Phenotypic Characterization and Isolation of Myeloid-Derived Suppressor Cells
Kerem Ben-Meir, Kerem Ben-Meir, Nira Twaik, Nira Twaik, Yaron Meirow, Yaron Meirow, Michal Baniyash, Michal Baniyash, Or Reuven, Or Reuven, Ivan Mikula Jr., Ivan Mikula Jr., Hadas Ashkenazi-Preiser, Hadas Ashkenazi-Preiser, Leonor Daniel, Leonor Daniel, Guy Kariv, Guy Kariv, Mahdi Kurd, Mahdi Kurd
chronic inflammation
flow cytometry
immunofluorescence imaging
immunosuppression
inflammatory mouse model
myeloid-derived suppressor cells
MDSC
patient MDSC
Abstract
Myeloid-derived suppressor cells (MDSCs) are heterogenous populations of immature myeloid cells that can be divided into two main subpopulations, polymorphonuclear (PMN) MDSCs and monocytic (M) MDSCs. These cells accumulate during chronic inflammation and induce immunosuppression evident in an array of pathologies such as cancer, inflammatory bowel disease, and infectious and autoimmune diseases. Herein, we describe methods to isolate and characterize MDSCs from various murine tissue, as well as to phenotype blood-derived MDSCs from patients. The protocols describe methods for isolation of total MDSCs and their subpopulations, for characterization, and for evaluation of their distribution within tissue, as well as for assessing their maturation stage by flow cytometry, immunofluorescence analyses, and Giemsa staining. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Single-cell suspension generation from different tissue
Alternate Protocol 1 : Single-cell suspension generation from subcutaneous melanoma tumors
Basic Protocol 2 : Characterization of MDSC phenotype
Basic Protocol 3 : Cell separation using magnetic beads: Separating pan-MDSCs or PMN-MDSC and M-MDSC subpopulations
Alternate Protocol 2 : Staining and preparing MDSCs for sorting
Support Protocol : PMN-MDSC and M-MDSC gating strategy in mouse
Basic Protocol 4 : Immunofluorescence analysis of MDSCs
Basic Protocol 5 : Handling human blood samples and characterizing human MDSCs
Alternate Protocol 3 : Flow cytometry staining of thawed human whole blood samples
INTRODUCTION
Myeloid-derived suppressor cells (MDSCs) are composed roughly of two subpopulations distinguished by their phenotype and activity. MDSC accumulation and associated immunosuppression are evident in diseases characterized by chronic inflammation. To study the biology and features of the different MDSC subpopulations during chronic inflammation, we established and optimized a murine model system, which features pathology-free chronic inflammation, is enriched with elevated levels of MDSCs, and leads to suppressed immune status. The model system is based on chronic exposure of mice to repeated immunization with heat-killed Mycobacterium tuberculosis emulsified in incomplete Freund's adjuvant, referred to as the Bacille Calmette–Guérin (BCG) model, described in detail in a Current Protocols article by Ben-Meir, Twaik, Meirow, & Baniyash (2022). To fully understand the contribution of MDSCs to the immunosuppressive environment induced during chronic inflammation, it is imperative to study MDSCs both in vivo and ex vivo. To this end, the BCG model represents an optimal model to study MDSCs, allowing for testing their accumulation, suppressive features, and functions without interference of a trigger featuring a specific pathology (Meirow et al., 2022; Sade-Feldman et al., 2013).
Herein, we present a set of protocols to identify and isolate MDSCs. The protocols were used successfully in numerous repetitive experiments and are in routine use by various laboratories, including our own.
Basic Protocol 1 consists of the methods used to obtain single-cell suspensions from different mouse tissue. These methods provide the starting point for most of the additional protocols described within this manuscript regarding isolating MDSCs and T cells and testing the function of MDSCs, among other methods. Although focusing mainly on cells obtained from tissue from the BCG mouse model, the steps can also be applied to generate single-cell suspensions from different mouse models and various tissue such as primary and secondary lymphatic organs, colons, and specific tumors, which are characterized by elevated levels of MDSCs.
Basic Protocol 2 focuses on the characterization of MDSC phenotypes using flow cytometry and microscopy, based on the single-cell suspensions obtained in Basic Protocol 1. The characterization of MDSCs uses both visualization and quantification of cells.
Basic Protocol 3 describes methods to isolate total MDSCs using the pan-MDSC marker Gr-1 and to isolate MDSC subpopulations using specific markers. Polymorphonuclear (PMN) MDSCs are isolated using magnetic bead separation in columns followed by labeling with anti-Ly6G. Monocytic (M) MDSCs are isolated using a commercial kit design for monocyte enrichment for CD11b+Ly6C+ cells. An additional method to isolate total MDSCs and their subpopulations is based on flow cytometry‒based sorting. The gating strategy for flow cytometry of MDSCs allows for distinguishing total MDSCs and MDSC subpopulations. The isolated MDSCs obtained following these protocols are suitable for evaluating their suppressive features, functions, and differentiation state (described in detail in a companion Current Protocols article by Reuven et al., 2022).
Basic Protocol 4 introduces an immunofluorescence method for the identification of MDSCs within tissue. This allows for the evaluation of the interaction between MDSCs and various other cells including immune cells in situ and for the assessment of their distribution in the examined tissue.
Basic Protocol 5 includes simple methods for characterizing MDSCs from fresh and frozen human blood samples. This protocol involves the handling of blood samples and the phenotyping of MDSCs using flow cytometry.
NOTE : All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
NOTE : All protocols involving humans and human samples must first be reviewed and approved by an Institutional Review Board (IRB) or must follow local guidelines for the use of human samples. All patients must provide informed consent.
NOTE : Perform procedures with cold solutions. Keep tissue and cells at 4°C until use unless stated otherwise. All solutions and equipment coming into contact with cells for culture must be sterile and endotoxin free, and steps should be performed using aseptic technique.
Basic Protocol 1: SINGLE-CELL SUSPENSION GENERATION FROM DIFFERENT TISSUE
Basic Protocol 1 describes the extraction of a single-cell suspension from different tissue of control and chronically inflamed mice (BCG model; described in detail in a Current Protocols article by Ben-Meir et al., 2022). This protocol represents the starting point for the various applications described herein. Extraction of cells can be performed under sterile conditions, if required when used for tissue culture.
This protocol has been used successfully with high reproducibility.
Materials
-
8- to 14-week-old C57BL/6 mouse
-
Erythrocyte lysis buffer (ELB; see recipe)
-
Dulbecco's phosphate-buffered saline (DPBS), without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
70% ethanol
-
0.4% trypan blue (e.g., Sigma-Aldrich, cat. no. T8154)
-
18-, 22-, and 25-G needle
-
1- and 5-ml syringe
-
0.5- and 1.5-ml microcentrifuge tube
-
Microcentrifuge
-
15-ml conical tube
-
Refrigerated centrifuge with swinging bucket rotor
-
6-cm culture dish (e.g., Thermo Scientific, cat. no. 150288)
-
70-μm mesh strainer (e.g., Fisher Scientific, cat. no. 22-363-548)
-
Hemocytometer (e.g., Fisher Scientific, cat. no. 02-671-6)
-
Straight surgical scissors
-
Curved surgical forceps
-
Additional reagents and equipment for mouse euthanasia (see Current Protocols article: Donovan & Brown, 2005)
Preparation of blood-derived single-cell suspension
Blood collection from submandibular vein
1.Anesthetize and restrain mouse with one hand, grabbing the scruff of the neck between the thumb and index finger with mouse bottom secured by the little finger holding the tail.
2.Insert 22-G needle caudal to the eye at the small freckle by the jawline with the other hand.
3.Collect required volume of blood into a 1.5-ml tube.
4.Put mouse back in home cage.
5.Let blood sit at room temperature for 30 min.
6.Centrifuge 10 min at 1000 × g , 4°C, and carefully transfer upper phase into a new tube.
Single-cell suspension for flow cytometry staining
7.Transfer 100 μl blood into a 15-ml conical tube filled with ELB and close cap. Flip tube twice to homogenize blood in ELB.
8.Centrifuge 8 min at 300 × g , 4°C. Discard most of the supernatant.
9.Wash pellet with 1 ml DPBS, and centrifuge as described in step 8.Discard supernatant.
10.Resuspend in 100 μl DPBS or volume appropriate for the pellet size.
Single-cell preparation from tissue
11.Euthanize mouse for excision of the desired tissue.
Splenocyte extraction
12.Wet fur using 70% ethanol to prevent fur from sticking to the spleen. Remove spleen and place in a 6-cm culture dish with cold DPBS. Remove as much fat and connective tissue as possible. In case of hair adhesion to the spleen, rinse and replace DPBS.
13.Homogenize spleen using the back of a 5-ml syringe plunger.
14.Collect cell suspension into 15-ml conical tube, and rinse culture dish with new DPBS to collect remaining cells.
15.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
16.Resuspend cell pellet with ELB, and immediately repeat step 15.
17.Resuspend cell pellet in DPBS, and strain through a 70-μm mesh strainer into a new 15-ml conical tube.
18.Count cells using a hemocytometer and trypan blue.
Bone marrow extraction
19.Remove leg and expose femur and tibia by trimming excess tissue. Carefully remove both edges of the bone near the joint by rotating the scissors back and forth. Remove as much muscle and connective tissue as possible.
20.Extract bone marrow cells from bones:
- Puncture bottom of a 0.5-ml tube with a 18-G needle. Place bone in tube with knee facing down and close cap.
Include a maximum of four bones per tube.
- Place closed 0.5-ml tube inside an open 1.5-ml tube, and add 150 μl DPBS. Centrifuge 15 s at 10,000 ×g, 4°C. Discard inner tube containing the bones.
At the bottom of the larger tube, a small cell pellet should be visible. For healthy mice, the cell pellet will be red, and the bone should be white. For inflamed mice, the cell pellet and bones should both be white.
21.Fill 1-ml syringe with 25-G needle with DPBS, and insert into the trimmed edge of the bone. Hold bone with forceps above a 1.5-ml tube, and flush out cells by pushing the plunger down. Repump syringe with the cell suspension from the tube, and pass it through the bone a few more times until all cells are depleted from the bone. Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
22.Resuspend pellet with 1 ml ELB, and centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
23.Resuspend cell pellet with 0.5 to 1.5 ml DPBS depending on the pellet size, and strain using mesh into a new 1.5-ml tube.
24.Count cells using a hemocytometer and trypan blue.
