, by clicking on the search examples provided below the search box ‘Yeast SIC1’).
Basic details of the protein entry
Protein names and functional descriptions, as well as organism names and taxonomic IDs, are automatically retrieved from UniProt. In the cross-references section, a link is provided to the UniProt page of the entry, and—to be able to evaluate the intrinsic disorder of the given protein—URLs to MobiDB and DisProt are provided (if available).
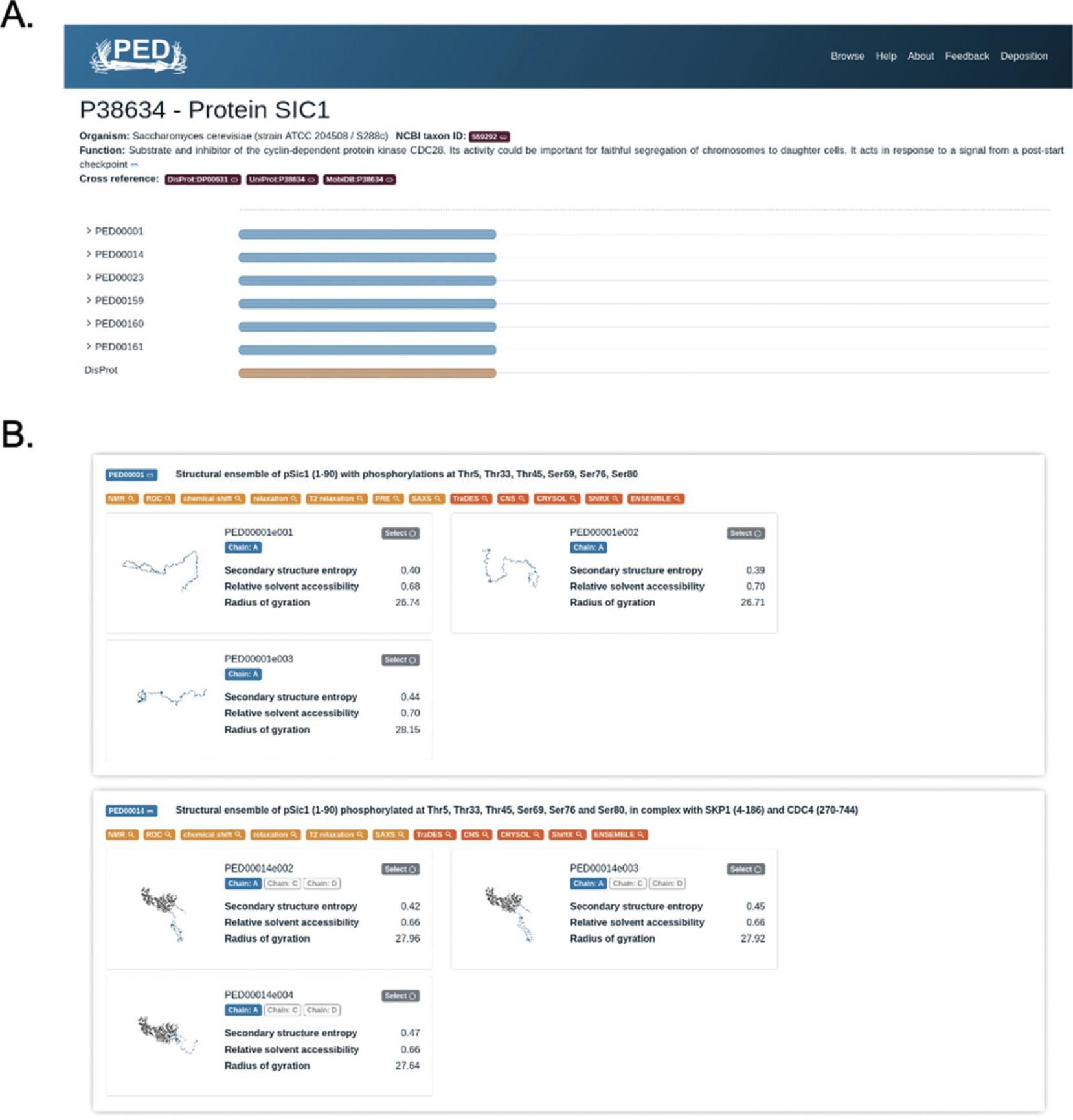
2.After navigating to the protein page of Sic1 as described in step 1, users can visualize the following details that are shown at the top (Fig. 4A):
-
UniProt ACC and protein name: P38634—Protein SIC1.
-
Organism and NCBI taxon ID: Saccharomyces cerevisiae (strain ATCC 204508 / S288c)—ID: 559292
-
Function: “Substrate and inhibitor of the cyclin-dependent protein kinase CDC28. Its activity could be important for faithful segregation of chromosomes to daughter cells. It acts in response to a signal from a post-start checkpoint” (derived from UniProt).
-
Cross-references: DisProt:DP00631, UniProt:P38634, MobiDB:P38634.
Feature viewer
A feature viewer, located in the middle of the page, indicates which regions are modeled by the deposited ensembles (Fig. 4A). The first element of the feature viewer is the protein sequence. Please note that for long proteins this may not be displayed with a font size large enough to be easily readable. The available entries and ensembles stored in the database for the given protein are listed below, grouped by entries (collapsed by default). Hovering the cursor over the regions will show the exact region modeled in the ensemble. If multiple replicas constitute an ensemble, those will be listed by clicking on the identifier of an entry. By clicking on the experimental IDs of ensemble replicas, a drop-down panel will display the location of post-translational modifications in the ensemble, the secondary structure content/entropy, and the solvent accessibility along the chain. Repeated clicking on the experimental ID will collapse the panel. The last element of the feature viewer is the experimentally verified disordered region(s) annotated in DisProt (if the protein has curated disorder).
3.Explore the multiple ensembles of Sic1 that are deposited, shown by blue regions annotated by the feature viewer (Fig. 4A). Hovering the mouse over the blue regions reveals the exact length of the regions, e.g., for Sic1 it ranges from amino acid 1 to 90 for all 12 ensemble models.
4.Visualize the IDs of the PED experimental-centric entries associated with the protein that are displayed on the left, e.g., the first three records are ensemble replicas of PED00001 (PED00001e001, PED00001e002, PED00001e003) and are followed by three other replicas of PED00014 (PED00014e001, PED00014e002, PED00014e003). By clicking on their experimental IDs, e.g., ‘PED00001e001’, the drop-down panel displays the following:
-
Post-translational modifications: Six phosphosites scattered around the sequence [3 phosphothreonine residues (TPO) and 3 phosphoserine residues (SEP)].
-
Secondary structure: Varying secondary structure entropy along the sequence corresponding to mostly coil-like conformers with low propensities to fold to short transient helices.
-
Relative accessible surface area: Very exposed overall surface area for the whole chain.
5.Observe a DisProt annotation that is available for region 1-90, covering the whole construct used in solving the ensemble.
List of ensembles with summary statistics
On the bottom of the page, the ensembles are listed on dedicated information cards with clickable PED identifiers (redirecting to the corresponding experimental entry pages), title of the ensemble, and clickable tags from the methods description (Fig. 4B). Their corresponding ensemble replicas have structural snapshots and summary statistics quantifying the secondary structure entropy, relative solvent accessibility, and radius of gyration. Clicking the tags will perform a search for the experimental entries with the selected tag associated with them. A detailed description on the origin of these tags can be found in the section “Exploring an experiment-centric page in PED,” below. Users interested in a single ensemble should click on the experimental entry identifier to be redirected to the corresponding ensemble entry.
