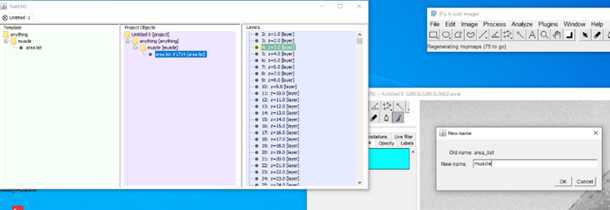
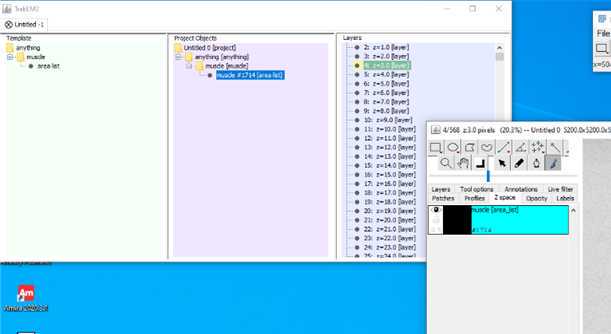
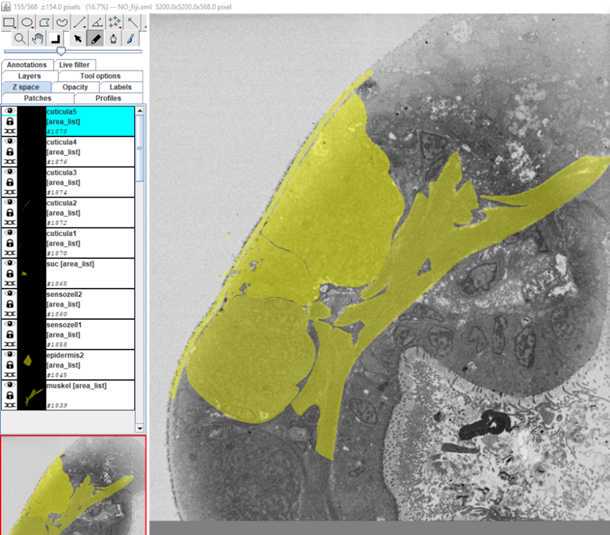
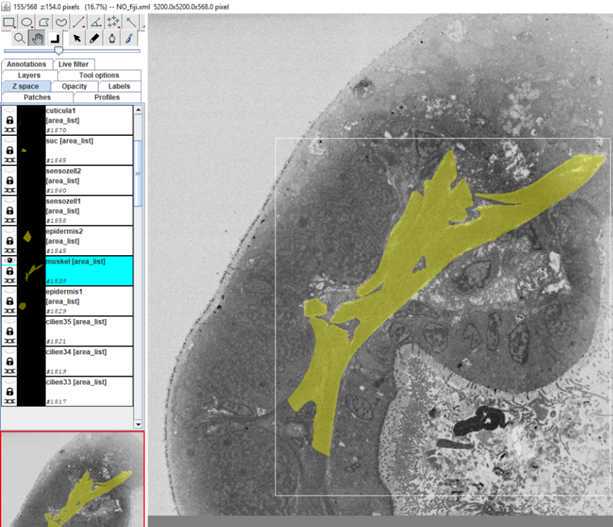
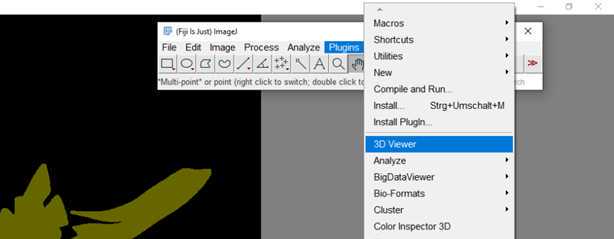
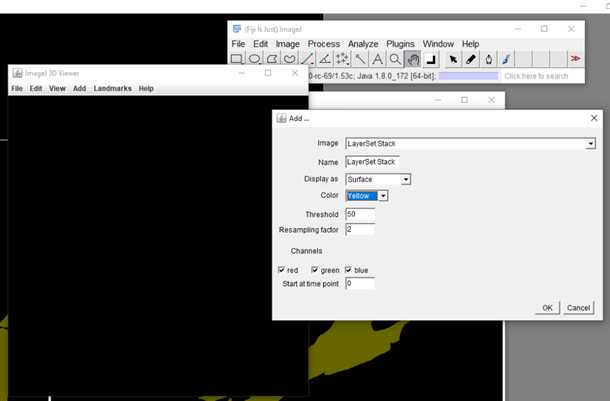
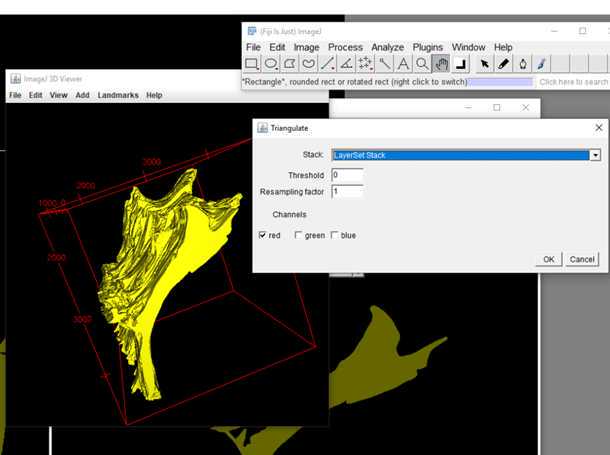
S1: Step-by-step-guide using image J in the work flow.