Alternate Protocol 1: SINGLE-CELL SUSPENSION GENERATION FROM SUBCUTANEOUS MELANOMA TUMORS
Alternate Protocol 1 is optimized for the preparation of single-cell suspensions from melanoma tumors in mice, without the use of digestive enzymes such as trypsin. Although these enzymes are very efficient in the disruption of cell-to-cell contacts, they may damage leukocyte cell surface molecules used as cell surface markers when characterizing cells within the tumor microenvironment using monoclonal antibodies for flow cytometry or immunofluorescence analyses. However, to prepare a single-cell suspension of B16-F10 melanoma cells grown in tissue culture, a brief digestion with trypsin B is advised.
This protocol describes a simple method for inducing B16-F10 melanoma tumors in mice by transplanting cells subcutaneously. The tumors can be excised and used to generate single-cell suspensions for additional testing.
Materials
-
B16-F10 cells (e.g., ATCC, cat. no. CRL-6475)
-
Complete Dulbecco's modified Eagle medium (DMEM; see recipe)
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
Heat-inactivated fetal calf serum (HI-FCS; e.g., Sigma-Aldrich, cat. no. F7524)
-
EDTA
-
0.25%/0.05% Trypsin-EDTA Solution B (e.g., Biological Industries, cat. no. 03-052-1A)
-
0.4% trypan blue (e.g., Sigma-Aldrich, cat. no. T8154)
-
8- to 14-week-old C57BL/6 mouse
-
Isoflurane
-
70% ethanol
-
ELB (see recipe)
-
10-cm culture dish
-
37°C incubator
-
15- and 50-ml conical tube
-
Centrifuge
-
40-, 70-, and 100-μm cell strainers (e.g., Fisher Scientific, cat. nos. 22-363-548 and 22-363-549)
-
Hemocytometer (e.g., Fisher Scientific, cat. no. 02-671-6)
-
Volatile anesthesia system
-
Electric shaver
-
Caliper
-
Culture dish
-
Surgical scissors
-
Scalpel
-
Forceps
-
25-ml serological pipette
-
2- or 5-ml syringe
-
Additional reagents and equipment for mouse euthanasia (see Current Protocols article: Donovan & Brown, 2005)
Cell preparation
1.Culture B16-F10 cells in DMEM until cell confluence reaches ∼80% in a 10-cm culture dish.
2.Discard medium and wash cells with 10 ml DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA. Discard DPBS.
3.Add 0.5 ml trypsin B to detach cells from the dish, and incubate for 3 min at 37°C.
4.Wash cells with 10 ml DMEM, and collect in a 15-ml tube.
5.Centrifuge tube 8 min at 300 × g , 4°C. Discard supernatant.
6.Resuspend pellet with 5 ml DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA.
7.Strain cells using a 40-μm cell strainer to generate a single-cell suspension.
8.Count cells using a hemocytometer and trypan blue. Adjust cell concentration to 0.25 × 106 cells/ml using sterile DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA.
9.Store cells at 4°C until injection. Strain cells with a 40-μm cell strainer before cell injection, as cells tends to aggregate.
Tumor cell injection
10.Anesthetize mouse using an isoflurane anesthesia setup.
11.Shave mouse fur at the flank using an electric shaver.
12.Disinfect skin at the injection site using 70% ethanol.
13.Inject 100 μl cell suspension (25 × 103 cells) subcutaneously into the mouse flank.
14.Follow mice for tumor development daily, and using a caliper measure tumor development and growth.
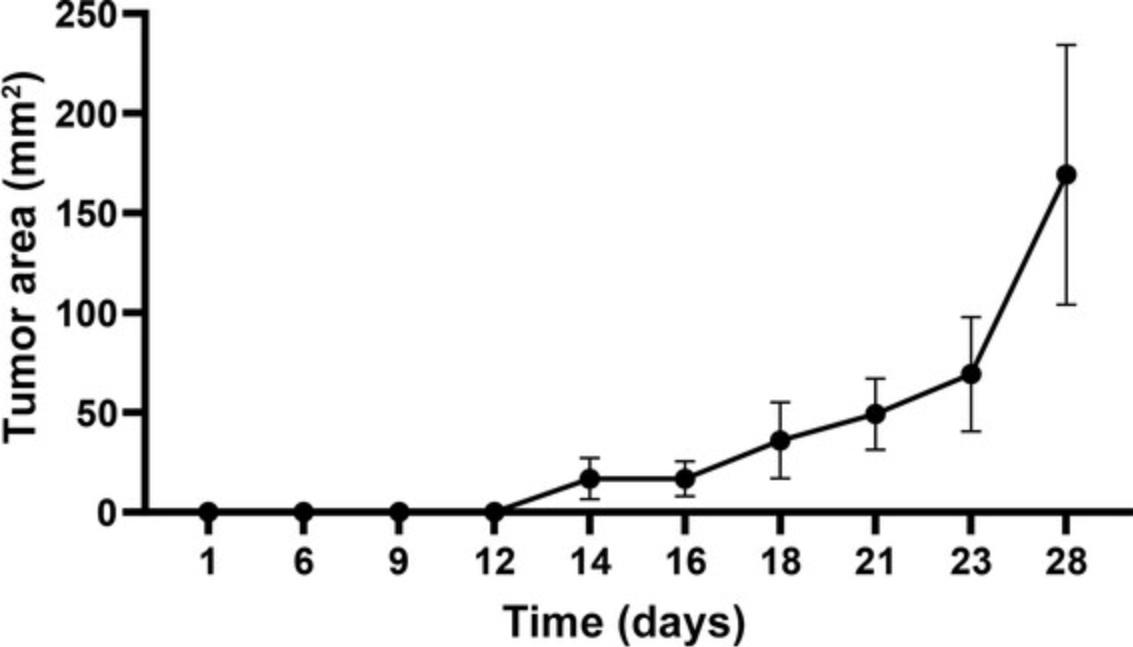
Extraction of cells from tumors
15.Euthanize mice and collect tumors. Transfer each tumor to a culture dish containing 5 ml DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA. Keep tumors intact with the surrounding skin.
16.Dissociate tumor mechanically using scissors, scalpel, and forceps.
17.Insert 100-μm cell strainer into a new 50-ml conical tube, and transfer fragmented tumor and DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA onto the cell strainer using a 25-ml pipette.
18.Using a 2- or 5-ml syringe plunger, mash tumors into the cell strainer, and wash with DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA. Repeat step until only the hard and undissociated tissue parts are left in the strainer.
19.Centrifuge 50-ml tubes 10 min at 300 × g , 4°C. Discard supernatant.
20.Resuspend tumor pellet in ELB.
21.Centrifuge 50-ml tubes 10 min at 300 × g , 4°C. Discard supernatant.
22.Resuspend tumor pellet in 20 ml DPBS supplemented with 1.5% (v/v) HI-FCS and 1 mM EDTA.
23.Insert 70-μm cell strainer into a new 50 ml conical tube, and transfer 20 ml cell suspension through the cell strainer.
Basic Protocol 2: CHARACTERIZATION OF MDSC PHENOTYPE
When conducting in vivo experiments using models of chronic inflammation, it is imperative to identify, track, and characterize MDSCs within the different tissue. The use of multicolor flow cytometry enables high-resolution analysis of the different MDSC subsets, according to specific cell surface markers. Basic Protocol 2 describes a simple and efficient preparation of samples for flow cytometry.
Table 1 details panels designed to characterize mouse and human MDSCs based on the expression of cell surface markers using flow cytometry. This protocol is suited for staining single-cell suspensions prepared from mouse tissue such as bone marrow, spleen, blood, lymph nodes, or tumors, as well as biopsies or other tissue types. Detection of pan-MDSCs or of PMN-MDSCs and M-MDSCs is performed using antibody panels 1 and 2, respectively, in Table 1.It is possible to add more markers to the panel for further subdivision of MDSC populations and for measurement of population-specific protein expression. All antibodies not mentioned in the Materials list must be optimized for the specific flow cytometry application.
| Panel designation | Required antibodiesa | PMN-MDSC phenotype | M-MDSC phenotype |
|---|---|---|---|
| Panel 1: Mouse pan-MDSC |
Gr-1 FITC CD11b APC CD16/32 unlabeled |
Not distinguishable Roughly Gr-1hi |
Not distinguishable Roughly Gr-1lo |
| Panel 2: Mouse PMN-/M-MDSC distinction |
Ly6C Alexa Fluor 700 Ly6G PE CD11b Pacific Blue Biotinylated lineage mix: Thy-1.2 biotin B220 biotin Ter119 biotin CD16/32 unlabeled Alexa Fluor 647‒conjugated streptavidin |
Lineage– Ly6Clo Ly6G+ CD11b+ | Lineage– Ly6Chi Ly6G– CD11b+ |
| Panel 3: Human PMN-/M-MDSC distinction |
CD14 PE/Cy7 CD15 FITC HLA-DR Pacific Blue CD33 PE CD11b APC Human TruStain FcX |
HLA-DR‒/lo CD33hi CD11b+ CD14– CD15+ | HLA-DR–/lo CD33hi CD11b+ CD14+ CD15– |
-
HLA-DR, human leukocyte antigen DR isotype; M, monocytic; MDSC, myeloid-derived suppressor cell; PMN, polymorphonuclear.
- a
Fluorophore panels can be modified as desired, according to the experiment and the equipment available.