6.Scroll down to the list of ensembles shown below the feature viewer. Here the ensembles shown in the feature viewer are displayed (Fig. 4B).
7.To explore a specific experiment-centric page, click on its experimental ID, e.g., PED00001 titled ’Structural ensemble of pSic1 (1-90) with phosphorylations at Thr5, Thr33, Thr45, Ser69, Ser76, Ser80.’ This will load the experimental entry page of PED00001. Go back.
8.To explore a specific experimental setup, click on the corresponding tag, e.g., ’RDC’ methods tag of PED00001. This will search for the list of PED experiment-centric pages where residual dipolar coupling NMR data was used to model or validate the ensembles. After being redirected to the Search page, users will see ’Experimental tag = RDC’ in the search criteria on top of the page. Go back.
Filtering the list of ensembles on the Protein page
If the available ensembles for a given protein exceed a certain number, manual browsing for relevant entries may require the user to scan a long list of ensembles. To this end, a filtering tool is implemented that enables the (case-insensitive) textual filtering of the entries. The textbox and its ‘Filter’ button are on the left side of the page, immediately above the ensemble info cards. For example, yeast Sic1 ensembles can be filtered either for a given phosphosite, like phosphothreonine in position 5, that returns experimental entries containing Thr5 in the title, or for a method, like “smFRET,” that returns a different subset of entries for Sic1 determined by this method. This is described in detail in the following steps of the protocol:
9.Click in the textbox of the filtering tool, type Thr5, and click on ’Filter’ After filtering out those that do not have this term, in the list of remaining hits the ensembles PED00001, PED00014 should be followed by PED000161.
10.Delete the text Thr and click on ’Filter’.
11.Click again in the textbox of the filtering tool and type smFRET, and click again on ‘Filter’. The top 3 ensembles displayed should be PED00159, PED00160 and PED000161, all containing the word smFRET in either the title or among their methods tags.
Exploring an experiment-centric page of PED
Basic details of the experimental entry
Experimental entry pages are identified with unique PED IDs in a format PED\d{5} with 5 digits. This PED ID is found on the top of the entry page along with the title of the ensemble determination experiment. This is followed by the name and contact info of data owners and the associated publication (PubMed ID or DOI also supplied).
12.Open the PED00001 experimental entry page (https://proteinensemble.org/PED00001) for the multi-phosphorylated Sic1 protein determined by NMR and SAXS measurements (Fig. 5A).

13.The name of the data owners matches the authors of the associated article published by Forman-Kay and collaborators (Mittag et al., 2010) (35). The entry has a PubMed ID (20399186) as a cross-reference to the associated publication.
The ensemble determination and modeling methodology is summarized in a three-way structure: ‘Experimental procedure’, ‘Structural ensemble calculation’, and, when available, ‘Molecular dynamics’. From these manually curated sections, searchable tags are automatically extracted with the use of a pre-compiled controlled vocabulary (to browse the hierarchical terms, see the Controlled vocabulary section on the About page of PED at https://proteinensemble.org/about). Furthermore, cross-references to the experimental raw data (SAXS: SASBDB, NMR: BMRB) and curated evidence of the disordered state from DisProt, when available, are provided.
14.In PED00001, users can see that only the ‘Experimental procedure’ and ‘Structural ensemble calculation’ aspects of the methods description were curated by five biocurators as relevant sections (Fig. 5A).
15.The tags extracted from these texts refer to the NMR measurements (‘NMR’, ‘RDC’, ‘chemical shift’, ‘relaxation’, ‘T2 relaxation’ and ‘PRE’), the SAXS measurement (‘SAXS’), and the software used to assign, process, fit, and evaluate the data (‘ShiftX’, ‘CRYSOL’, ‘CNS’, ‘TraDES’ and ‘ENSEMBLE’). By clicking on these tags, the user will be redirected to the Browse page and shown the hits for the ensembles having the given tags assigned (as described in Exploring a protein-centric page of PED; step 8, above).
16.The NMR experimental raw data is available in BMRB with ID:16659, thus cross-referenced in PED00001.
17.PED00001 also has a cross-reference to DisProt that points to the curated disorder pieces of evidence of Sic1 (Saccharomyces cerevisiae) (Fig. 5A).
Feature viewer
A feature viewer is provided on the experimental entry page (bottom of Fig. 5A) to visualize the basic features of the chains that compose the assembly in the ensemble model. The first element of the feature viewer is the protein sequence corresponding to the construct modeled in the deposited PDB-type structure files. This is followed by the fragments of the construct that may indicate the presence of cloning/purification/fluorescent, etc., tags and describe the build-up of fusion constructs. Post-translational modifications (PTMs), mutations, and other deviations from the specified construct are described in the next line. Hovering the mouse over the PTM/mutation will provide more information on the type of modification/mutation and the associated changes. The last two elements of the feature viewer display the secondary structure and relative solvent accessibility (RSA) of the chain in the same way as described in the previous section. If the ensemble has more than one chain, these elements are repeated for each chain.
18.The construct of PED00001 is built up from a single chain (Chain ‘A’) of two fragments: a dipeptide tag with sequence ‘GS’ and a fragment corresponding to the region 1-90 of yeast Sic1 (O14558) (Fig. 5A).
19.The construct has six phosphosites scattered around the sequence [3 phosphothreonine residues (TPO) and 3 phosphoserine residues (SEP)] as mentioned in the previous section.
20.Secondary structure and RSA annotation works the same way as described for the Protein page (Exploring a protein-centric page of PED; step 4b-c, above).
Deposited ensembles
For each of the ensemble replicas modeled and deposited by the authors, the replica ID (PED\d{5}e\d{3}) and the number of conformers in the ensemble are displayed along with the chain names. A structure viewer shows (maximum) 10 conformers of the ensemble sampled with different compactness values. The active chain—tag in blue color—is shown in the structure viewer as well, while the other chains are colored light gray. Chain statistics with regard to secondary structure entropy, RSA, and average radius of gyration (Rg), as well as an Rg distribution box plot and Ramachandran maps, are displayed for the active chain.
21.Scroll down to the deposited ensembles of PED00001 (Fig. 5B).
22.Compare the number of models in the three deposited conformational ensemble replicas: all are modeled by 10-11 conformers.
23.The summary statistics for the three replicas are very similar (secondary structure entropy: 0.39-0.44; RSA: 0.68-0.70; average Rg: 26.71-28.15 Å).
24.Based on the Rg distributions, users can see that the box plots are sampling almost the same distribution, i.e., the differences between the replicas are not significant (Fig. 5B).
25.Very similar tendencies can also be seen for the Ramachandran maps located next to the structure viewers.
Support Protocol 2: DOWNLOADING OPTIONS
Several download options are available in PED. Users can download a specific conformational ensemble or a set of selected ensembles from the dedicated protein page of a protein (downloading ensembles via the Protein page). Moreover, from each experiment-centric page, it is possible to download the deposited ensemble—including multiple replicas, if available (download ensembles via the Experimental entry page)—and the validation report of the entry (download the validation report via the Experimental entry page).
Necessary Resources
Hardware
- While PED works best on laptop or desktop computers, it is also accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
No input data are required
Downloading ensembles via the Protein page
Users can choose to download all ensembles associated with a specific protein as follows
1a. Open a web browser and connect to PED at https://proteinensemble.org/.
