S-2 SOIL PROCESSING
REDI-NET Consortium
Disclaimer
This work is supported by the US Army Medical Research and Development Command under Contract No.W81XWH-21-C-0001, W81XWH-22-C-0093 and HT9425-23-C-0059. The views, opinions and/or findings contained in this report are those of the author(s) and should not be construed as an official Department of the Army or Navy position, policy or decision unless so designated by other documentation.
Abstract
This protocol details about soil processing.
Before start
BEFORE START
- Clean the work surfaces with RNaseZap, then wipe the surfaces with 70% molecular biology grade ethanol to remove additional contaminants.
- Transfer 0.1 mm zirconium oxide beads (2 spoons, Appendix 1) to Clear RINO brand 1.5 ml screw-cap microcentrifuge tubes.*
- For the first time use of IndiMag pathogen kit, add 100% ethanol to Buffer AW1 and AW2, and add 100% isopropanol to ACB as indicated on the bottles ( Optional if using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit ).
- Buffer ATL may form precipitates upon storage. If necessary, warm to
56°Cuntil the precipitates have fully dissolved. Prepare buffer ATL-DX: add100µLReagent DX to15mLBuffer ATL. If smaller amounts are needed, transfer1.5mLof Buffer ATL into a sterile 2 ml vial and add10µLReagent DX. Mix well, after addition of Reagent DX. After preparation, the mixture is stable for 6 months atRoom temperature(15-25°C)** - MagAttract Suspension G from IndiMag pathogen kit needs to be vortexed thoroughly for
0h 3m 0s(before first use) or0h 1m 0s(before subsequent uses) to ensure that the magnetic silica particles are fully resuspended.**Note*If using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit, transfer all the 0.1 mm beating beads into a new clear RINO tube brand 1.5 mL screw-cap microcentrifuge tube. **Optional if using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit .
Steps
1. SAMPLE LYSIS
Add 320µL of 1x PBS and 80µL ATL-DX Buffer to the bead tubes prepared on step 3 of Before Start section under the Guidelines & Warnings tab.
0.25g soil sample and place it into each prepared bead tube. Record the weight. Include a positive control for each batch of samples: transfer 37.5µL ZymoBIOMICS Microbial Community Standard Material and µL EBV, and 100µL HIV standard into a tube from step 3 of Before Start section. Add 162.5µL 1xPBS.
Include a negative control for each batch of samples: a bead tube from step 3 of Before Start section with 320µL cold sterile 1xPBS only.
Add dry ice into the cooling compartment of Bullet Blender and then load the all bead tubes (samples and controls).
Set the speed at 12 and time at 3. Press Start.
Let the samples settle for 0h 1m 0s and then repeat step 6.
2. INSTRUMENT SET UP
Confirm 96 deep-well magnetic heads and 96 well deep-well heat blocks are being used.
Ensure the program IndiMag_Pathogen_KF_Flex_4wash or the program has been downloaded and loaded onto the KingFisher Flex instrument.
3. SET UP THE PROCESSING PLATES
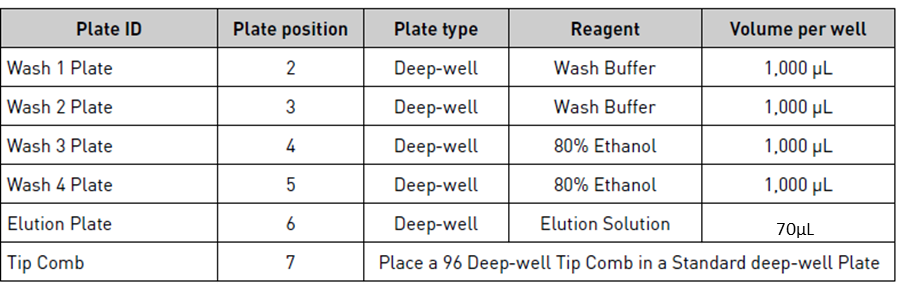
Set up the Wash, Elution, and Tip Comb Plates outside the instrument according to the following table.
