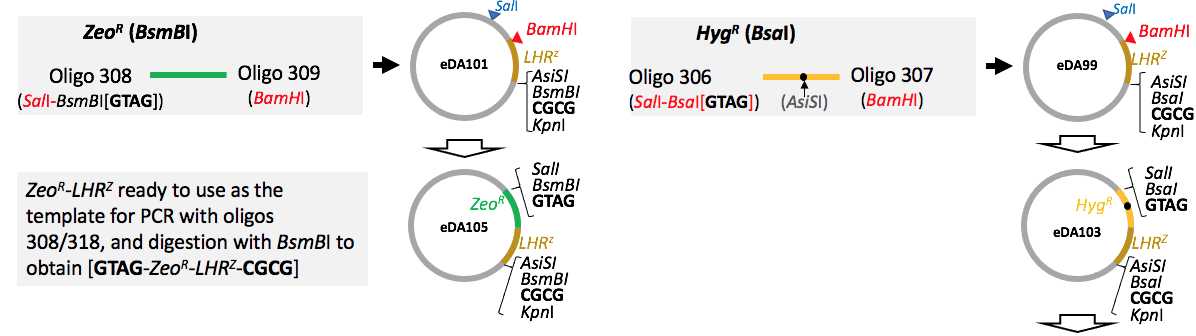
Preparation of parts HygR-LHRZ and ZeoR-LHRZ
Dariusz Abramczyk
Abstract
This cloning protocol is for preparation tandem pair HygR-LHRZ and ZeoR-LHRZ as a part of combinatory assembly system for preparation of integration/insertion arrays.
https://www.protocols.io/edit/nanochromosome-arrays-combinatory-assembly-cprkvm4w
Either the HygR-LHRZ or the ZeoR-LHRZ tandem pairs are designed to be use for all arrays and are displayed on 3'-end each of arrays.
During inchworming/swapping HR double cross-over recombination process, the antibiotic selection marker is replaced in every HR event while the LHRZ works as a homology recombination region used during all inchworming/swapping rounds.
(see https://www.protocols.io/edit/nanochromosome-arrays-combinatory-assembly-cprkvm4w)
Both parts are AsiSI free (this is for an optional excise site the cassette from the plasmid to isolate an insertion array before Pichia transformation)

Steps
Cloning and preparation of HygR-LHRZ and ZeoR-LHRZ, integrative/insertion array lasting 3'-part
PCR preparation part LHRZ for cloning with KpnI/BamHI (3'-end of LHRZ with
BsaI or BsmBI RE site delivered by an oligo)
2h 0m 0s
| A | B | C |
|---|---|---|
| components | LHRZ for (BsmBI) | LHRZ for (BsaI) |
| eDA8 (125pg/uL) | 1 | 1 |
| oligo 310 | 1.25 | 1.25 |
| oligo317 | 1.25 | |
| oligo 318 | 1.25 | |
| PrimeStar Premix 2x | 25 | |
| water | up to 50 |
LHR(non-coding DNA) delivered from the DNA library design by dr Valentin Zulkover and synthesised by prof Adele Marston Adele Marston lab
| A | B | C |
|---|---|---|
| 310 (F) | LHRZ (BamHI | CCGGATCCCCGTGTAGGTCTACAAACTGG |
| 318 (R) | LHRZ (KpnI-BsmBI-AsiSI) | GCGGTACCCGTCTCAcgcgGCGATCGCCCTGCAGGTTGAGTTGGCGAAGGTGCG |
| 317 (R) | LHRZ (KpnI-BsaI-AsiSI) | GGGGTACCGGTCTCAcgcgGCGATCGCCCTGCAGGTTGAGTTGGCGAAGGTGCG |
| A | B | C |
|---|---|---|
| Program | time | number of cycles |
| 98oC | 10 sec | 33 x |
| 55oC | 10sec | |
| 68oC | 1min 30 sec | |
| 4oC | hold |
0h 30m 0s
PCR clean-up QIAquick PCR Purification Kit (Qiagen)QIAquick PCR Purification Kit
Elution with water 50uL
KpnI and BamHI digestion of PCR product and pUC19 (all enzymes NEB) BamHI/ KpnI
| A | B | C | D |
|---|---|---|---|
| 10 x CutSmart | 8 | 8 | 8 |
| BamHI-HF (20u/uL) | 1.5 | 1.5 | 1 |
| KpnI-HF (20u/uL) | 1.5 | 1.5 | 1 |
| PCR LHRZ 310/317 | 50 | ||
| PCR LHRZ 310/318 | 50 | ||
| pUC19 298 ng/uL | 10 | ||