2a. Look for a specific protein in PED. Please refer to steps 1, 2, 3b, or 3f of “Performing a text search” in Basic Protocol 1.
3a. To download all the available ensembles for a given protein, click on the green ‘Download all’ button on the right side above the ensemble info cards, just below the feature viewer.
4a. Save the compressed TAR (.tar) file.
Alternatively, it is possible to download a selected set of ensembles associated with a specific protein
1b. Open a web browser and connect to PED at https://proteinensemble.org/.
2b. Search for a specific protein in PED. Please refer to steps 1, 2, 3b, or 3f of “Performing a text search” in Basic Protocol 1.
3b. A gray ‘Select’ button is available for each ensemble replica on the top right corner of the replica panels. It is possible to find more than one replica panel inside the info cards dedicated to each PED experiment-centric entry. This option enables the users to download a manually specified selection of ensembles.
4b. Select the ensembles of interest by clicking on the respective gray “Select” buttons.
5b. When at least one ensemble is selected, the ‘Download selected’ button on the right side below the sequence viewer turns from gray to green.
6b. Click the ‘Download selected’ button and save the compressed TAR (.tar) file.
Downloading ensembles via the Experimental entry page
1c. Open a web browser and connect to PED at https://proteinensemble.org/.
2c. Search for a specific experiment-centric entry in PED. Please refer to steps 1, 2, 3a—alternatively 3b or 3f—of “Performing a text search” in Basic Protocol 1.
3c. Scroll down to the deposited ensembles of the Experimental entry page.
4c. Click on the green ‘Download’ button on the right side of the ensemble replica.
5c. Select ‘Ensemble in PDB (compressed)’ to download the PDB models. Alternatively, click on the ‘Sampled sub-ensemble (max 10 conformations, mmCIF)’ button to download a smaller sampled set of conformers.
6c. Save the compressed TAR (.tar) file. After uncompression, the file extension should indicate a PDB file (.pdb).
7c. Steps 4c-6c should be repeated for as many replicas as needed.
Downloading the validation report via the Experimental entry page
1d. Open a web browser and connect to PED at https://proteinensemble.org/.
2d. Search for a specific experiment-centric entry in PED. Please refer to steps 1, 2, 3a—alternatively 3b or 3—of “Performing a text search” in Basic Protocol 1.
3d. Click on the green ‘Validation report’ download button on the top right side of the page to download the PDF report.
4d. Save the PDF (.pdf) file.
Support Protocol 3: UNDERSTANDING THE VALIDATION REPORT—THE SIC1 USE CASE
PED provides validation reports for all fully processed experimental entries with completed manual curation pipeline. The validation pipeline is an automated set of program scripts that evaluate the consistency of the metadata curation (e.g., automatically inferred differences between the constructs annotated and found in the models) and the completeness and accuracy of the PDB files (e.g., models should have the same sequence). The biocurators of PED are responsible for ensuring that all entries meet the expected quality standards by requesting changes in the models and metadata and re-running the validation pipeline until no errors are encountered.
Besides providing quality control, the validation report also generates structural insights by plotting distributions of structural features of the ensemble, such as secondary structure propensities, radii of gyration, and pairwise model similarity.
In the following, it is demonstrated how users of the database can interpret the elements of the validation report.
Necessary Resources
Hardware
- While PED works best on laptop or desktop computers, it is also accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
No input data are required
Validation reports can be accessed and downloaded for each ensemble replica from the experimental entry pages. This report is compiled in a single file that summarizes the metadata for an entry and gives a compact and a detailed evaluation for the ensemble content and quality.
- 1.Visit the experimental page of PED00001 (https://proteinensemble.org/PED00001).
- 2.Scroll down the deposited ensemble section and click the green ‘Download’ button on the right side of the ensemble replicas.
-
Select the ‘Validation report’ option. You will be given a PDF file with the report.
The metadata contains the identifier and title of the entry, the corresponding publication, the cross-references, the release date and the name of the depositors, as well as the number of ensemble replicas. In the ‘Overall structure of ensembles’ section, the status, the number of conformer models, and the level of modeling is provided for each ensemble. The level of model resolution might be limited to a very coarse alpha-carbon (CA)−only backbone representation, although this is contraindicated. Current status codes include ‘Uploaded’, ‘Rejected’, ‘Validated’, ‘Processed’, and ‘Completed’. ‘Uploaded’ is assigned to newly uploaded ensembles awaiting validation and ‘Rejected’ to ensembles returning with errors during the validation process. ‘Validated’ is assigned to format-validated ensembles and ‘Processed’ to ensembles that are both validated and assessed using Molprobity, DSSP, and other in-house scripts. Finally, ‘Completed’ is shown for fully processed ensembles for which all the data plots are available. Full validation reports are only generated for ‘Completed’ ensembles.
The content of the ensembles is listed by chain fragments, also displaying the corresponding sequence and the corresponding UniProt accessions (if available). Missing, mutated, and modified residues are displayed separately, along with small-molecule ligands.
- The metadata reports that the entry PED00001 has three multi-phosphorylated Sic1 ensembles in the free state (Mittag et al., 2010), released in 2012.
- It has two cross-references: one for DisProt DP00631, another for BMRB 16659.
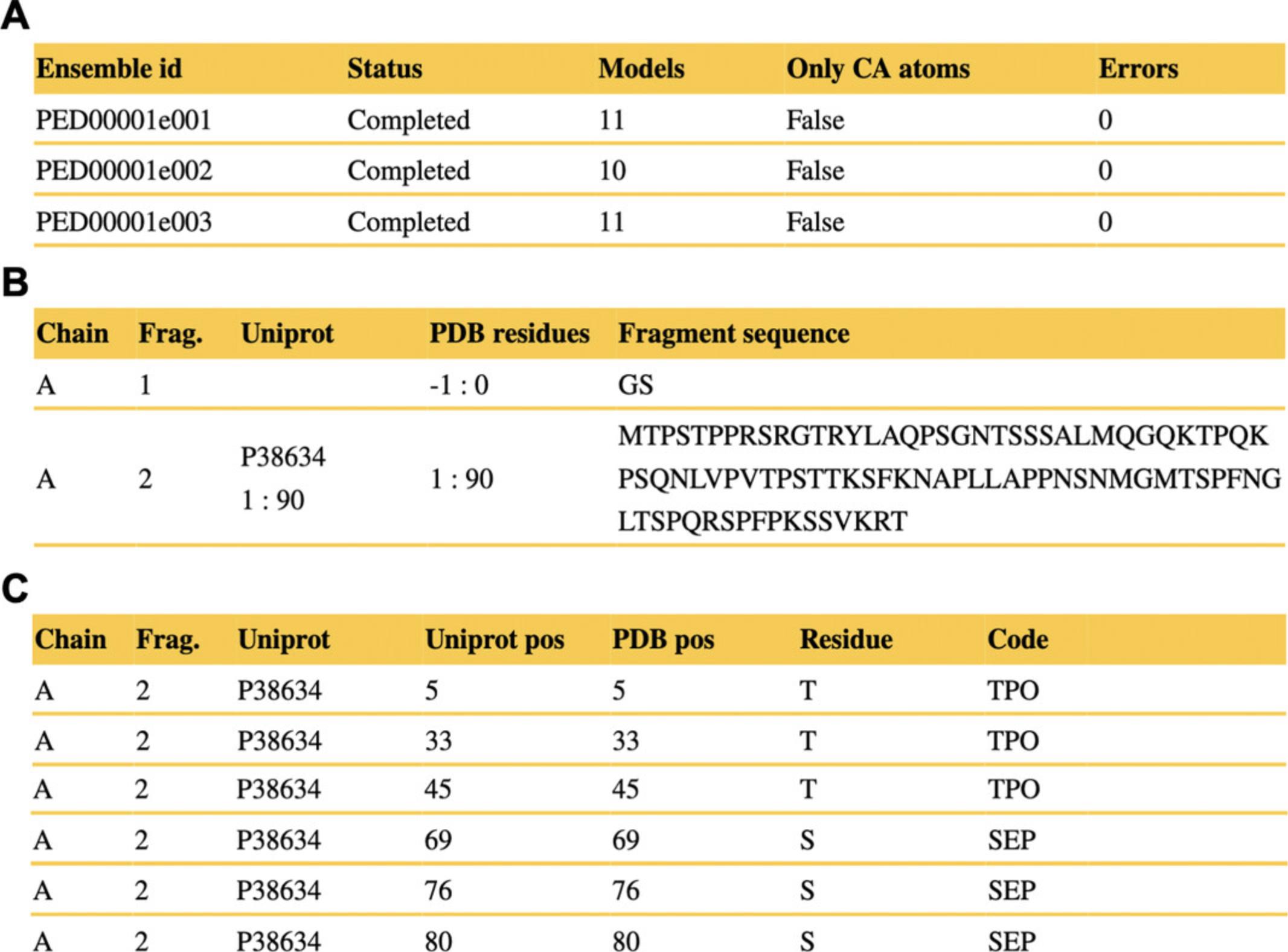
- The three replicas (PED00001e001, e002, e003) are all ‘Completed’ all-atom models (validation ran without error), comprising 11, 10, and 11 models, respectively (Fig. 6A).