Paul Kalke, Conrad Helm
Published: 2022-04-28 DOI: 10.17504/protocols.io.b2x9qfr6
Abstract
How to use ImageJ in our 3D-workflow from data manipulation, import, segmentation and export as OBJ-file.
Steps
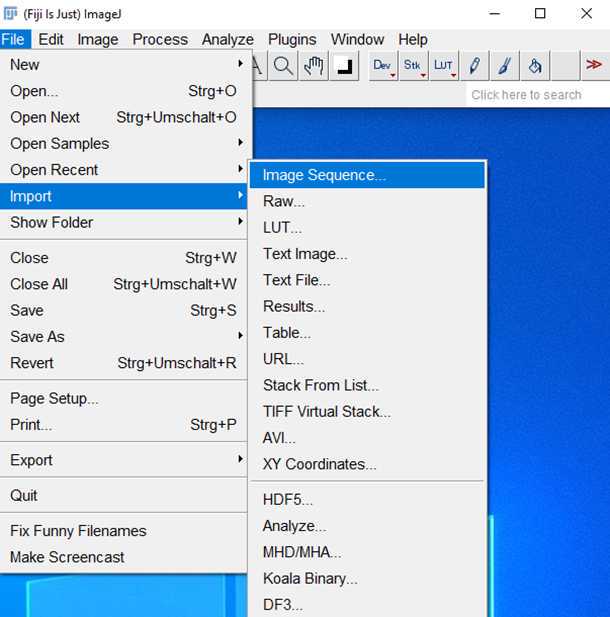
1.
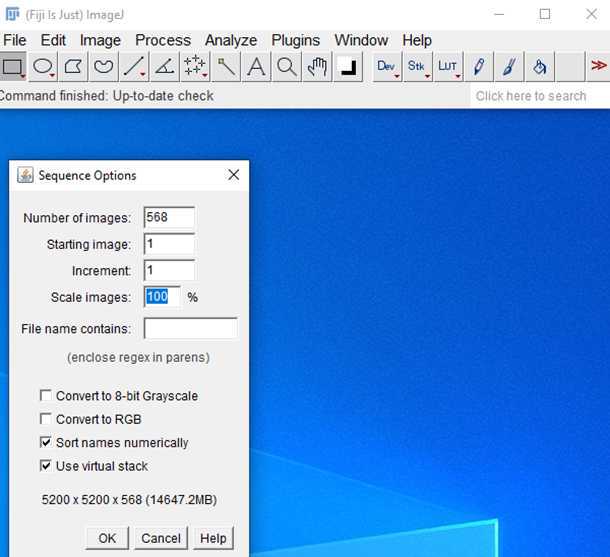
2.
3.
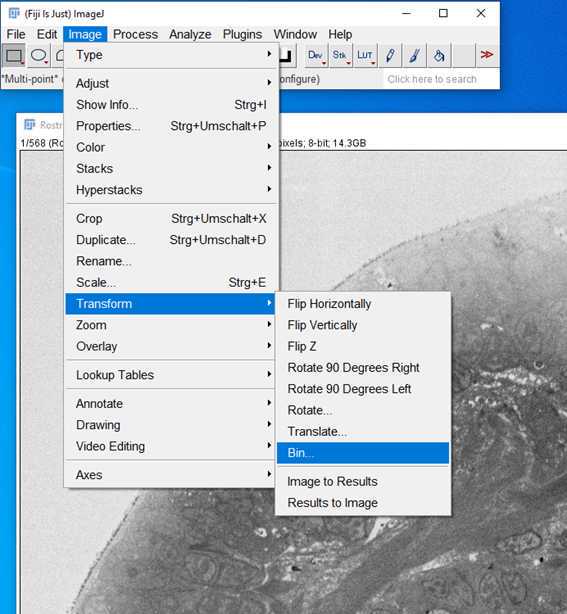
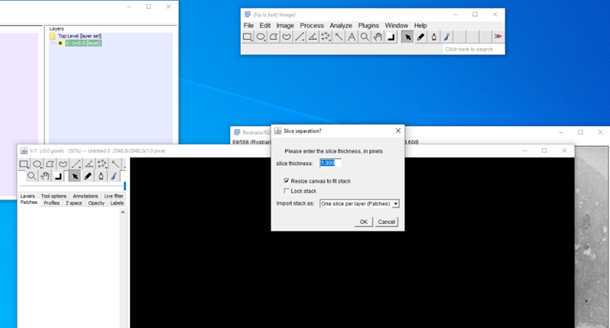
5.
7.
9.