Materials
-
Fluorescence-activated cell sorting (FACS) buffer (see recipe)
-
Antibodies of interest:
- Anti-mouse Ly6G PE, clone 1A8 (e.g., BioLegend, cat. no. 127608)
- Anti-mouse Ly6C Alexa Fluor 700, clone HK1.4 (e.g., BioLegend, cat. no. 128024)
- Anti-mouse Thy-1.2 biotin, clone 30-H12 (e.g., BioLegend, cat. no. 105304)
- Anti-mouse B220 biotin, clone RA3-6B2 (e.g., BioLegend, cat. no. 103204)
- Anti-mouse Ter119 biotin, clone TER-119 (e.g., BioLegend, cat. no. 116204)
- Anti-mouse CD11b Pacific Blue, clone M1/70 (e.g., BioLegend, cat. no. 101224)
- Anti-mouse Ly6C/G (Gr-1) FITC, clone RB6-8C5 (e.g., BioLegend, cat. no. 108406)
- Purified anti-mouse CD16/32 antibody, Fc blocker, clone 93 (e.g., BioLegend, cat. no. 101302)
- Optional: Anti-mouse CD45.2 PE/Cy7, clone 104 (e.g., BioLegend, cat. no. 109830)
- Anti-Ly6C PE, clone HK1.4 (e.g., BioLegend, cat. no. 128007)
- Anti-Ly6G Alexa Fluor 647, clone 1A8 (e.g., BioLegend, cat. no. 127609)
- Anti-CD11b FITC, clone M1/70 (e.g., BioLegend, cat. no. 101205)
-
Single-cell suspension of tissue type of interest (see Basic Protocol 1 or Alternate Protocol 1)
-
Streptavidin Alexa Fluor 647 (e.g., Jackson ImmunoResearch Laboratories, cat. no. 016-600-084)
-
0.4% trypan blue (e.g., Sigma-Aldrich, cat. no. T8154)
-
1% (w/v) paraformaldehyde in DPBS (e.g., Thermo Fisher Scientific, cat. no. AAJ61899AK)
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
Permeabilization buffer (see recipe)
-
DAPI (e.g., Invitrogen, cat. no. D1306)
-
Methanol (e.g., Fisher Scientific, cat. no. M/4062/17)
-
Giemsa stain, modified (e.g., Sigma-Aldrich, cat. no. GS500)
-
Xylene-based mounting medium (e.g., Millipore Sigma, cat. no. 1079610500)
-
96-well U-bottom plate, tissue culture surface (e.g., Thermo Scientific, cat. no. 163320)
-
Refrigerated centrifuge equipped with swinging bucket rotor compatible with 96-well plates
-
60-µm nylon mesh (e.g., Millipore Sigma, cat. no. NY6000010)
-
Multicolor flow cytometer with plate loader module (e.g., Beckman Coulter CytoFLEX V-B-R or equivalent)
-
Hemocytometer (e.g., Fisher Scientific, cat. no. 02-671-6)
-
1.5-ml microcentrifuge tubes
-
Refrigerated microcentrifuge with fixed-bucket rotor
-
40-μm cell strainer
-
Flow cytometer equipped with 405-, 488-, 642-nm lasers (e.g., Amnis ImageStreamX Mk II)
-
Superfrost plus slides (e.g., Thermo Scientific, cat. no. J1800AMNZ)
-
Cytocentrifuge: SLEE Cellspin I
-
Coplin jar
-
Coverslips
-
Microscope
Flow cytometry for MDSC phenotyping
1.Prepare suggested antibody mix in FACS buffer:
- 0.5 μg/ml anti-Ly6G PE
- 1 μg/ml anti-Ly6C Alexa Fluor 700
- 0.5 μg/ml anti-Thy-1.2 biotin
- 0.5 μg/ml anti-B220 biotin
- 0.5 μg/ml anti-Ter119 biotin
- 0.5 μg/ml anti-CD11b Pacific Blue
- 0.5 μg/ml anti-Gr-1 FITC
- 1 μg/ml unlabeled anti-CD16/32.
Calculate the volume of antibody mix to be prepared by multiplying the number of sample wells to be stained by 50 µl antibody mix for each well. It is suggested to prepare enough for three extra samples.
Choose the suitable panel from Table 1: antibody panel 1 for pan-MDSC staining or antibody panel 2 for PMN- or M-MDSC staining. For both panels, always add the unlabeled anti-CD16/32 for blocking Fc receptors.
It is recommended to add anti-CD45.2 to the mix when staining cell suspensions of nonhematopoietic or lymphoid tissue to discriminate immune cells from other cell types.
Prepare samples stained with each antibody separately to compensate for overlapping channels to remove channel signal leakage before staining and analyzing samples stained for the entire antibody panel.
Prepare an unstained sample to determine any autofluorescence of the samples.
2.Plate 0.2–2.0 × 106 total cells of the desired tissue (blood, spleen, bone marrow, or tumor) per well in a 96-well U-bottom plate.
3.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant. To discard supernatant, lay a folded towel paper on a flat surface. In one motion, turn plate upside-down strongly into a waste container, and dry leftovers by gently pressing the inverted plate on the towel paper. Turn plate upright only after dried.
4.Use a multichannel pipettor to resuspend all wells in 50 µl antibody mix for staining, and incubate for 30 min at 4°C in the dark.
5.Add 200 µl FACS buffer to each well and mix gently.
6.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
7.Prepare streptavidin staining mix at a final concentration of 1 µg/ml streptavidin Alexa Fluor 647 in FACS buffer.
8.Perform secondary staining when using biotinylated antibodies by resuspending all wells with 50 µl streptavidin staining mix.
9.Incubate for 15 min at 4°C in the dark.
10.Add 200 µl FACS buffer to each well and mix gently.
11.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
12.Resuspend all wells in 200 to 250 µl FACS buffer, and filter into new empty wells using a multichannel pipettor and 60-µm nylon mesh to avoid clogging the flow cytometer nasal.
13.Analyze using a flow cytometer.
ImageStream analysis of MDSC populations
Image stream analysis using ImageStream software is a technique that couples flow cytometry and fluorescence microscopy and enables distinguishing between MDSC subpopulations based on the expression of cell surface markers, morphology, nucleus structure, and cell size. Analysis of ImageStream data using the IDEAS software allows for using different algorithms to analyze the change in cell morphology, cell cycle, colocalization of molecules, and engulfing of extracellular particles.
Steps 14 through 27 provide a simple method to use a prepared cell population (e.g., splenocytes or bone marrow as described in Basic Protocol 1) for ImageStream analysis. It allows the analysis and visualization of cells in order to determine frequency of PMN- and M-MDSCs, as well as intermediate subpopulations. This method was used successfully to characterize the response of MDSCs to the presence of bacteria, as measured by altered cell morphology upon interaction and engulfment of the bacteria.
14.Prepare splenocytes, bone marrow, or blood single-cell suspension as described in Basic Protocol 1.
15.Count cells using a hemocytometer and trypan blue. Adjust concentration to 5 × 106 cells/ml.
16.Divide 5 × 106 cells in 1.5-ml tubes for each treatment planned.
17.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
18.Fix cells by suspending in 300 μl of 1% (w/v) paraformaldehyde in DPBS and incubating at 4°C for 15 min protected from light.
19.Wash cells from fixation by adding 500 μl cold DPBS to each sample and repeating centrifugation as described in step 17.
20.Stain cells using antibodies for cell surface markers. For single-stained samples, use only one fluorophore per sample.
21.Wash cells from unbound antibody by adding 500 μl cold DPBS to each sample and repeating centrifugation as described in step 17.
22.Permeabilize cells using 300 μl permeabilization buffer for 20 min in the dark at room temperature.
23.To visualize the cell nucleus, stain cells by suspending in 300 μl of 1 μg/ml DAPI in DPBS for 10 min at room temperature, protected from light.
24.Strain cells through 40-μm mesh to ensure cells are in single-cell suspension.
25.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
26.Suspend cells at a concentration of 5 × 106 cells/ml in DPBS.
27.Read samples using the ImageStream machine. First read a sample containing all fluorophores used in your experiment. Adjust laser intensities, and make sure to read all samples with the same values so as not to bias results.
Giemsa staining to assess cell maturity
Under normal conditions, MDSCs represent the progenitor cells for dendritic cells, macrophages, and neutrophils, whereas under chronic inflammation immature MDSCs leave the bone marrow and can be found in the periphery. The increased demand for myeloid cells during an inflammatory response leads to enhanced myelopoiesis that is indicative of inflammation severity. Giemsa staining solution contains the pigments methylene blue, azure, and eosin, which allows for the identification of different hematopoietic cells located in different tissue. This protocol provides a method to assess the maturity of the cells as an indication of an ongoing inflammation.
28.Prepare single-cell suspension of splenocytes or bone marrow as described in Basic Protocol 1.
29.Count cells using a hemocytometer and trypan blue. Adjust cell concentration to 1 × 106 cells/ml.
30.Divide 0.2 × 106 cells in a 1.5-ml tube for each treatment planned.
31.Mark slides appropriately, and load into Cellspin machine. To do so, open the cellclip, and insert microscope slide with labeled side facing forward. Insert filter card and cell funnel, and fix assembly using the brackets. Then load assembly into the rotor.
32.Load samples into the appropriate cell funnel, making sure that the correct sample is loaded on the appropriately marked slide.
33.Set Cellspin machine to centrifuge 5 min at 100 × g , and start centrifugation.
34.Disassemble and remove slides carefully from rotor.
35.Incubate slides containing cells in 100% methanol for 5 min at room temperature, and let slides air dry for a few minutes.
36.Prepare Giemsa staining solution by diluting 1 vol. Giemsa solution with 20 vol. deionized distilled water (1:20 dilution; 0.02% [w/v] final). Prepare enough staining solution to use 100 μl per slide.
37.Stain slides for 20 to 40 min at room temperature using diluted Giemsa staining solution.
38.Wash slides thoroughly in a Coplin jar containing 20 ml DPBS three times.
39.Let slides air dry, and mount using xylene-based mounting solution by adding a small drop of mounting solution onto the sample and pressing a coverslip on gently. Allow slides to cure at room temperature for 30 min before inspection on a microscope.