- The models are composed of a single chain (chain ‘A’) constructed from two fragments: a remnant Gly-Ser tag-linker dipeptide, and the main fragment corresponding to the 90-residue-long N-terminal IDR of Sic1 (UniProt ACC: P38634) as shown in Figure 6B.
- Phosphorylated serines (SEP) and threonines (TPO) of the Sic1 ensembles are listed in Figure 6C.
The summary statistics component of the validation lists stereochemical descriptors (relative number of clashes, covalent bond length/angle, beta-carbon and rotamer outliers, and Ramachandran mapping), solvent accessibility (global and per residue) and compactness measures (radius of gyration, maximum dimension), and model similarity calculations (pairwise RMSDs) for all ensemble replicas (Fig. 7). For each measure, the distribution of the scores obtained for the respective number of conformer models is characterized by the mean, standard deviation, minimum and maximum scores, and quartiles.
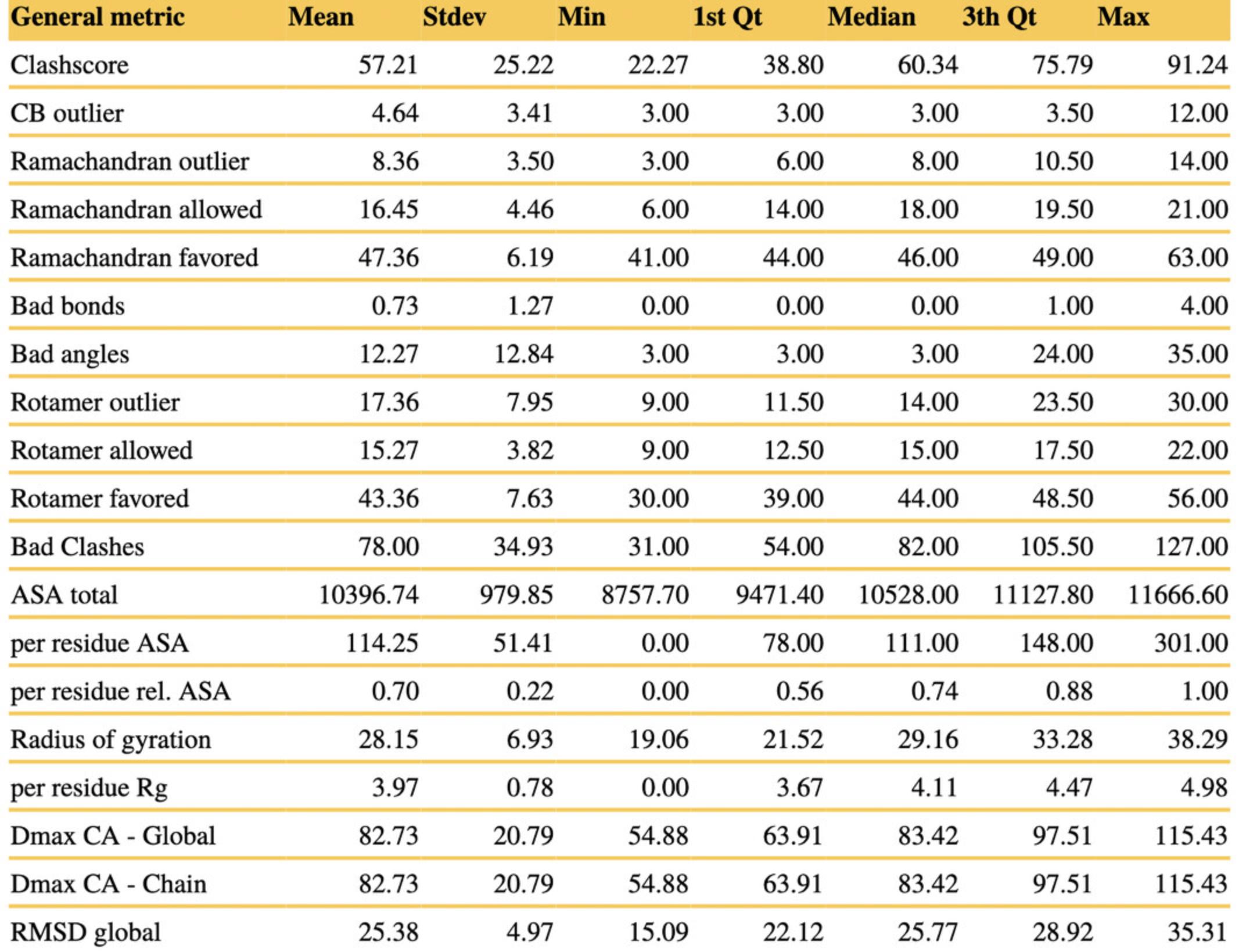
In Figure 7, the summary statistics table is shown for PED0001e003 as an example:
- The three ensemble replicas exhibit only minor differences in the distributions of scores. As visible from the descriptive statistics of the metrics, models of IDP ensembles may have more stereochemical outliers (covalent and dihedral angle, beta-carbon outliers, atomic clashes) than well-refined crystal structures of globular proteins in the PDB (wwwPDB Consortium, 2019).
- The 90-residue-long Sic1 IDR of PED0001e003 has a mean solvent-accessible surface area of 10400 Å2 and an average radius of gyration of 28 Å.
- The mean RMSD for pairwise model comparisons exceeds 25 Å, which signifies a very heterogeneous ensemble, although roto-translational superimposition−based RMSD tends to be exaggerated for IDPs (Lazar et al., 2020).
The validation report ends with the lists of the exact stereochemical issues, the hydrogen-bonding and dihedral angle-based secondary structure assignment, a more detailed local and global accessibility and compactness analysis, and pairwise model RMSD matrix given for each ensemble.
