Protocol for RNA isolation and RT‐PCR confirmation for Neomycin gene expression in transformed Bodo saltans
Fatma Gomaa, Zhu-Hong Li, Roberto Docampo, Virginia Edgcomb
Abstract
Developing transfection protocol for Bodo saltans , using SaCas9/sgRNA ribonucleoprotein (RNP) complex in conjunction with DNA repair template to disrupt the Paraflagellar rod 2 gene ( BsPFR2 ) and increase the efficiency of targeted homologous recombination when a repair template DNA is provided. The exogenous repair template is double stranded DNA and it consists of eGFP fused with the drug selection gene nptII/neo and flanked by 500 bp of the untranslated regions (UTRs) upstream and downstream of the targeted BsPFR2 as homologous repair arms.
Steps
-Total RNA was isolated from 4 months old transformed clones and wild type Bodo saltans cultures using the Quick-RNA Miniprep Kit (Zymo Research) according to the manufacturer’s instructions.
-RNA underwent two rounds of DNAse treatments. First, we performed an on-column DNAse digestion, following the protocol of the Quick-RNA Miniprep Kit (Zymo Research). Second, DNAse digestion was performed on the purified RNA using gDNA wipeout buffer and the QuantiTect Reverse Transcription Kit (Qiagen, Germany), in the following 14 µl reaction: 2 µl of gDNA wipeout buffer 7x, 6 µl of RNA, and 6 µl of RNase-free water.
-cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen, Germany) according to the manufacturer’s instructions, the RT Primer Mix and the 5X Quantiscript RT buffer were mixed in 1:4 ratio in a 20 µl reaction that included 14 µl of RNA template (post-DNA elimination).
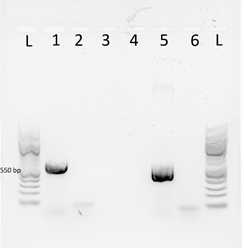
-cDNA was then amplified in a 25 µl PCR reaction, with primers targeting the Neomycin gene; the Neo F primer (5’-CGCTTGGGTGGAGAGGCTATTCGGCTATGAC 3’) and Neo R primer (5’-TCCACCATGATATTCGGCAAGCAGGCATC -3’).
-Control PCR reactions was performed using RNA template without the RT to verify the absence of the DNA from the transformed B. saltans cell.
-PCR products were visualized by gel electrophoresis, with purified plasmid as a positive control for the PCR. Amplified PCR products at the expected size of 550-pb were documented (Figure 1).