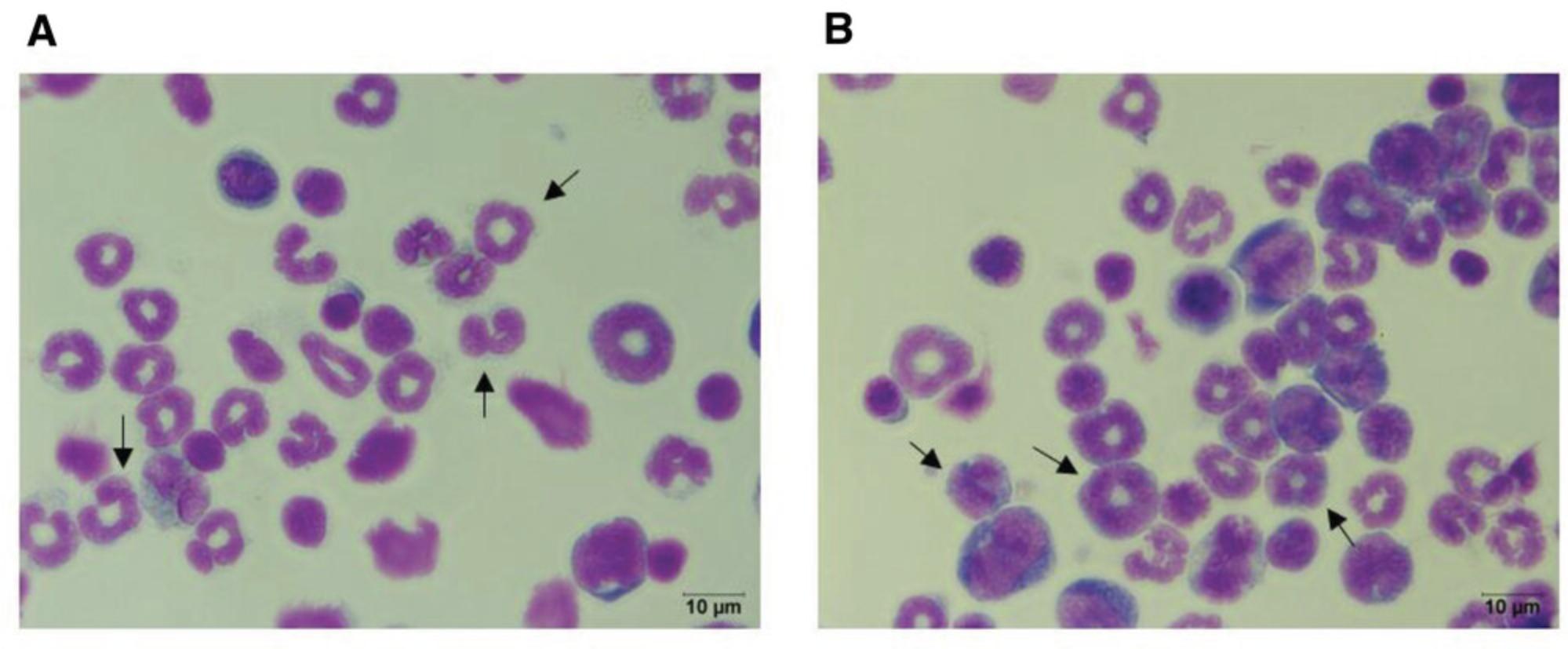
Basic Protocol 3: CELL SEPARATION USING MAGNETIC BEADS: SEPARATING PAN-MDSCs OR PMN-MDSC AND M-MDSC SUBPOPULATIONS
Basic Protocol 3 has been used in our laboratory for many years and uses commercial kits and reagents optimized for maximum efficiency and minimum monetary expense. The first time working with these protocols, we highly recommend analyzing each fraction by FACS throughout the process to validate efficient separation and purity.
Materials
-
Splenocyte or bone marrow single-cell suspension (see Basic Protocol 1)
-
Magnetic-activated cell sorting (MACS) buffer (see recipe)
-
Antibodies for positive selection:
- For Ly6G+ selection: biotin anti-mouse Ly-6G antibody, clone 1A8 (e.g., BioLegend, cat. no. 127604)
- For Gr-1+ selection: biotin anti-mouse Ly-6G/Ly-6C (Gr-1) antibody, clone RB6-8C5 (e.g., BioLegend, cat. no. 108404)
- Purified anti-mouse CD16/32 antibody, clone 93 (e.g., BioLegend, cat. no. 101301.)
-
Streptavidin microbeads (e.g., Miltenyi Biotec, cat. no. 130-048-101)
-
0.4% trypan blue (e.g., Sigma-Aldrich, cat. no. T8154)
-
EasySep buffer (see recipe; e.g., StemCell Technologies, cat. no. 20144)
-
EasySep Monocyte Isolation kit (e.g., StemCell technologies, cat. no. 19861) containing:
- Normal rat serum
- Component A
- Component B
- RapidSphere beads
-
LS magnetic separation columns (e.g., Miltenyi Biotec, cat. no. 130-042-401)
-
Microcentrifuge
-
Magnet and metal stand (e.g., Miltenyi Biotec, cat. nos. 130-042-302 and 130-042-303)
-
15-ml conical tubes
-
Hemocytometer (e.g., Fisher Scientific, cat. no. 02-671-6)
-
4-ml tubes
-
Vortex mixer
Isolation of Ly6G+ or Gr-1+ cells
Before beginning: Whether it is Ly6G+ or Gr-1+ separation, it is important to have an estimate of the desired cell percentages within your sample.
Preparation of single-cell suspension for cell separation
1.Calculate number of LS columns to be used according to your sample, considering that the LS column capacity is ∼100 × 106 marked cells.
2.Prepare splenocytes or bone marrow single-cell suspension as described in Basic Protocol 1.
3.Centrifuge 8 min at 300 × g , 4°C. Discard supernatant.
4.Prepare antibody stock: For each column, mix 200 μl MACS buffer with 4.5 μl of 0.5 mg/ml biotinylated antibody of choice and 4.5 μl of 0.5 mg/ml purified anti-CD16/32.Mix thoroughly by pipetting.
5.Resuspend cells pellet in 200 μl antibody mix per column.
6.Incubate for 30 min at 4°C, gently resuspending every 10 min.
7.Wash with up to 10 times the volume of MACS buffer, and repeat step 3.
8.Prepare streptavidin beads mixture: For each column used, mix 160 μl MACS buffer with 40 μl streptavidin spheres.
9.Resuspend cell pellet in 200 μl streptavidin bead mixture. Mix thoroughly by pipetting.
10.Incubate 45 min at 4°C, gently resuspending every 10 min.
11.Wash with up to 10 times the volume of MACS buffer, and repeat step 3.
12.Resuspend cells in 500 μl MACS buffer for each column used.
Magnetic cell separation
13.Insert column onto magnet stand, and continue protocol according to manual, as also described in steps 14 to 20.
14.Prewash column using 3 ml MACS buffer. Wait until the flow stops.
15.Load 500 μl cells onto the center of the column. Wait until the flow stops.
16.Gently add 3 ml MACS buffer onto the center of the column. Wait until the flow stops.
17.Repeat washing step twice (three washes total).
18.Elute positive cell fraction: Remove column from magnet, and insert column into a new 15-ml tube.
19.Add 5 ml MACS buffer onto the column center; using the plunger, press hard to elute cells into the 15-ml tube.
20.For negative selection of M-MDSCs from the effluent fraction, count effluent cells using a hemocytometer and trypan blue, and continue to M-MDSC separation (step 21).
M-MDSC separation
M-MDSCs can be separated directly from single-cell suspensions of homogenized tissue or from the effluent fraction from Ly6G+ magnetic column separation. As monocytes are not the major cell within the spleen or bone marrow, separating them from the effluent, which is depleted of Ly6G+, is beneficial to the purity of the final obtained cells and will consume less kit reagents. Steps 21 to 33 describe the isolation of monocytes according to the manufacture's protocol.
21.Prepare total bone marrow single-cell suspensions (as described in Basic Protocol 1) or effluent cells from Ly6G+ cell magnetic separation (as described in steps 1 to 20).
22.Count cells using a hemocytometer and trypan blue, and adjust cell concentration to 100 × 106 cells/ml in EasySep buffer.
23.Transfer 0.5 to 2 ml cells to a 4-ml tube.
24.Add 50 μl normal rat serum for each 1 ml sample.
25.Mix components A and B at a 1:1 ratio, and incubate at room temperature for 5 min.
26.Add 100 μl A+B mixture per 1 ml sample to cells. Pipette gently with 1-ml pipettor, and incubate at 2°C to 8°C for 5 min.
27.Vortex RapidSpheres beads for 30 s until solution appears homogeneous.
28.Add 100 μl RapidSpheres beads for each 1 ml sample. Pipette gently with a 1-ml pipettor, and incubate at 2°C to 8°C for 3 min.
29.Add EasySep buffer to a volume of 2.5 ml, and pipette gently with a 1-ml pipettor.
30.Transfer sample tube without cap to the magnet, and incubate for 5 min.
31.Collect cells gently using a Pasteur pipette, and transfer to a new 4-ml tube.
32.Incubate in magnet for 5 min.
33.Gently collect cells, and transfer to a 15-ml conical tube.
Alternate Protocol 2: STAINING AND PREPARING MDSCs FOR SORTING
Although similar in principle, staining and preparing samples for sorting of MDSCs have critical additional steps. These are designed to prevent cell clumping and sorter clogging and maximize the efficiency and yield of the sorting process. This protocol is suitable for sorting mouse total MDSC populations and MDSC subsets after completing the staining protocol described in Basic Protocol 2 (also see Table 1, antibody panels 1 and 2).
Materials
-
Single-cell suspension of tissue of interest (see Basic Protocol 1 or Alternate Protocol 1)
-
0.4% trypan blue (e.g., Sigma-Aldrich, cat. no. T8154)
-
Antibody mix (see Basic Protocol 2 steps 1 to 13)
-
RPMI 1640 full medium (see recipe)
-
Sorting medium (see recipe)
-
Collection medium (see recipe)
-
Hemocytometer (e.g., Fisher Scientific, cat. no. 02-671-6)
-
Centrifuge
-
15-ml polypropylene conical tubes (e.g., Miniplast, cat. no. 835-015-40-111)
-
40-µm nylon cell strainer (e.g., Corning, cat. no. 431750)
-
Multicolor cell sorter (e.g., BD ARIA III or equivalent)
Preparation of single-cell suspension for sorting
1.Prepare single-cell suspension of the desired tissue as described in Basic Protocol 1.
2.Count cells using a hemocytometer and trypan blue, and calculate total number of cells in the sample.
3.Prepare antibody mix in RPMI 1640 full medium (see Basic Protocol 2 steps 1 through 13).
4.Centrifuge sample in a 15-ml tube 10 min at 300 × g , 4°C, to pellet cells.
5.Gently discard supernatant using a pipette, and resuspend in antibody mix solution.
6.Incubate at 4°C in the dark for 30 min.
7.Fill tube to 15 ml with RPMI 1640 full medium, and centrifuge as described in step 4.
8.Discard supernatant gently, and resuspend cells in sorting medium.
Sorting and handling of sorted cells
9.Prepare 15-ml tubes with 3 ml collection medium for the designated cell population sorted from each sample.
10.Strain sample into a new 15-ml tube using a 40-µm cell strainer shortly before sorting.
11.Sort populations of interest into 15-ml collection tubes (see gating strategy in the Support Protocol).
12.Centrifuge collection tubes 20 min at 350 × g , 4°C, to pellet sorted cells.
13.Carefully aspirate supernatant using a pipette. Leave at least 1 ml supernatant above the pellet to avoid loss of cells.