Elements of the detailed stereochemical evaluation:
- Clashscore (clashes per 1000 atoms) histogram
- Distribution (box plot) of the seriousness of clashes (overlap in Å) (Fig. 8A)
- Distributions (3 box plots) for the 3 Ramachandran classes (favored, allowed, outlier) (Fig. 8B)
- Per-ensemble Ramachandran maps (General, Gly, Ile/Val, Pre-Pro, Trans-Pro, Cis-Pro)
- Distribution (box plot) of the beta-carbon deviation
- Distributions (3 box plots) for the rotamer classification (favored, allowed, outlier)
- Distributions (2 box plots) for the covalent bond length and angle outliers
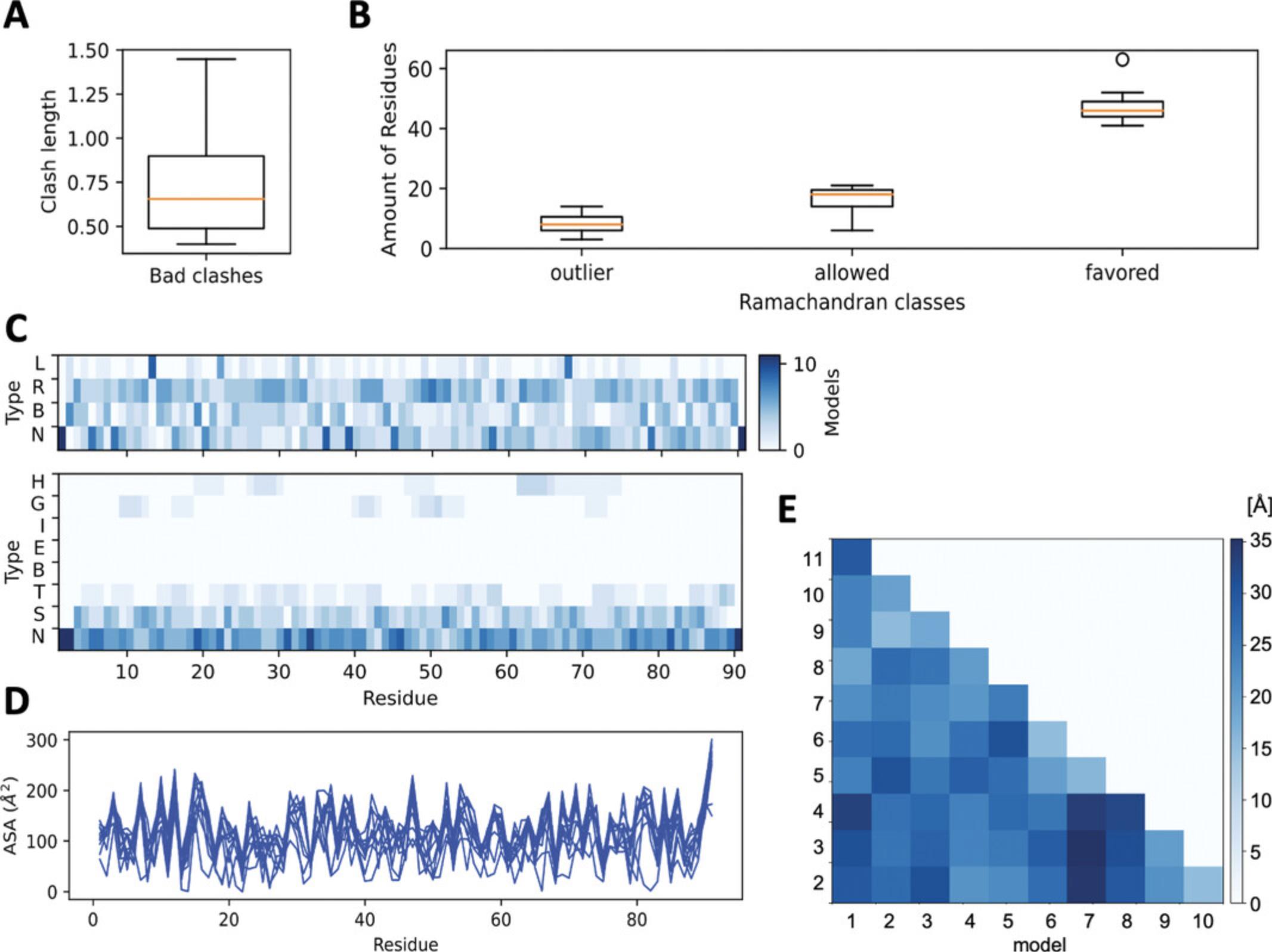
Secondary-structure propensity based on the assignment (Fig. 8C):
- Per-residue propensity based on the dihedral angles (heatmap of left-handed ‘L’ and right-handed ‘R’ helical propensity, beta-strand ‘B’ propensity and polyproline structure propensity ‘N’)
- Per-residue propensity based on the DSSP (heatmap of alpha helix ‘H’, 310 helix ‘G’, Pi-helix ‘I’, beta-strand ‘E’, isolated beta-bridge ‘B’, turn ‘T’, and bend ‘S’ propensities; ‘N’ stands for no structure)
Solvent-accessible surface area calculations:
- Global ASA histogram
- Per-residue ASA spectra for each model (multiple spectra by line plot) (Fig. 8D)
- Per-residue (Gly-X-Gly tripeptide-based) relative ASA spectra (not displayed for modified residues)
Compactness analysis:
- Global Rg distribution (histogram)
- Local compactness calculated by a pentapeptide sliding-window Rg measure (multiple spectra by line plot)
- CA-based Dmax distribution (histogram and box plot)
RMSD matrix for pairwise model comparisons:
- RMSD matrix of pairwise model dissimilarities shown as a heatmap (darker shades representing larger RMSD values) (Fig. 8E)
Basic Protocol 3: SUBMITTING NEW CONFORMATIONAL ENSEMBLES TO PED
Research groups interested in contributing their conformational ensembles to PED are encouraged to contact the PED biocuration team at https://proteinensemble.org/feedback to plan and discuss the deposition process with an expert biocurator. Conformational ensembles can be deposited in PED at any stage of the corresponding publication, e.g., manuscript in preparation or submitted or published article. Authors submitting conformational ensembles from manuscripts in preparation will be provided with a temporary URL containing a token for limited access, suitable for private dissemination with journal reviewers and editors. Ensembles deposited from manuscripts in preparation or from any stage of pre-publication will become publicly available starting from the next release of the database or after approval by the author.
A detailed description of the required information about the ensemble is available to authors from the Deposition page of PED (https://proteinensemble.org/deposition). To this end, a template spreadsheet can be downloaded from the Deposition page and—once completed—submitted to the PED biocuration team. Here we describe in detail the information required to submit a new conformational ensemble to PED.
Necessary Resources
Hardware
- Laptop or desktop computer, in order to be able to complete the template spreadsheet for deposition and to submit PDB file(s) of conformational ensembles. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
Completed template spreadsheet for deposition and one or more PDB files of the conformational ensemble
Authors’ data contact:
- “Authors’ name”: a list of all the authors who participated in the generation of the conformational ensemble, including the last and corresponding authors of the associated publication. The name of each author should be provided in the format of first name followed by last name (if any).
- “Authors’ email”: institutional emails of all the authors who participated in the generation of the conformational ensemble, including those of the last and corresponding authors of the associated publication.
- “ORCID”: ORCID identifiers of all the authors who participated in the generation of the conformational ensemble, including those of the last and corresponding authors of the associated publication.