| A | B | C | D | E |
|---|---|---|---|---|
| Plate ID | Plate position | Plate type | Reagent | Volume per well |
| Tip comb | 7 | Place a 96 Deep-well Tip comb in a deep-well plate | ||
| Elution | 6 | Deep-Well | Nuclease-free water | 75 µL |
| Wash 4 | 5 | Deep-Well | 100% ethanol | 750 µL |
| Wash 3 | 4 | Deep-Well | 80% ethanol | 750 µL |
| Wash 2 | 3 | Deep-Well | Buffer AW2 | 700 µL |
| Wash 1 | 2 | Deep-Well | Buffer AW1 | 700 µL |
| Sample | 1 | Sample Lysate | Lysate and lysis buffer | 990 µL |
4. EXTRACTION
Centrifuge the bead tubes with lysate from step 7 for 12000x g.
Add 20µL of Proteinase K into wells (based on number of samples) of a new Deep-well plate.
Transfer 270µL supernatant without any particle carryover to the wells of the Deep-well plate containing proteinase K. This plate becomes the Sample Plate.
Add 135µL Buffer VXL, 540µL Buffer ACB, and 20µL MagAttract Suspension G to each sample in the sample plate. For multiple samples, make a master mix with 10% overage. Invert slowly to mix the master mix, avoid foaming (can be mixed on Hula mixer for 2 min). Add 695µL mixture to each sample (Optional if using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit).
Select the program IndiMag_Pathogen_KF_Flex_4wash or the program MagMAX_Microbiome_Liquid_Buccal_Flex on the instrument according to the kit used.
Start the run, then load the prepared plates into position when prompted by the instrument.
5. BIND, WASH AND ELUTE (Only if using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit)
Vortex magnetic beads vigorously and for each sample, transfer 20µL beads to 500µL Binding Buffer. Make the master mix for multiple samples with 10% overage. Mix the master mix by inverting, then place the master mix on a rocker until use (Do not vortex).
When prompted (approximately 20 minutes after the start of the protocol), remove the Sample plate from the instrument.
Invert Binding Bead mix prepared in step 17 to mix, then add 520µL to each sample in the Sample Plate. Remix the Binding Bead mix frequently to ensure even distribution of beads to all samples.
Place the Sample Plate back onto the instrument, then start the run.
6. QUANTIFICATION AND STORAGE
After the running protocol is completed (~35 minutes), immediately remove the elution plate from the instrument and cover the plate or transfer the eluate to the final tube or plate of choice for final storage.
In a 0.6 mL microcentrifuge tube, use 3µL total nucleic acid for DNA and RNA concentration measurement using Qubit 4 Fluorometer following manufacturer instructions.
Proceed with sample testing following the REDI-NET SOP S-4 Soil Testing or store at -20°C for less than 2 weeks.
7. INSTRUMENT SET UP
Confirm 12-tip magnetic heads and 12 well deep-well heat blocks are being used.
Ensure the program IndiMag_Pathogen_KF_Duo_4wash has been downloaded and loaded onto the KingFisher Duo Prime instrument.