14.Resuspend pellets in the 1 ml supernatant, and combine collection tubes containing cells of the same population from the same sample if more than one tube was used.
15.Top combined tubes with RPMI 1640 full medium, and centrifuge 20 min at 350 × g , 4°C, to wash cells.
16.Carefully aspirate supernatant using a pipette, and resuspend in 1 ml RPMI 1640 full medium. Count cells using a hemocytometer and trypan blue.
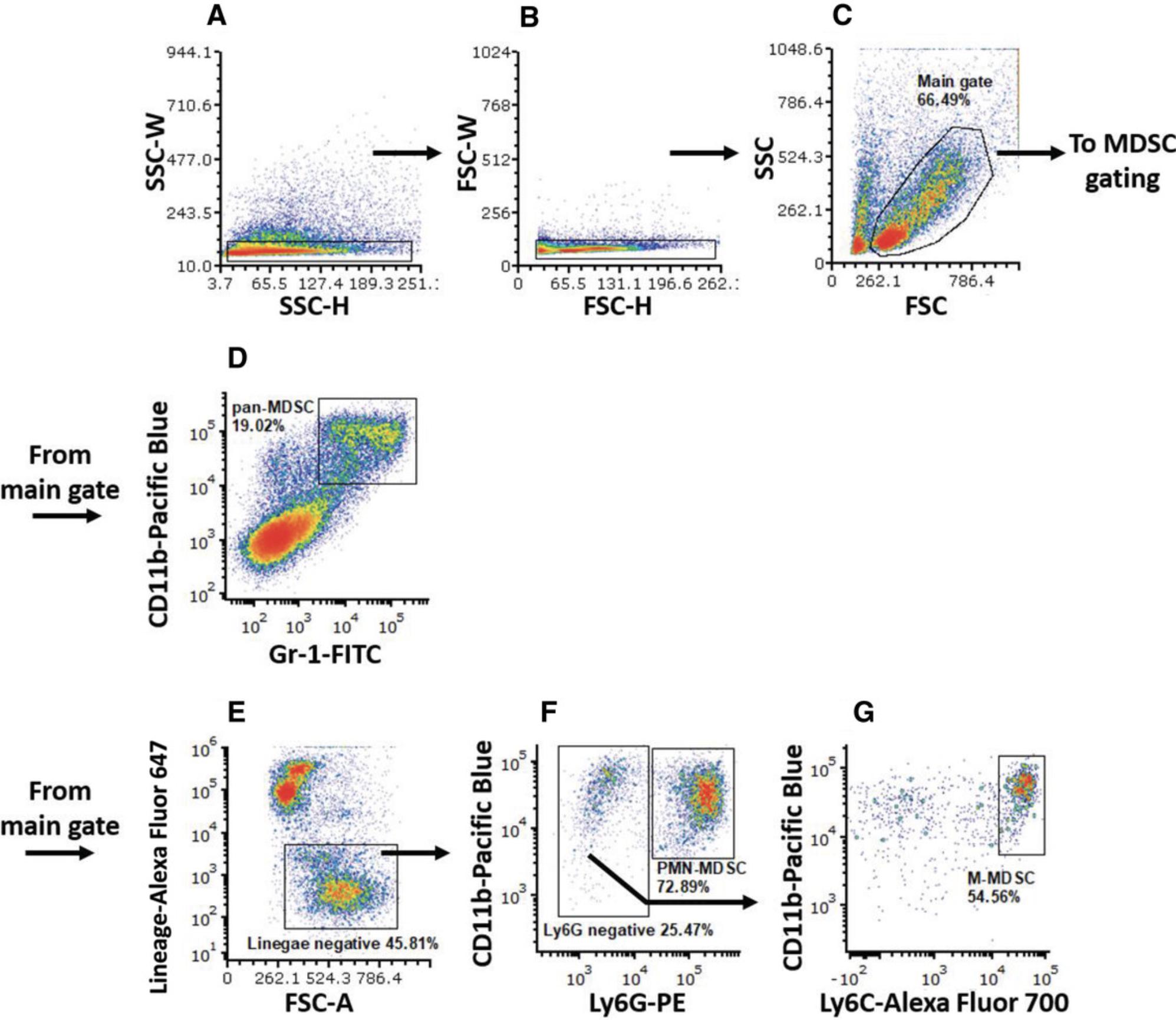
Support Protocol: PMN-MDSC and M-MDSC GATING STRATEGY IN MOUSE
Analysis of flow cytometry data can be performed in many ways. The strategy presented here is applicable to most commonly used flow cytometers and program suites. The following protocol is suitable for tracking mouse and human PMN- and M-MDSC subsets using panels 1, 2, and 3 in Table 1 and the staining procedure described in Basic Protocol 2.
Materials
- Flow cytometry analysis software (e.g., FCS Express version 6, DeNovo software, or equivalent)
Doublet discrimination
1.Plot SSC-W on the y -axis against SSC-H on the x -axis.
2.Gate main population scattered along the x -axis but low on the y -axis (SSC gate; Fig. 2A).
3.Plot FSC-W (y -axis) against FSC-H (x -axis) of the cells gated in the SSC gate.
4.Gate main population scattered along the x -axis but low on the y -axis (FSC gate; Fig. 2B).
Main gate
5.Continue with cells in the SSC gate. Plot SSC-A on the y -axis against FSC-A on the x -axis.
6.Gate live cell population (main gate; Fig. 2C).
Gating mouse pan-MDSCs
7.Continue with cells in the main gate. Plot CD11b parameter (Pacific Blue channel) on the y -axis against Gr-1 parameter (PE channel) on the x -axis, and gate Gr-1+CD11b+ (Gr-1, CD11b double positive) pan-MDSCs (Fig. 2D).
Gating mouse MDSCs subsets
8.Continue with cells in the main gate. Plot lineage parameter (Alexa Fluor 647 channel) on the y -axis against FSC on the x -axis, and gate lineage negative (lineage‒) population (Fig. 2D).
9.PMN-MDSC gating : Continue with cells in the lineage‒ gate. Plot CD11b (Pacific Blue channel) on the y -axis and Ly6G (PE channel) on the x -axis. Gate Ly6G+CD11b+ (Ly6G positive, CD11b positive) PMN-MDSCs (Fig. 2E). Use the following formula to get the actual percentage of PMN-MDSCs out of the live cells in the sample:
\begin{eqnarray*} &&{\rm{Percentage\ PMN \hbox{-} MDSC\ (of\ live\ cells)}}\\\nonumber &&\quad = \frac{{{\rm{percent\ lineag}}{{\rm{e}}}^ - }}{{{\rm{100}}}} {\rm{ \times \ percent\ Ly6}}{{\rm{G}}}^{\rm{ + }}{\rm{CD11}}{{\rm{b}}}^{\rm{ + }}. \end{eqnarray*}
10.Gate Ly6G‒ cells, and continue with them to the next plot.
11.M-MDSC gating : Plot CD11b on the y -axis and Ly6C (Alexa Fluor 700 channel) parameter on the x -axis. Gate Ly6ChiCD11b+ (Ly6C high, CD11b positive) M-MDSCs (Fig. 2F). Use the following formula to get the actual percentage of M-MDSCs out of the live cells in the sample:
\begin{eqnarray*} &&{\rm{Percentage\ M \hbox{-} MDSC\ (of\ live\ cells)}}\\\nonumber &&\quad = \frac{{{\rm{percent\ lineag}}{{\rm{e}}}^ - }}{{{\rm{100}}}}{\rm{ \times }}\frac{{{\rm{percent\ Ly6}}{{\rm{G}}}^ - }}{{{\rm{100}}}} {\rm{ \times \ percent\ Ly6}}{{\rm{C}}}^{{\rm{hi}}}{\rm{CD11}}{{\rm{b}}}^{\rm{ + }}. \end{eqnarray*}
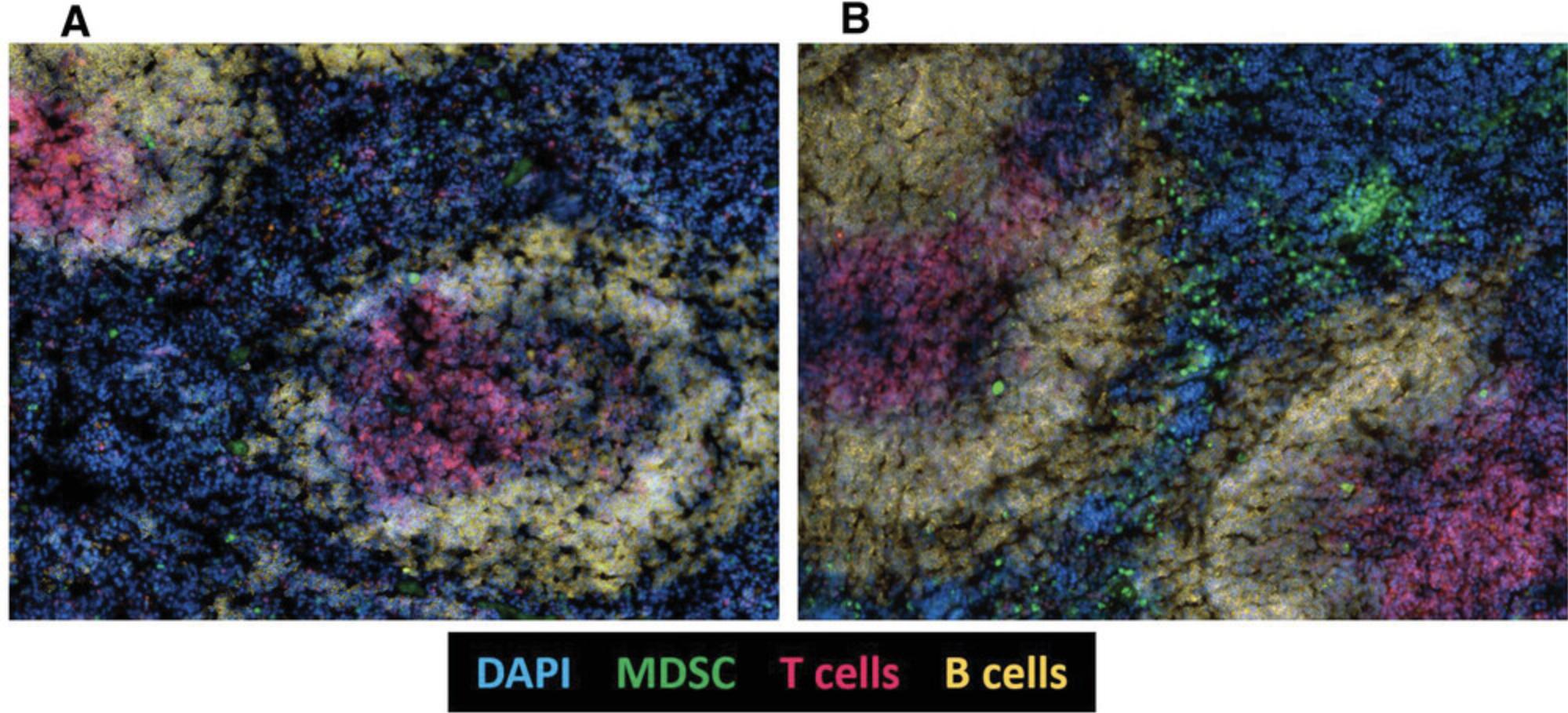
Basic Protocol 4: IMMUNOFLUORESCENCE ANALYSIS OF MDSCs
Immunofluorescence analysis of tissue sections allows for the detection of cell distribution in vivo , as well as the assessment of interaction among different cells types. Accordingly, this protocol enables the assessment of MDSC distribution in vivo in different tissue such as the colon, spleen, and lymph nodes. By combining various labeled antibodies, it is possible to image the different subpopulations of MDSCs, and the obtained results are complementary to those acquired by flow cytometry, which shows levels of the cells within the tissue.