Publication:
- “Publication status”: available options are “in preparation,” “pre-print,” “submitted,” “accepted,” “published.”
- “Publication identifier”: if the publication is already published, please provide us with the corresponding identifier, i.e., PubMed identifier and/or DOI. If both a PubMed identifier and a DOI are available, please provide them both, separated by a comma.
Conformational ensemble's experimental setup:
- “Description of the ensemble”: a short but detailed description of the deposited ensemble, specifying if the protein is wild-type or a mutant—if so, what kind of mutant—the boundaries of the protein(s) in the ensemble (using UniProt numbering), the post-translational modifications (PTMs), if any, and other details useful to characterize the conformational ensemble, e.g., presence of cofactors. The entry title must not be the title of the associated publication; instead, it should clearly describe the submitted ensemble in a compact format, e.g., “Structural ensemble of pSic1 (1-90) phosphorylated at Thr5, Thr33, Thr45, Ser69, Ser76 and Ser80.”
- “Experimental procedure”: a detailed description—not longer than 10 sentences—of the experimental procedure, e.g., the corresponding NMR or SAXS measurements.
- “Conformational ensemble calculation”: a detailed description—not longer than 10 sentences—of the conformational ensemble calculations.
- “Molecular dynamics calculation”: a detailed description—not longer than 10 sentences—of the molecular dynamics calculations, if available.
- “Expression organism”: name of the expression organism, if available, e.g., Escherichia coli BL21(DE3). Skip this field in case of cell-free protein synthesis.
- “Expression organism NCBI Taxon ID”: NCBI Taxon identifier of the expression organism used, that can be retrieved from NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy), e.g., the NCBI Taxon ID of Escherichia coli BL21(DE3) is 469008. Skip this in case of cell-free protein synthesis.
- “Raw experimental data xrefs”: cross-references to raw experimental data deposited in BMRB (https://bmrb.io/) for NMR data and SASBDB (https://www.sasbdb.org/) for small-angle scattering data (if any). If more than one cross-reference is available, they should be added separated by commas.
- “Chain description”: UniProt accession number of the protein chain/fragment of the conformational ensemble, e.g., “P04637” for the human P53. If the provided protein is an isoform and not the canonical sequence, the corresponding UniProt identifier should be provided.
- “Sequence boundaries”: start and end positions of the submitted protein fragment as per the corresponding UniProt sequence, e.g., “1-90” of pSic1, UniProt ACC:P38634.
- “Tags”: specification of tags inside the ensemble model construct. The sequence of the tag must be specified, along with its position on the construct, e.g., N-terminal or C-terminal, e.g., position: 1-2, sequence: “GS.”
PDB files:
Authors are encouraged to provide a single compressed PDB file for each submitted conformational ensemble. However, multiple PDB files will be accepted and merged by the PED biocuration team. The PDB file(s) should be in standard format (http://www.wwpdb.org/documentation/file-format-content/format33/v3.3.html), i.e., all heavy atoms, side chains, and chain names should be present in order to be able to pass the validation pipeline. Authors willing to provide replicas (equally good solutions) of the submitted conformational ensemble should submit each replica corresponding to a separate PDB file.
In case conformers’ weights are available for a weighted ensemble, authors should submit them as a separate text file.
Basic Protocol 4: PROVIDING FEEDBACK IN PED
Feedback on site experience and technical issues, along with questions related to the data deposited in PED, can be submitted using the PED Feedback form (https://proteinensemble.org/feedback) (Fig. 9).
Necessary Resources
Hardware
- While PED works best on laptop or desktop computers, it is also easily accessible from smartphones and tablets. An active and stable internet connection is required.
Software
- Internet browser. The browsing was optimized for Firefox (http://www.mozilla.org/firefox/), Google Chrome (http://www.google.com/chrome/), and Safari (http://www.apple.com/safari/).
Input data
No input data are required.
1.Open a web browser and connect to PED at https://proteinensemble.org/.
2.Click on the “Feedback” button on the top-right of the PED navigation bar.
3.Provide your contact information, name, and email address, in the corresponding boxes.
4.Add a subject of your message in the dedicated field, e.g., “feedback on site experience.”
5.Use the “Message” box to add a detailed feedback, comment or question. The minimum length of the message should be 15 characters.
6.Click on the green “Send” button to submit your feedback, comment or question to the PED team.
GUIDELINES FOR UNDERSTANDING RESULTS
PED is a manually curated primary database for the deposition of conformational ensembles of protein systems with substantial intrinsic disorder or flexibility. In PED, a dedicated team of biocurators manually screens published literature to retrieve new conformational ensembles of IDPs and, at the same time, works together with data owners to ensure the high quality of annotated entries. Therefore, the PED biocuration team manually curates metadata describing each conformational ensemble and ensures that the PDB files of each submitted ensemble satisfy the required standards of the database. PED is a hybrid-layout database, i.e., its entries are centered around the experiments of ensemble determination (Experimental entry pages), but this site also compiles protein-centric pages to annotate all ensembles to different regions of the same protein from a given organism (Protein pages).
To fully understand the data presented in PED with the methods summarized in the structured textual descriptions (Experimental procedure, Structural ensemble calculation, Molecular dynamics) on Experimental entry pages, one has to understand the basics of solution scattering and NMR experiments, as well as the fundamental principles of ensemble modeling techniques. Links to the source publications are provided for users to easily access the supplementary information and motivation of the ensemble modeling experiment. Besides SAXS and NMR, PED also has a significant number of smFRET-based entries (Dong et al., 2017; Fuertes et al., 2017; Gomes et al., 2020; Lincoff et al., 2020) that are all combined with SAXS and/or NMR. In the future, we anticipate receiving ensembles generated by other methods as well, including circular dichroism (CD; Nagy, Igaev, Jones, Hoffmann, & Grubmüller, 2019), high-speed atomic force microscopy (HS-AFM; Kodera et al., 2021), electrospray ionization mass spectrometry (ESI-MS; D'Urzo et al., 2015), and electron paramagnetic resonance (EPR) spectroscopy (Karthikeyan et al., 2018). Upon submission of multiple CD-based ensembles, we will integrate cross-referencing with deposition databases for CD spectra such as PCDDB (Whitmore, Miles, Mavridis, Janes, & Wallace, 2017). Among our further future goals, we have secured the availability of the database by mirroring. Moreover, we plan to integrate the ens_dRMS (Lazar et al., 2020) intramolecular distance-based metric to complement the superimposition-based RMSD measures for ensemble comparison. To stay up to date with the recent developments implemented on PED, follow our official Twitter page @ProteinEnsemble (https://twitter.com/proteinensemble).