| water | up to 80 -> |
RE digestion reactions
37°C 4h 0m 0s
0h 30m 0s
PCR clean-up QIAquick PCR Purification Kit (Qiagen)QIAquick PCR Purification Kit
Elution with water 50uL
Checking DNA concentration
LHRZ 310/317 (BamHI/KpnI) - ~100ng/ul
LHRZ 310/318 (BamHI/KpnI) - ~100ng/ul
Ligation reactions T4 DNA ligase (NEB)
| A | B | C |
|---|---|---|
| pUC B/K ~100ng/uL) BamHI/KpnI | 1 | 1 |
| T4 DNA ligase | 1 | 1 |
| LHRZ (100ng/ul) B/K(310/317) | 5.5 | |
| 10 X buffer | 1 | 1 |
| water | up to 10 -> | |
| LHRZ (100ng/ul)(310/318) B/K | 5.5 |
2h 0m 0s
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL 37°C
1h 0m 0s
Several colonies obtained on both agar plates. Single colonies transferred on new LB +100ug/mL ampicillin plate, growth overnight and a single colonies submitted for bacterial colony PCR.
see protocol colony PCR
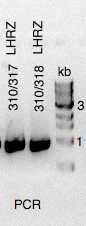
Material loaded on agarose gel for verification
Positively verified clones re-cultured in LB + 100 ug/ml
Plasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
PCR verified and Sanger sequencing intermediate plasmid LHRZ(BsmBI)-pUC19 called eDA101eda101.dna
PCR verified and Sanger sequencing intermediate plasmid LHRZ(BsaI)-pUC19 called eDA99eDA99.dna
PCR amplification of Zeocin expression cassette (BsaI-free) for cloning with BsmBI
Zeocin and hygromycin cassettes originally derived from plasmids pGS-GnTI and pGS-GnTII (Glycoswitch) glycoswitch plasmids
| A | B | C |
|---|---|---|
| eDA48 (175 (ng/uL) | 1 | |
| eDA78 (125pg/uL) | 1 | |
| oligo 308 | 1.25 | |
| oligo309 | 1.25 | |
| oligo 306 | 1.25 | |
| oligo 307 | 1.25 | |
| PrimeStar Premix 2x | 25 | 25 |
| water | up to 50 | up to 50uL |
| A | B | C |
|---|---|---|
| 308 (F) | Zeocin cassette (SalI-BsmBI) overhang GTAG | CTGTCGACCGTCTCAgtagCCCACACACCATAGCTTCAAAATG |
| 309(R) | Zeocin cassette (BamHI) | GGGATCCGCAAATTAAAGCCTTCGAGCG |
| 306 (F) | HygR (SalI-BsaI) | CCTGTCGACGGTCTCAgtagGACATGGAGGCCCAGAATACCC |
| 307 (R) | HygR (BamHI) | GCGGATCCCAGTATAGCGACCAGCATTCACATAC |
0h 30m 0s
PCR clean-up QIAquick PCR Purification Kit (Qiagen)QIAquick PCR Purification Kit
Elution with water 50uL
SalI and BamHI digestion of PCR product and pUC19 (all enzymes NEB)BamHI
| A | B | C | D | E |
|---|---|---|---|---|
| 10x cutsmart buffer NEB | 8 | 8 | 8 | 8 |
| BamHI-HF (20u/uL) | 1.5 | 1.5 | 1.5 | 1.5 |
| eDA99 70 ng/uL | 30 | |||
| eDA101 81 ng/uL | 30 | |||