Materials
-
Mouse tissue of interest
-
4% (w/v) formalin
-
Optimal Cutting Temperature (OCT) medium (e.g., Fisher Scientific, cat. no. 23-730-625)
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
-
4% (w/v) paraformaldehyde in PBS (e.g., Thermo Fisher Scientific, cat. no. AAJ61899AK)
-
Blocking buffer (see recipe)
-
Antibodies:
- APC anti-mouse/human CD11b antibody, clone M1/70 (e.g., BioLegend, cat. no. 101211)
- FITC anti-mouse Ly6C antibody, clone HK1.4 (e.g., BioLegend, cat. no. 128005)
- PE anti-mouse Ly-6G antibody, clone 1A8 (e.g., BioLegend, cat. no. 127607)
- Purified anti-mouse CD16/32 antibody, clone 93 (e.g., BioLegend, cat. no. 101302)
-
Tris-buffered saline containing Tween 20 (TBST; see recipe)
-
DAPI (e.g., Invitrogen, cat. no. D1306)
-
Fluorescence mounting medium (e.g., ProLong Gold Antifade Mountant)
-
Clear nail polish
-
Peel away disposable cryomold (e.g., Fisher Scientific, cat. no. 50-189-9236)
-
Cyrostat
-
Microscope slides (e.g., Fisher Scientific, cat. no. 12-550-15)
-
Hydrophobic pen (e.g., Sigma-Aldrich, cat. no. Z377821)
-
Dark humid chamber (i.e., slides storage box containing ∼5 ml DPBS)
-
Glass coverslip, no. 1 thickness (e.g., Millipore Sigma, cat. no. C8181)
-
Immunofluorescence microscope
Tissue freezing and sectioning
1.Fix spleen using 4% formalin overnight at room temperature.
2.Mark cryomold designated for tissue, and fill with OCT medium. Transfer formalin-fixed tissue to mold.
3.Freeze cryomold containing the tissue at −80°C.
4.Section blocks using cryostat to 7-μm-thick sections, and adhere sections to microscope slide.
Tissue staining (day 1)
These steps provide a description for staining using direct immunofluorescence with conjugated antibodies.
5.Thaw desired slides for staining at room temperature for 5 to 10 min.
6.Mark slide around tissue with hydrophobic pen. Allow to dry for 1 to 3 min.
7.Rehydrate slides by pipetting 150 μl DPBS onto the tissue for 5 min.
8.Discard DPBS from slide, tilting the slide to allow DPBS to drip off.
9.Add 100 μl of 4% (w/v) paraformaldehyde onto the slide without disturbing the tissue. Incubate for 10 min at room temperature in a dark, humid chamber.
10.Discard liquid by tilting the slides on a paper towel.
11.Wash with DPBS for 5 min twice.
12.Add 200 μl blocking buffer, and incubate for 60 to 90 min at room temperature in a dark, humid chamber.
13.Prepare antibody mix in blocking buffer:
- 1 μg/ml anti-CD11b APC
- 1 μg/ml anti-Ly6C FITC
- 1 μg/ml anti-Ly6G PE
- 0.5 μg/ml unlabeled anti-CD16/32.
Prepare enough to add at least 100 μl antibody mix to each slide. In some cases in which the hydrophobic frame is larger, an additional volume is required to cover the tissue.
14.Discard liquid by tilting the slides.
15.Add 100 μl antibody mix solution.
16.Incubate at 4°C overnight (16 hr) in a dark, humid chamber.
Tissue staining (day 2)
17.Wash slides with TBST three times for 10 min per wash.
18.Prepare 1 μg/ml DAPI in DPBS.
19.Discard liquid by tilting the slides.
20.Add 100 μl DAPI, and incubate for 10 min at room temperature in a dark, humid chamber.
21.Wash slides with TBST two times for 5 min per wash.
22.Mount slides using fluorescence mounting medium, and cover with glass coverslip.
23.Seal coverslip to slide with nail polish.
24.Let slide cure at room temperature for 10 min in the dark.
25.Analyze slides using an immunofluorescence microscope.
Basic Protocol 5: HANDLING HUMAN BLOOD SAMPLES AND CHARACTERIZING HUMAN MDSCs
For tracking MDSCs in human blood samples, it is imperative to use whole blood samples containing all M and PMN cells. This protocol is designed for freezing, storing, and thawing human whole blood samples for later analyses.
Materials
-
Freeze buffer (see recipe)
-
Isopropyl alcohol, analytical grade (e.g., Frutarom, cat. no. 2355531200)
-
RPMI 1640 full medium (see recipe)
-
DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A) or FACS buffer (see recipe)
-
4-ml sodium heparin–coated vacutainers (e.g., BD, cat. no. 367871)
-
15-ml tubes
-
1.8-ml screw cap cryotubes (e.g., Thermo Fisher, cat. no. 368632)
-
Freezing container (e.g., Mr. Frosty Freezing Container; Thermo Fisher, cat. no. 5100-0001)
-
Centrifuge with swinging bucket rotor compatible with 15-ml tubes
-
37°C water bath
-
Liquid nitrogen Dewar vapor phase storage tank (e.g., MVE cryosystem 4000 or equivalent)
-
Small liquid nitrogen–compatible container
-
Long forceps
Freezing whole blood
1.Draw blood using vacuum tubes (vacutainers) coated with heparin for anticoagulation.
2.Assess total amount of blood to be frozen in each sample using a 5-ml pipette, and transfer each sample into a 15-ml tube.
3.Prepare and label cryotubes for aliquoting.
4.Using a 5-ml pipette, add freeze buffer to each 15-ml tube containing blood at 1:1 ratio (blood:freeze buffer). Mix well and place on ice until aliquoting.
5.Dispense aliquots to the labeled cryotubes using a pipettor and 1000-µl filter tips.
6.Keep aliquots on ice for 10 to 15 min.
7.Prepare freezing container filled with isopropyl alcohol at room temperature according to the manufacturer's instructions.
8.Place aliquots inside freezing container at room temperature, and freeze at −80°C.
Thawing whole blood samples
9.Warm centrifuge with swinging bucket rotor to 25°C.
10.For each aliquot to be thawed, prepare and label 15-ml tube with 14 ml RPMI 1640 full medium.
11.Warm tubes with RPMI 1640 full medium to 37°C in a water bath.
12.Fill a small container with liquid nitrogen, and put frozen samples inside.
13.Select a sample from the liquid nitrogen using long forceps, and take the correspondingly labeled 15-ml tube with warm RPMI 1640 full medium.
14.Carefully uncap cryotube, and dispose cap in a biohazard-compatible container.
15.With a 1000-µl pipettor and filter tip, draw 1 ml warm culture medium from the 15-ml tube.
16.Without touching the frozen buffer in the cryotube, release warm medium to thaw the sample. Gently mix with slow up and down motions, avoiding bubbles. Transfer medium with thawed cells back to the 15-ml tube. Repeat this process several times until the entire sample is thawed and transferred to the 15-ml tube.
17.Centrifuge samples 10 min at 300 × g , 25°C, to pellet cells. Discard supernatant.
18.Resuspend thawed cells in room temperature DPBS or FACS buffer or in RPMI 1640 full medium if in vitro tissue culture assays are performed. Stain cells promptly.
Alternate Protocol 3: FLOW CYTOMETRY STAINING OF THAWED HUMAN WHOLE BLOOD SAMPLES
Immunostaining human blood samples requires additional blocking reagents to prevent nonspecific binding. Moreover, although freezing and thawing human blood samples depletes most erythrocytes, remaining erythrocytes may disrupt flow cytometry data acquisition. This protocol describes all necessary blocking, erythrocyte lysis, and staining steps required for specific staining of MDSC subsets in frozen human whole blood samples. This protocol is suited for antibody panel 1 in Table 1.
Materials
-
Antibodies:
- Anti-human leukocyte antigen DR isotype (HLA-DR) Pacific Blue, clone LN3 (e.g., BioLegend, cat. no. 327016)
- Anti-human CD33 PE, clone WM53 (e.g., BioLegend, cat. no. 303404)
- Anti-human CD14 PE/Cy7, clone 63D3 (e.g., BioLegend, cat. no. 367112)
- Anti-human CD15 FITC, clone W6D3 (e.g., BioLegend, cat. no. 323003)
- Anti-human CD11b APC, clone ICRF44 (e.g., BioLegend, cat. no. 301350)
- Human TruStain FcX, Fc receptor blocking solution (e.g., BioLegend, cat. no. 422302)
-
FACS buffer (see recipe)
-
Human whole blood sample (see Basic Protocol 5)
-
10× 1-Step Fix/Lyse Solution (e.g., Thermo Fisher Scientific, cat. no. 00-5333-54)
-
Centrifuge with swinging bucket rotor compatible with 96-well plates
-
96-well U-bottom plate, tissue culture surface (e.g., Thermo Scientific, cat. no. 163320)
-
60-µm nylon mesh (e.g., Millipore-Sigma, cat. no. NY6000010)
-
Flow cytometer and analysis software
Staining of thawed human blood cells
1.Prewarm centrifuge to 25°C.