LIST OF ABBREVIATIONS
| ACC | accession number |
| API | application programming interface |
| ASA | accessible surface area |
| BMRB | Biological Magnetic Resonance Data Bank |
| CA | alpha-carbon |
| CB | beta-carbon |
| CD | circular dichroism spectroscopy |
| CV | controlled vocabulary |
| Dmax | maximum dimension |
| DOI | digital object identifier |
| EPR | electron paramagnetic resonance |
| ESI-MS | electrospray ionization mass spectrometry |
| FRET | Förster resonance energy transfer |
| HS-AFM | high-speed atomic force microscopy |
| IDP | intrinsically disordered protein |
| IDR | intrinsically disordered region |
| JSON | JavaScript object notation |
| MD | molecular dynamics |
| NMR | nuclear magnetic resonance |
| PCCD | Protein Circular Dichroism Data Bank |
| PDB | Protein Data Bank |
| PED | Protein Ensemble Database |
| PMID | PubMed ID |
| PTM | post-translational modifications |
| REST | representational state transfer |
| Rg | radius of gyration |
| RMSD | root-mean-square deviation |
| RSA | relative solvent accessibility |
| SASBDB | Small Angle Scattering Biological Data Bank |
| SAXS | small-angle X-ray scattering |
| SEP | phosphoserine |
| Ser | serine |
| smFRET | single-molecule FRET |
| Thr | threonine |
| TPO | phosphothreonine |
| URL | uniform resource locator |
| x-ref | cross-reference |
AUTHOR CONTRIBUTIONS
Federica Quaglia : conceptualization; data curation; formal analysis; writing original draft. Tamas Lazar : conceptualization; data curation; formal analysis; writing original draft. András Hatos : software; writing original draft. Peter Tompa : supervision; funding acquisition; writing original draft. Damiano Piovesan : supervision; writing original draft. Silvio Tosatto : funding acquisition; project administration; supervision; writing original draft. All authors contributed to the writing of the submitted version of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the Protein Ensemble Database (PED) at https://proteinensemble.org/.
LITERATURE CITED
- Bernadó, P., & Svergun, D. I. (2012). Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Molecular BioSystems , 8(1), 151–167. doi: 10.1039/c1mb05275f.
- Cavalli, A., Camilloni, C., & Vendruscolo, M. (2013). Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. The Journal of Chemical Physics , 138(9), 094112. doi: 10.1063/1.4793625.
- Chan-Yao-Chong, M., Deville, C., Pinet, L., van Heijenoort, C., Durand, D., & Ha-Duong, T. (2019). Structural characterization of N-WASP domain V using MD simulations with NMR and SAXS data. Biophysical Journal , 116(7), 1216–1227. doi: 10.1016/j.bpj.2019.02.015.
- Chan-Yao-Chong, M., Marsin, S., Quevillon-Cheruel, S., Durand, D., & Ha-Duong, T. (2020). Structural ensemble and biological activity of DciA intrinsically disordered region. Journal of Structural Biology , 212(1), 107573. doi: 10.1016/j.jsb.2020.107573.
- Davey, N. E., Babu, M. M., Blackledge, M., Bridge, A., Capella-Gutierrez, S., Dosztanyi, Z. … Tosatto, S. C. E. (2019). An intrinsically disordered proteins community for ELIXIR. F1000Research , 8, ELIXIR–1753. doi: 10.12688/f1000research.20136.1.
- Dong, X., Gong, Z., Lu, Y.-B., Liu, K., Qin, L.-Y., Ran, M.-L. … Tang, C. (2017). Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch. Proceedings of the National Academy of Sciences of the United States of America , 114(26), 6770–6775. doi: 10.1073/pnas.1705718114.
- Dunker, A. K., Brown, C. J., David Lawson, J., Iakoucheva, L. M., & Obradović, Z. (2002). Intrinsic disorder and protein function. Biochemistry , 41(21), 6573–6582. doi: 10.1021/bi012159+.
- Dunker, A. K., Keith Dunker, A., David Lawson, J., Brown, C. J., Williams, R. M., Romero, P. … Obradovic, Z. (2001). Intrinsically disordered protein. Journal of Molecular Graphics and Modelling , 19(1), 26–59, doi: 10.1016/S1093-3263(00)00138-8.
- D'Urzo, A., Konijnenberg, A., Rossetti, G., Habchi, J., Li, J., Carloni, P., … Grandori, R. (2015). molecular basis for structural heterogeneity of an intrinsically disordered protein bound to a partner by combined ESI-IM-MS and modeling. Journal of The American Society for Mass Spectrometry , 26(3), 472–481, doi: 10.1007/s13361-014-1048-z.
- Feldman, H. J., & Hogue, C. W. V. (2002). Probabilistic sampling of protein conformations: New hope for Brute force? Proteins , 46(1), 8–23. doi: 10.1002/prot.1163.
- Fisher, C. K., Huang, A., & Stultz, C. M. (2010). Modeling intrinsically disordered proteins with Bayesian statistics. Journal of the American Chemical Society , 132(42), 14919–14927. doi: 10.1021/ja105832g.
- Fuertes, G., Banterle, N., Ruff, K. M., Chowdhury, A., Mercadante, D., Koehler, C. … Lemke, E. A. (2017). Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proceedings of the National Academy of Sciences of the United States of America , 114(31), E6342–E6351. doi: 10.1073/pnas.1704692114.
- Gomes, G.-N. W., Krzeminski, M., Namini, A., Martin, E. W., Mittag, T., Head-Gordon, T., … Gradinaru, C. C. (2020). Conformational ensembles of an intrinsically disordered protein consistent with NMR, SAXS, and single-molecule FRET. Journal of the American Chemical Society , 142(37), 15697–15710. doi: 10.1021/jacs.0c02088.
- Hatos, A., Hajdu-Soltész, B., Monzon, A. M., Palopoli, N., Álvarez, L., Aykac-Fas, B., … Piovesan, D. (2019). DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Research , 48(D1), D269–D276. doi: 10.1093/nar/gkz975.
- Jensen, M. R., Ruigrok, R. W. H., & Blackledge, M. (2013). Describing intrinsically disordered proteins at atomic resolution by NMR. Current Opinion in Structural Biology , 23(3), 426–435. doi: 10.1016/j.sbi.2013.02.007.
- Karthikeyan, G., Bonucci, A., Casano, G., Gerbaud, G., Abel, S., Thomé, V., … Mileo, E. (2018). A bioresistant nitroxide spin label for in-cell EPR spectroscopy: In vitro and in oocytes protein structural dynamics studies. Angewandte Chemie International Edition , 57(5), 1366–1370. doi: 10.1002/anie.201710184.
- Kikhney, A. G., Borges, C. R., Molodenskiy, D. S., Jeffries, C. Y. M., & Svergun, D. I. (2020). SASBDB: Towards an automatically curated and validated repository for biological scattering data. Protein Science , 29(1), 66–75. doi: 10.1002/pro.3731.
- Kodera, N., Noshiro, D., Dora, S. K., Mori, T., Habchi, J., Blocquel, D. … Ando, T. (2021). Structural and dynamics analysis of intrinsically disordered proteins by high-speed atomic force microscopy. Nature Nanotechnology , 16(2), 181–189. doi: 10.1038/s41565-020-00798-9.
- Köfinger, J., Stelzl, L. S., Reuter, K., Allande, C., Reichel, K., & Hummer, G. (2019). Efficient ensemble refinement by reweighting. Journal of Chemical Theory and Computation , 15(5), 3390–3401, doi: 10.1021/acs.jctc.8b01231.
- Krzeminski, M., Marsh, J. A., Neale, C., Choy, W.-Y., & Forman-Kay, J. D. (2013). Characterization of disordered proteins with ENSEMBLE. Bioinformatics , 29(3): 398–399, doi: 10.1093/bioinformatics/bts701.
- Lazar, T., Guharoy, M., Vranken, W., Rauscher, S., Wodak, S. J., & Tompa, P. (2020). Distance-based metrics for comparing conformational ensembles of intrinsically disordered proteins. Biophysical Journal , 118(12), 2952–2965. doi: 10.1016/j.bpj.2020.05.015.
- Lazar, T., Martínez-Pérez, E., Quaglia, F., Hatos, A., Chemes, L. B., Iserte, J. A. … Babu M. M. (2021). PED in 2021: A major update of the Protein Ensemble Database for intrinsically disordered proteins. Nucleic Acids Research , 49(D1), D404–D411.