| PCR hygR 306/307 | 50 | |||
| PCR zeo 308/309 | 50 | |||
| SalI-HF (20u/uL) | 1.5 | 1.5 | 1.5 | 1.5 |
| water | up to 80 -> |
`37°C` `4h 0m 0s`
0h 30m 0s
PCR clean-up QIAquick PCR Purification Kit (Qiagen)QIAquick PCR Purification Kit
Elution with water 50uL
Ligation reactions T4 DNA ligase (NEB)
| A | B | C |
|---|---|---|
| T4 DNA ligase | 1 | 1 |
| 10 X buffer | 1 | 1 |
| eDA101 (S/B) | 3 | |
| zeo 308/309 (S/B) | 5 | |
| eDA99 (S/B) | 3 | |
| hygR 306/307 (S/B) | 5 | |
| water | up to 10 -> |
2h 0m 0s
E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB + 100ug/mL carbenicillin and growth at 37°C
1h 0m 0s
.Several colonies obtained on both agar plates. Single colonies transferred on new LB +100ug/mL ampicillin plate, growth overnight and a single colonies submitted for bacterial colony PCR
see protocol colony PCR
(oligo 131/40 for verification ZeoR-LHRC-puC see plasmid map eda105.dna
(oligo 306/42 for verification HygR-LHRC-puC see plasmid map eda103.dna
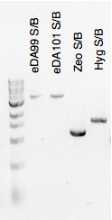
Material loaded on agarose gel for verification
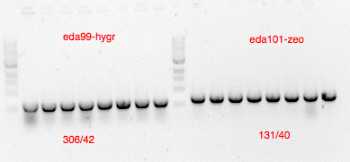
Positively verified clones re-cultured in LB ampicillin for miniprep (plasmid preparation)
Plasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
PCR verified and Sanger sequencing intermediate plasmid ZeoR-LHRC-puC see plasmid eda105.dna
Tandem paired ZeoR-LHRZ part (BsmBI-free, AsiSI-free,) ready for cloning with BsmBI as a PCR template for a use in the protocol
https://www.protocols.io/edit/nanochromosome-arrays-combinatory-assembly-cprkvm4w
PCR verified and Sanger sequencing intermediate plasmid HygR-LHRC-puC see plasmid map eda103.dna The part is BsaI-free but contains internal AsiSI in hph gene , requires mutagenesis
Mutagenesis of AsiSI in eDA103 plasmid by Gibson assembly
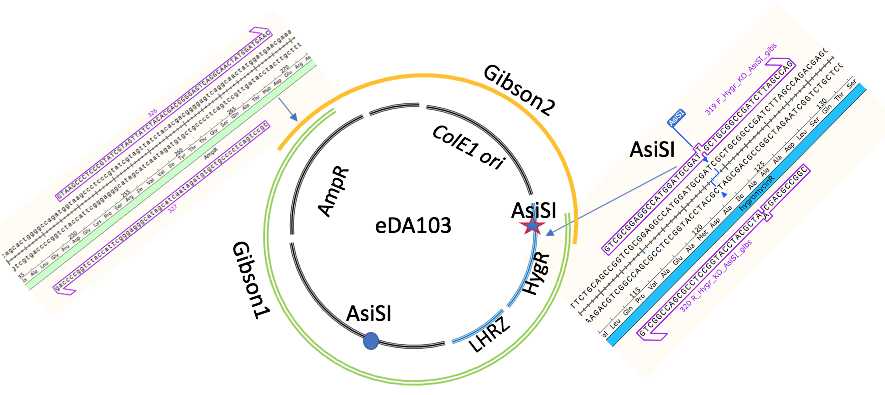
1h 30m 0s PCR amplification for further Gibson assembly reaction
PrimeStarHS 2x premix (Takara) High Fidelity
| A | B | C |
|---|---|---|
| water | up to 50 uL | for Gibson 1 |
| Primestar2x | 25 | 25 |
| oligos 327/320 | 1.25/1.25 | |
| oligos 319/326 | 1.25/1.25 | |
| eDA103 (74pg/uL) | 1.5 | 1.5 |
| A | B | C |
|---|---|---|
| 98C | 10sec | 33x |
| 55C | 10 sec | |
| 68C | 2min 30 sec | |
| 4C | hold |
PCR reaction treatment with DpnI for 0h 30m 0s at 37°C
0h 30m 0s PCR clean-up QIAquick PCR Purification Kit (Qiagen)QIAquick PCR Purification Kit
Elution with water 50uL.