2.Prepare antibodies mix in FACS buffer as follows, and keep in the dark at room temperature:
- 1 μg/ml anti-HLA-DR Pacific Blue
- 1 μg/ml anti-CD33 PE
- 0.5 μg/ml anti-CD14 PE/Cy7
- 0.8 μg/ml anti-CD15 FITC
- 1 μg/ml anti-CD11b APC
- 1:100 human TruStain FcX.
3.Dispense human whole blood samples in a 96-well U-bottom plate.
4.Centrifuge 5 min at 300 × g , room temperature. Discard supernatant.
5.Add 50 µl antibody mix or control medium (for unstained samples) to all samples and mix well. Incubate for 30 min in the dark at room temperature.
6.Mix 1 part 10× 1-Step Fix/Lyse Solution with 9 parts deionized water to make a 1× solution.
7.Add 200 µl of 1× 1-Step Fix/Lyse Solution to all wells and mix well. Incubate for 20 min in the dark at room temperature.
8.Centrifuge 5 min at 500 × g , room temperature. Resuspend in 200 µl FACS buffer.
9.Centrifuge 5 min at 500 × g , room temperature. Resuspend in 200 µl FACS buffer, and filter samples into an empty well of a 96-well plate using a 60-µm nylon mesh.
Gating human MDSC subsets
10.Continue with cells in the main gate. Plot HLA-DR parameter (Pacific Blue channel) on the y -axis against the FSC parameter on the x -axis, and gate HLA-DR–/lo (HLA-DR negative/low) cells (Fig. 5A).
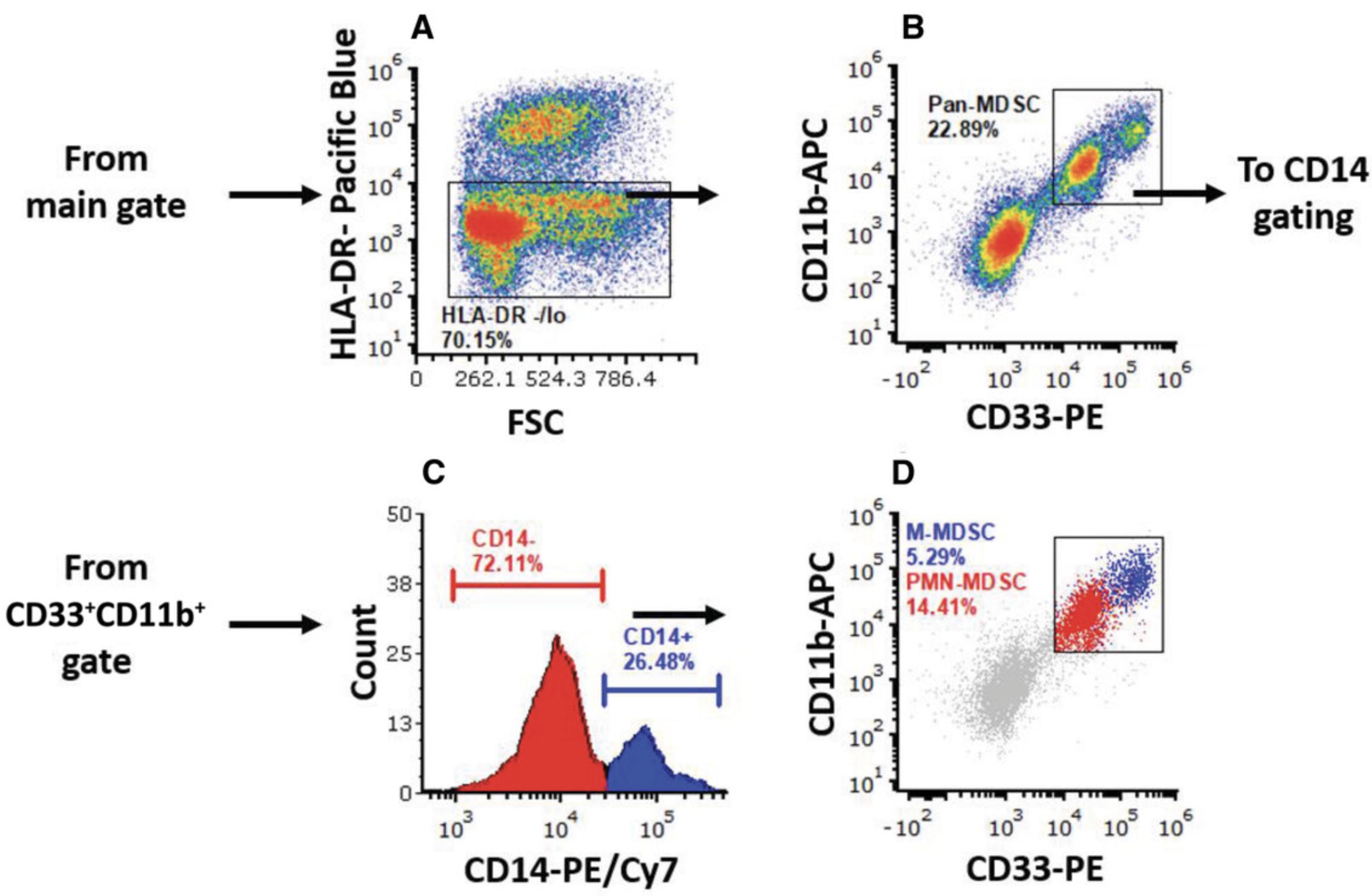
11.Continue with HLA-DR–/lo cells. Plot CD11b parameter (APC channel) on the y -axis and CD33 parameter (PE channel) on the x -axis, and gate CD33+CD11b+ cells (Fig. 5B).
12.Continue with HLA-DR–/loCD33+CD11b+ cells. Plot CD14 parameter (PE/Cy7 channel) using a histogram. Place markers over the CD14– population (PMN-MDSCs) and over the CD14+ positive population (M-MDSCs; Fig. 5C).
REAGENTS AND SOLUTIONS
Blocking buffer
- 3335 μl DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
- 1500 μl of 10% (w/v) bovine serum albumin (BSA), fraction V, ≥98% (3% [w/v] final; e.g., MP Biomedicals, cat. no. 02160069-CF) in DPBS
- 150 μl heat-inactivated normal goat serum (3% [v/v] final; e.g., Abcam, cat. no. ab7481)
- 50 μl of 10% (v/v) Triton X-100 (0.1% [v/v] final; e.g., Sigma-Aldrich, cat. no. X100) in DPBS
- Prepare fresh before use
Collection medium
- RPMI 1640 or Minimal Essential Medium (MEM-α)
- 12% to 15% (v/v) HI-FCS (e.g., Sigma-Aldrich, cat. no. F7524)
- 150 U/ml penicillin, 0.15 mg/ml streptomycin (e.g., Biological Industries, cat. no. 03-031-1B)
- Store at 4°C for up to 1 month
DMEM, complete
- 500 ml DMEM medium, high glucose, no glutamine (e.g., Biological Industries, cat. no. 01-055-1A)
- 40 ml HI-FCS (8% [v/v] final; e.g., Sigma-Aldrich, cat. no. F7524) filtered through 0.45-µm mesh
- 5 ml of 100 mM L-glutamine (2 mM final; e.g., Biological Industries, cat. no. 03-020-1B)
- 5 ml of 10,000 U/ml penicillin, 10 mg/ml streptomycin (e.g., Biological Industries, cat. no. 03-031-1B)
- Store at 4°C for up to 1 month
EasySep buffer
- 49 ml DPBS
- 1 ml HI-FCS (2% [v/v] final; e.g., Sigma-Aldrich, cat. no. F7524)
- 100 μl of 0.5 M EDTA, pH 8 (1 mM final; e.g., Biological Industries, cat. no. 01-862-1B)
- Strain through a 0.45-μm filter into a 50-ml tube
- Store at 4°C for up to 1 month
ELB
- 4.011 g ammonium chloride (150 mM final; e.g., Sigma-Aldrich, cat. no. 213330)
- 500.6 mg potassium bicarbonate (10 mM final; e.g., Sigma-Aldrich, cat. no. 237205)
- 80 μl of 0.5 M EDTA, pH 8 (80 μm final; e.g., Biological Industries, cat. no. 01-862-1B)
- 500 ml deionized distilled water
- Adjust pH to 7.2 using HCl
- Store at room temperature for up to 1 year
FACS buffer
- 50 ml of 10× DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-5A)
- 5 ml of 0.1 M sodium azide (e.g., Sigma-Aldrich, cat. no. 199931)
- 7.5 ml HI-FCS (e.g., Sigma-Aldrich, cat. no. F7524) filtered through 0.45-µm mesh
- 437.5 ml double distilled water
- Store at 4°C for up to 1 month
Freeze buffer
- 2 ml DMSO (20% [v/v] final; e.g., Sigma-Aldrich, cat. no. D4540)
- 8 ml HI-FCS (80% [v/v] final; e.g., Sigma-Aldrich, cat. no. F7524)
- Filter through 0.45-µm filter
- Prepare fresh and store at 4°C until use
MACS buffer
- 100 ml DPBS, without Ca2+ or Mg2+ (e.g., Biological Industries, cat. no. 02-023-1A)
- 0.5 g BSA (0.5% [w/v] final; e.g., MP Biomedicals, cat. no. 02160069-CF)
- 400 μl of 0.5 M EDTA, pH 8 (2 mM final; e.g., Biological Industries, cat. no. 01-862-1B)
- Store at 4°C up to a month
Different BSA products may contain some concentration of serum-derived biotin. We therefore recommend using BSA from MP Biomedicals (cat. no. 02160069-CF) or otherwise to optimize the use of other BSA products.