- van der Lee, R., Buljan, M., Lang, B., Weatheritt, R. J., Daughdrill, G. W., Keith Dunker, A. … Babu, M. M. (2014). Classification of intrinsically disordered regions and proteins. Chemical Reviews , 114(13), 6589–6631, doi: 10.1021/cr400525m.
- Lincoff, J., Haghighatlari, M., Krzeminski, M., Teixeira, J. M. C., Gomes, G.-N. W., Gradinaru, C. C. … Head-Gordon, T. (2020). Extended experimental inferential structure determination method in determining the structural ensembles of disordered protein states. Communications Chemistry , 3, 74. doi: 10.1038/s42004-020-0323-0.
- Marsh, J. A., & Forman-Kay, J. D. (2012). Ensemble modeling of protein disordered states: Experimental restraint contributions and validation. Proteins , 80(2), 556–572. doi: 10.1002/prot.23220.
- Mittag, T., Marsh, J., Grishaev, A., Orlicky, S., Lin, H., Sicheri, F., … Forman-Kay, J. D. (2010). Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure , 18(4), 494–506. doi: 10.1016/j.str.2010.01.020.
- Nagy, G., Igaev, M., Jones, N. C., Hoffmann, S. V., & Grubmüller, H. (2019). SESCA: Predicting circular dichroism spectra from protein molecular structures. Journal of Chemical Theory and Computation , 15(9), 5087–5102. doi: 10.1021/acs.jctc.9b00203.
- Nath, A., Sammalkorpi, M., DeWitt, D. C., Trexler, A. J., Elbaum-Garfinkle, S., O'Hern, C. S., & Rhoades, E. (2012). The conformational ensembles of α-synuclein and tau: Combining single-molecule FRET and simulations. Biophysical Journal , 103(9), 1940–1949. doi: 10.1016/j.bpj.2012.09.032.
- Orchard, S., Ammari, M., Aranda, B., & Breuza, L. (2014). The MIntAct project—IntAct as a common curation platform for 11 Molecular Interaction Databases. Nucleic Acids , 42, D358–D363. doi: 10.1093/nar/gkt1115.
- Ozenne, V., Bauer, F., Salmon, L., Huang, J.-R., Jensen, M. R., Segard, S., … Blackledge, M. (2012). Flexible-Meccano: A tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics , 28(11), 1463–1470. doi: 10.1093/bioinformatics/bts172.
- Piovesan, D., Tabaro, F., Mičetić, I., Necci, M., Quaglia, F., Oldfield, C. J., … Tosatto, S. C. (2017). DisProt 7.0: A major update of the database of disordered proteins. Nucleic Acids Research , 45(D1), D219–D227. doi: 10.1093/nar/gkw1056.
- Rangan, R., Bonomi, M., Heller, G. T., Cesari, A., Bussi, G., & Vendruscolo, M. (2018). Determination of structural ensembles of proteins: Restraining vs reweighting. Journal of Chemical Theory and Computation , 14(12), 6632–6641. doi: 10.1021/acs.jctc.8b00738.
- Rauscher, S., Gapsys, V., Gajda, M. J., Zweckstetter, M., de Groot, B. L., & Grubmüller, H. (2015). Structural ensembles of intrinsically disordered proteins depend strongly on force field: A comparison to experiment. Journal of Chemical Theory and Computation , 11(11), 5513–5524, doi: 10.1021/acs.jctc.5b00736.
- Robustelli, P., Piana, S., & Shaw, D. E. (2018). Developing a molecular dynamics force field for both folded and disordered protein states. Proceedings of the National Academy of Sciences of the United States of America , 115(21), E4758–E4766. doi: 10.1073/pnas.1800690115.
- Romero, P. R., Kobayashi, N., Wedell, J. R., Baskaran, K., Iwata, T., Yokochi, M., … Markley, J. L. (2020). BioMagResBank (BMRB) as a resource for structural biology. Methods in Molecular Biology , 2112, 187–218. doi: 10.1007/978-1-0716-0270-6_14.
- Schramm, A., Bignon, C., Brocca, S., Grandori, R., Santambrogio, C., & Longhi, S. (2019). An Arsenal of methods for the experimental characterization of intrinsically disordered proteins—How to choose and combine them? Archives of Biochemistry and Biophysics , 676, 108055, doi: 10.1016/j.abb.2019.07.020.
- Tompa, P. (2002). Intrinsically unstructured proteins. Trends in Biochemical Sciences , 27(10), 527–533. doi: 10.1016/S0968-0004(02)02169-2.
- Tompa, P. (2011). Unstructural biology coming of age. Current Opinion in Structural Biology , 21(3), 419–425. doi: 10.1016/j.sbi.2011.03.012.
- UniProt Consortium. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research , 47(D1), D506–D515. doi: 10.1093/nar/gky1049.
- Varadi, M., Kosol, S., Lebrun, P., Valentini, E., Blackledge, M., Dunker Keith, A., … Tompa, P. (2014). pE-DB: A database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Research , 42(Database issue), D326–D335. doi: 10.1093/nar/gkt960.
- Varadi, M., & Tompa, P. (2015). The Protein Ensemble Database. Advances in Experimental Medicine and Biology , 870, 335–349. doi: 10.1007/978-3-319-20164-1_11.
- White, J. (2020). PubMed 2.0. Medical Reference Services Quarterly , 39(4), 382–387, doi: 10.1080/02763869.2020.1826228.
- Whitmore, L., Miles, A. J., Mavridis, L., Janes, R. W., & Wallace, B. A. (2017). PCDDB: New developments at the Protein Circular Dichroism Data Bank. Nucleic Acids Research , 45(D1), D303–307. doi: 10.1093/nar/gkw796.
- Wright, P. E., & Dyson, H. J. (1999). Intrinsically Unstructured Proteins: Re-Assessing the protein structure-function paradigm. Journal of Molecular Biology , 293(2), 321–331. doi: 10.1006/jmbi.1999.3110.
- wwwPDB Consortium. (2019). Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Research , 47(D1), D520–D528. doi: 10.1093/nar/gky949.
Citing Literature
Number of times cited according to CrossRef: 3
- Prakash Kulkarni, Vitor B. P. Leite, Susmita Roy, Supriyo Bhattacharyya, Atish Mohanty, Srisairam Achuthan, Divyoj Singh, Rajeswari Appadurai, Govindan Rangarajan, Keith Weninger, John Orban, Anand Srivastava, Mohit Kumar Jolly, Jose N. Onuchic, Vladimir N. Uversky, Ravi Salgia, Intrinsically disordered proteins: Ensembles at the limits of Anfinsen's dogma, Biophysics Reviews, 10.1063/5.0080512, 3 , 1, (011306), (2022).
- Ruth Nussinov, Mingzhen Zhang, Ryan Maloney, Yonglan Liu, Chung-Jung Tsai, Hyunbum Jang, Allostery: Allosteric Cancer Drivers and Innovative Allosteric Drugs, Journal of Molecular Biology, 10.1016/j.jmb.2022.167569, 434 , 17, (167569), (2022).
- Zita Harmat, Dániel Dudola, Zoltán Gáspári, DIPEND: An Open-Source Pipeline to Generate Ensembles of Disordered Segments Using Neighbor-Dependent Backbone Preferences, Biomolecules, 10.3390/biom11101505, 11 , 10, (1505), (2021).