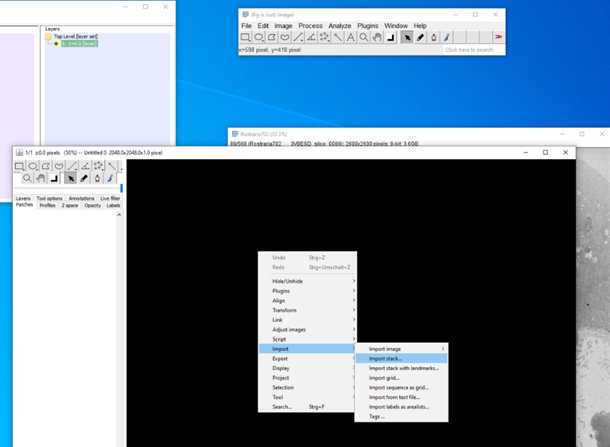
10.
12. 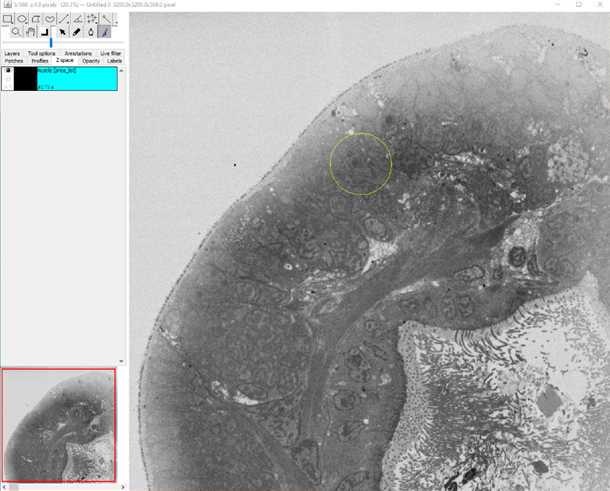

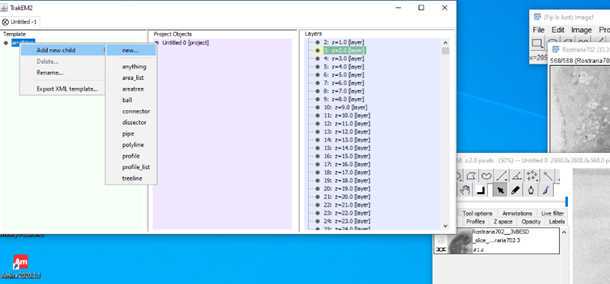
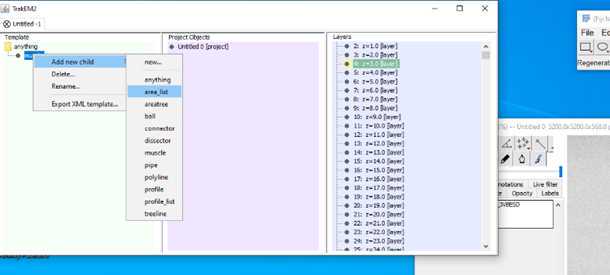
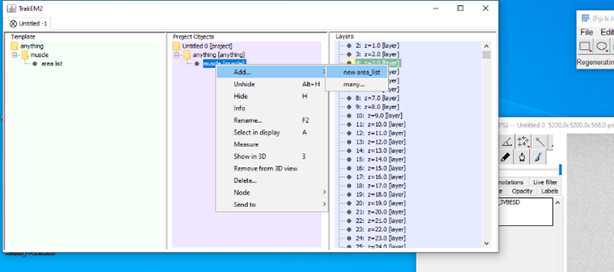
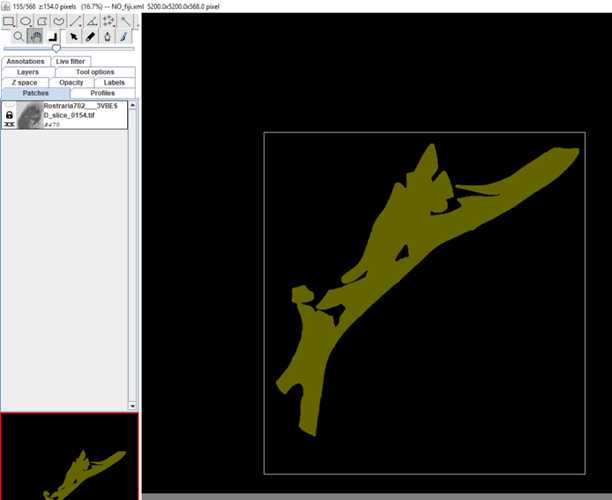
•Now start to segment your structure
•Make sure the brush tool in the main tool bar is checked
•Hot keys and how to reconstruct:
-Mouse wheel (mw); , and . : scrolling through data set
-Ctrl + mw; + and - : zoom in and out
-Shift + mw: size of pen
-Left mouse button (l mb): mark
-Shift + l mb: fill
-Alt + l mb: erase
-Alt + shift + l mb: delete all on slice
-Right mouse button (r mb) – Area: for all kind of interpolations
13.
14.
15.
17.
18.