Permeabilization solution
- 45 ml FACS buffer
- 5 ml of 1% saponin Quillaja sp. (0.1% [w/v] final; e.g., Sigma-Aldrich, cat. no. S4521) in DPBS
- Prepare fresh before use
RPMI 1640 full medium
- 500 ml RPMI 1640 medium (e.g., Biological Industries, cat. no. 01-100-1A)
- 40 ml HI-FCS (8% [v/v] final; e.g., Sigma-Aldrich, cat. no. F7524) filtered through 0.45-µm mesh
- 5 ml of 200 mM L-glutamine (2 mM final; e.g., Biological Industries, cat. no. 03-020-1B)
- 5 ml of 10,000 U/ml penicillin, 10 mg/ml streptomycin (e.g., Biological Industries, cat. no. 03-031-1B)
- 0.5 ml of 50 mM 2-mercaptoethanol (e.g., Thermo Fisher, cat. no. 31350010)
- 5 ml of 1 M HEPES (10 mM final; e.g., Biological Industries, cat. no. 03-025-1B)
- Store at 4°C for up to 1 month
Sorting medium
- RPMI 1640 full medium (see recipe)
- 5 mM EDTA (e.g., Biological Industries, cat. no. 01-862-1B)
- Optional: 100 µg/ml DNAse I
- Prepare fresh before use
TBST
- 500 ml DPBS
- 150 μl of 10% Tween 20 (0.03% [v/v] final; e.g., Sigma-Aldrich, cat.no. P1379)
- Store at room temperature for up to 3 months
Mix the solution gently by rotating the bottle, being careful not to lather the solution.
COMMENTARY
Background Information
MDSCs represent a population of heterogeneous immature cells related to the myeloid lineage. These cells possess a potent immunosuppressive activity affecting the features and function of various immune cells such as natural killer, B, dendritic, and T cells. Under normal conditions, immature myeloid cells reside within the bone marrow and give rise to differentiated cells including macrophages, neutrophils, and dendritic cells that are found in the periphery and that support immune responses. In contrast, under pathologic conditions characterized by chronic inflammation, these cells undergo differentiation arrest, expansion in the bone marrow, and polarization toward immunosuppressive cells found in the periphery and site of inflammation (Ben-Meir, Twaik, & Baniyash, 2018). Major advances in recent years emphasize the involvement of MDSCs in inflammation and cancer, making MDSCs one of the main obstacles affecting the success of anticancer therapies. (Sade-Feldman et al., 2016). Thus, identifying novel markers for diagnosis, prognosis, and targeting is imperative for improving anticancer therapies. MDSCs have been reported in various human cancer types and in mouse models depicting a specific pattern regarding: (1) the increase in total amount during disease progression; (2) the diversity and plasticity of MDSC subpopulations; (3) their increased suppressive features and functions; and (4) their distinct homing into specific tissue (Ashkenazi-Preiser, Mikula, & Baniyash, 2021; Hou, Hou, Huang, Lei, & Chen, 2020).
In general, the protocols presented herein can be divided into methods for the generation of a single-cell suspension from different tissue, isolation of MDSC subpopulations (PMN-MDSCs and M-MDSCs), and phenotypic characterization using flow cytometry and imaging. Moreover, the protocols allow for studying the biology of MDSCs in relation to their distribution in vivo in different tissue.
Critical Parameters and Troubleshooting
In order to study MDSCs in a tumor-bearing mouse model, we chose to present the melanoma model. In our laboratory, we use the B16-F10 melanoma cell line, which forms very aggressive and highly proliferative tumors when transplanted subcutaneously (in immune-competent C57BL/6 mice). These tumors tend to overgrow and become necrotic in a short period of time, causing acute inflammation. In order to generate tumors that lead to chronic inflammation associated with angiogenesis in the tumor with reduced occurrence of necrosis, we recommend optimizing the number of cells to be injected to the lowest number that will lead to slowly growing tumors. We implant 25 × 103 B16-F10 cells, and tumors are usually evident in 70% of mice within 10 days, reaching an endpoint at day 25 to 30.
When planning sorting experiments, it should be noted that the nozzle size affects the sorting rate; when using smaller size nozzle, sorting is slower. Also, the nozzle size has a great effect on cell sorting, as a 70-µm nozzle is suitable for sorting mouse bone marrow, spleen, or blood cells, and an 85-µm nozzle is suitable for human blood cells. For cells extracted from murine or human tumors, colon, skin, or other tissue that need to be enzymatically digested and that contain large and "sticky" cells, use a 100- or 120-µm nozzle to avoid clogs. To prevent loss of cells and to aid recovery of cells post sorting, in cases where a low number of cells are being sorted, it is recommended to precoat the collection tube with a protein. This can be done by filling the collection tubes with 5% BSA or 20% FCS diluted in PBS and incubating overnight at 4°C. Coated collection tubes should be emptied and filled with collection buffer before sorting.
For obtaining the best results using the immunofluorescence methods, several parameters have to be optimized beforehand regarding the origin of the stained tissue, the tissue type, and the different antibodies to be used. Some tissue, such as of the colon, tend to have greater autofluorescence than other tissue, which should be taken into consideration. Blocking is important to avoid nonspecific antibody binding that could be achieved using serum, proteins (e.g., BSA), or a combination of the two. Usually, when using serum for blocking, it is common to add serum derived from the host of the secondary antibody used within the experiment. Antibody concentration should be properly tittered, as low antibody concentrations will result in a lack of staining signal, whereas high concentrations will lead to nonspecific binding, high background, and antibody precipitation. Furthermore, when using biotin-streptavidin binding, blocking of endogenous biotins will improve the signal-to-noise ratio for some tissue.
Time Considerations
Before conducting an experiment, it should be taken into consideration that various protocols may require different time investments to achieve the results. Complex experiments that involve the isolation of various cells types from different organs might take more time than simple experiments. In order to isolate MDSCs from BCG mice (described in detail in a Current Protocols article by Ben-Meir et al., 2022), steps to generate the model should first be conducted. The model lasts 3 weeks before endpoint is reached, and thus experiments using such mice should be carefully planned. In general, the time required to complete the protocols changes in relation to the number of mice used. For example, generation of single-cell suspensions from different tissue can require up to 2 hr depending on the tissue of interest if using up to six mice per group. More time will be required if more mice are used. Preparation for FACS analysis and cell sorting is described within the protocols, but the reading time of samples for flow cytometry of cell sorting could require hours to reach the desired cell number. Whether using software to analyze flow cytometry data or a microscope to analyze staining of tissues, the time required depends on the results to be obtained.
Acknowledgments
The authors gratefully acknowledge the support of the Society of Research Associates of the Lautenberg Center and the Harold B. Abramson Chair in Immunology. They also thank the grant support from the Israel Science Foundation, the Israeli Ministry of Health, the Israel Cancer Research Fund, the Israel Ministry of Science and Technology, the Gross Foundation, the Bruce and Baila Waldholtz funds, and the Joseph and Matilda Melnick Funds.
Author Contributions
Or Reuven : data curation, formal analysis, methodology, validation, writing—original draft, review, and editing; Ivan Mikula : formal analysis, methodology, validation, writing—original draft; Hadas Ashkenazi-Preiser : formal analysis, methodology, validation, writing—original draft; Nira Twaik : formal analysis, methodology, validation, writing—original draft; Kerem Ben-Meir : formal analysis, methodology, validation, writing—original draft; Yaron Meirow : formal analysis, methodology, validation, writing—original draft; Leonor Daniel : validation, writing—review and editing; Guy Kariv : validation; Mahdi Kurd : validation; Michal Baniyash : conceptualization, funding acquisition, project administration, supervision, validation, writing—review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data, tools, and materials that support the protocols of this article (or their source) are available from the corresponding author upon reasonable request.
Literature Cited
- Ashkenazi-Preiser, H., Mikula, I., & Baniyash, M. (2021). The diverse roles of myeloid derived suppressor cells in mucosal immunity. Cellular Immunology , 365, 104361. doi: 10.1016/j.cellimm.2021.104361
- Ben-Meir, K., Twaik, N., & Baniyash, M. (2018). Plasticity and biological diversity of myeloid derived suppressor cells. Current Opinion in Immunology , 51, 154–161. doi: 10.1016/j.coi.2018.03.015
- Ben-Meir, K., Twaik, N., Meirow, Y., & Baniyash, M. (2022). An in vivo mouse model for chronic inflammation–induced immune suppression: A “factory” for myeloid-derived suppressor cells (MDSCs). Current Protocols , 2, e558. doi: 10.1002/cpz1.558
- Donovan, J., & Brown, P. (2005). Euthanasia. Current Protocols in Neuroscience , 33, A.4H.1–A.4H.4. doi: 10.1002/0471142301.nsa04hs33
- Hou, A., Hou, K., Huang, Q., Lei, Y., & Chen, W. (2020). Targeting myeloid-derived suppressor cell, a promising strategy to overcome resistance to immune checkpoint inhibitors. Frontiers in Immunology , 11, 783. doi: 10.3389/fimmu.2020.00783
- Meirow, Y., Jovanovic, M., Zur, Y., Habib, J., Colombo, D. F., Twaik, N., … Baniyash, M. (2022). Specific inflammatory osteoclast precursors induced during chronic inflammation give rise to highly active osteoclasts associated with inflammatory bone loss. Bone Research , 10, 36. doi: 10.1038/s41413-022-00206-z
- Reuven, O., Mikula, I., Jr., Ashkenazi-Preiser, H., Twaik, N., Ben-Meir, K., Meirow, Y., … Baniyash, M. (2022). Functional assays evaluating immunosuppression mediated by myeloid-derived suppressor cells. Current Protocols , 2, e557. doi: 10.1002/cpz1/557
- Sade-Feldman, M., Kanterman, J., Ish-Shalom, E., Elnekave, M., Horwitz, E., & Baniyash, M. (2013). Tumor necrosis factor-α blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity , 38, 541–554. doi: 10.1016/j.immuni.2013.02.007
- Sade-Feldman, M., Kanterman, J., Klieger, Y., Ish-Shalom, E., Olga, M., Saragovi, A., … Baniyash, M. (2016). Clinical significance of circulating CD33+CD11b+HLA-DR− myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clinical Cancer Research , 22, 5661–5672. doi: 10.1158/1078-0432.CCR-15-3104
Citing Literature
Number of times cited according to CrossRef: 2
- Kerem Ben‐Meir, Nira Twaik, Yaron Meirow, Michal Baniyash, An In Vivo Mouse Model for Chronic Inflammation–Induced Immune Suppression: A “Factory” for Myeloid‐Derived Suppressor Cells (MDSCs), Current Protocols, 10.1002/cpz1.558, 2 , 10, (2022).
- Or Reuven, Ivan Mikula, Hadas Ashkenazi‐Preiser, Nira Twaik, Kerem Ben‐Meir, Yaron Meirow, Leonor Daniel, Guy Kariv, Mahdi Kurd, Michal Baniyash, Functional Assays Evaluating Immunosuppression Mediated by Myeloid‐Derived Suppressor Cells, Current Protocols, 10.1002/cpz1.557, 2 , 10, (2022).