8. SET UP THE SAMPLE PLATE AND ELUTION STRIP
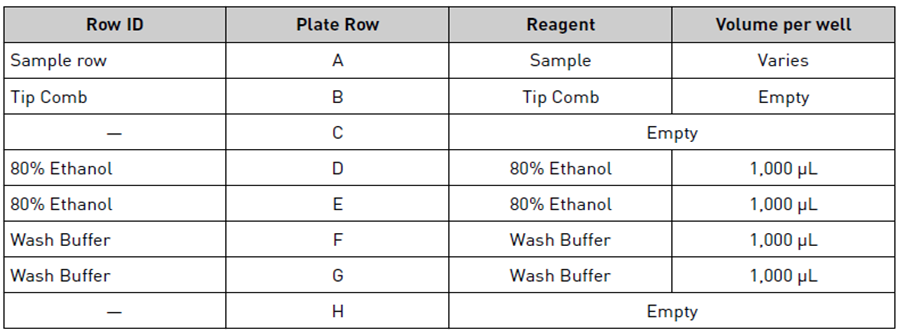
Set up the Sample Plate according to the table below:
| A | B | C | D |
|---|---|---|---|
| Row ID | Plate Row | Reagent | Volume per well |
| Sample row | A | Lysate and lysis buffer | 985 µL |
| Wash 1 | B | Buffer AW1 | 700 µL |
| Wash 2 | C | Buffer AW2 | 700 µL |
| Wash 3 | D | 80% ethanol | 750 µL |
| Wash 4 | E | 100% ethanol | 750 µL |
| Tip Comb | F | Tip comb | |
| G | Empty | ||
| H |
Set up the Elution Strip according to the table below:
| A | B | C | D |
|---|---|---|---|
| Row ID | Plate Row | Reagent | |
| Elution | A | Nuclease-free water | 75 µL |
9. EXTRACTION
Centrifuge the bead tubes with lysate from Sample Lysis step 7 for 12000x g.
Add 20µL of Proteinase K into wells (based on number of samples) of a new Deep-well plate.
Transfer 270µL supernatant without any particle carryover to the wells of the Deep-well plate containing proteinase K. This plate becomes the Sample Plate.
Add 135µL Buffer VXL, 540µL Buffer ACB, and 20µL MagAttract Suspension G to each sample in the sample plate. For multiple samples, make a master mix with 10% overage. Invert slowly to mix the master mix, avoid foaming. Add 695µL mixture to each sample (Optional if using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit).
Select the program IndiMag_Pathogen_KF_Duo_4wash or the program MagMAX_Microbiome_Soil_Duo on the instrument.
Start the run, then load the prepared plates into position when prompted by the instrument.
10. BIND, WASH AND ELUTE(Only if using the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit)
Vortex magnetic beads vigorously and for each sample, transfer 20µL beads to 500µL Binding Buffer. Make master mix for multiple samples with 10% overage. Mix the master mix by inverting, then place the master mix on a rocker until use (Do not vortex).
When prompted (approximately 0h 20m 0s after the start of the protocol), remove the Sample plate from the instrument.
Invert Binding Bead mix prepared in step 35 to mix, then add 520µL to each sample in the Sample Plate. Remix the Binding Bead mix frequently to ensure even distribution of beads to all samples.
Place the Sample Plate back onto the instrument, then start the run.
11. QUANTIFICATION AND STORAGE
After the running protocol is completed (~ 0h 35m 0s), immediately remove the elution plate from the instrument and cover the plate or transfer the eluate to the final tube or plate of choice for final storage.
In a 0.6 mL microcentrifuge tube, use 3µL total nucleic acid for DNA and RNA concentration measurement using Qubit 4 Fluorometer following manufacturer instructions.
Proceed with sample testing following the REDI-NET SOP S-4 Soil Testing or store at -20°C for less than 2 weeks.
APPENDIX 2. DNA and RNA Measurement using QUBIT FLUOROMETER 4.0
DNA quantification:
According to the volume of sample used, add the 1xHS dsDNA Qubit Assay for a final volume of 200µL (i.e., if using 3µL of sample, add 197µL of 1x HS dsDNA Qubit Assay. Vortex for 5 - 10 seconds, then Incubate for 0h 2m 0s at Room temperature before reading.
RNA Quantification:
In a new microcentrifuge tube/falcon tube (depending on the number of samples processed), prepare a working solution of the Qubit HS RNA Assay:
| A | B | C |
|---|---|---|
| Reagents | Volume/sample | Volume for n+1 sample |
| Qubit RNA HS Assay buffer | 199 µL | …. µL |
| Qubit RNA HS Assay Dye | 1 µL | …. µL |
In a new 0.6 ml tube, mix 197µL of Qubit HS RNA Assay working solution and 3µL of the sample. Vortex for 5 - 10 seconds, then incubate for 0h 2m 0s at Room temperature before reading.