DNA Molar concentration of purified PCR products was calculated using bioline conventor DNA concentration convertor
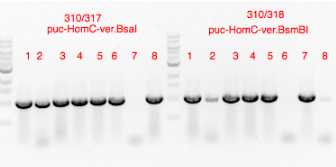
Gibson 1 fragment 86 ng/uL
Gibson 2 fragment 46 ng/ul
PCR products verified on 1% agarose gel (4 uL loaded)
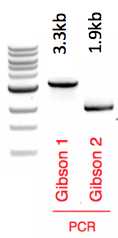
Gibson assembly of 3.3kb fragment 1 and 1.9kb fragment 2
0h 30m 0s 37°C
| A | B |
|---|---|
| Gibson1 319/326 (3.3kb) 40nM 3.3 kb | 1 |
| Gibson2 327/320 (2.05 kb) 40nM | 1 |
| Gibson 2x mix | 2.5 |
| water | 0.5 |
Gibson assembly protocol protocol NEB Gibson Assembly Master Mix
Immediately after Gibson reaction is E.coli transformation (DH5alpha chemically LiAc competent cells)E.coli chemical competent cells
Selection on LB +100ug/mL ampicillin and growth at 37°C
Single colonies verified by colony PCR
1h 0m 0s
Several colonies obtained on both agar plates. Single colonies transferred on new LB +100ug/mL ampicillin plate, growth overnight and a single colonies submitted for bacterial colony PCR
see protocol colony PCR
Oligos 41 and 307 used for primary verification.
Oligo sequences available in the plasmid eDA115 map eda115.dna
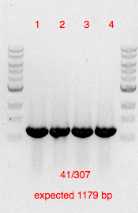
Positively verified clones re-cultured in LB + 100 ug/m
Plasmid isolation (miniprep) Qiagen miniprep 1h 0m 0s
Restriction digestion verification. Plasmid eDA115 eda115.dna digested with AsiSI (NEB)
| A | B | C |
|---|---|---|
| AsiSI | 1 | 1 |
| eDA115 326 ng/uL | 3 | |
| eDA103 103 ng/uL | 8 | |
| 10 x CutSmart | 3 | 3 |
| water | up to 30 -> |
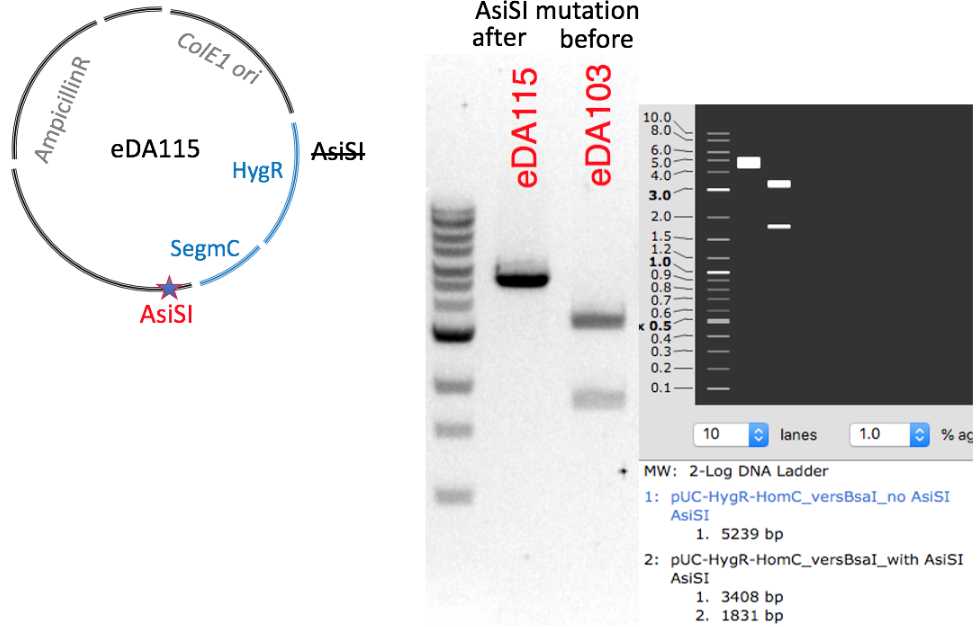
Tandem paired Hyg-LHRZ part (BsaI-free, AsiSI-free,) ready for cloning with BsaI as a PCR template for a use in the protocol
https://www.protocols.io/edit/nanochromosome-arrays-combinatory-assembly-cprkvm4w