Evaluating the Clinical and Immune Responses to Spotted Fever Rickettsioses in the Guinea Pig-Tick-Rickettsia System
John V. Stokes, John V. Stokes, Michael L. Levin, Michael L. Levin, Claire E. Cross, Claire E. Cross, Anne-Marie L. Ross, Anne-Marie L. Ross, Alyssa N. Snellgrove, Alyssa N. Snellgrove, Bridget V. Willeford, Bridget V. Willeford, Navatha Alugubelly, Navatha Alugubelly, Andrea S. Varela-Stokes, Andrea S. Varela-Stokes
Abstract
The guinea pig was the original animal model developed for investigating spotted fever rickettsiosis (SFR). This model system has persisted on account of the guinea pig's conduciveness to tick transmission of SFR agents and ability to recapitulate SFR in humans through clinical signs that include fever, unthriftiness, and in some cases the development of an eschar. The guinea pig is the smallest animal model for SFR that allows the collection of multiple blood and skin samples antemortem for longitudinal studies. This unit provides the basic protocols necessary to establish, maintain, and utilize a guinea pig-tick-Rickettsia model for monitoring the course of infection and immune response to an infection by spotted fever group Rickettsia (SFGR) that can be studied at biosafety level 2 (BSL-2) and arthropod containment level 2 (ACL-2); adaptations must be made for BSL-3 agents. The protocols cover methods for tick feeding and colony development, laboratory infection of ticks, tick transmission of Rickettsia to guinea pigs, and monitoring of the course of infection through clinical signs, rickettsial burden, and immune response. It should be feasible to adapt these methods to study other tick-borne pathogens. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Tick transmission of SFGR to guinea pigs
Support Protocol 1 : Laboratory infection of ticks by injection
Alternate Protocol 1 : Needle inoculation of SFGR to guinea pigs
Basic Protocol 2 : Monitoring the course of guinea pig rickettsial infection: clinical signs
Basic Protocol 3 : Monitoring the course of guinea pig rickettsial infection: collection of biological specimens
Support Protocol 2 : Guinea pig anesthesia
Basic Protocol 4 : Monitoring rickettsial burden in guinea pigs by multiplex qPCR
Basic Protocol 5 : Monitoring guinea pig immune response to infection: blood leukocytes by flow cytometry
Basic Protocol 6 : Monitoring immune response to guinea pig rickettsial infection: leukocyte infiltration of skin at the tick bite site by flow cytometry
Basic Protocol 7 : Monitoring the immune response to guinea pig rickettsial infection: antibody titer by ELISA
Support Protocol 4 : Coating ELISA Plates
Alternate Protocol 2 : Monitoring immune response to guinea pig rickettsial infection: antibody titer by immunofluorescence assay
INTRODUCTION
The mouse remains the most common biomedical model for studying rickettsial infection and rickettsiosis. Its modest maintenance cost and the availability of murine immunological reagents make the mouse a perennial favorite of many researchers despite the differences between the immune systems of common murine models and humans (Mestas & Hughes, 2004), which diminishes their relevance as a human disease model. In contrast, the guinea pig has an immune system that more closely approximates a human's (Broad_Institute, 2022; Padilla-Carlin, McMurray, & Hickey, 2008), making it a more relevant biomedical model for studying tick-borne and other infectious disease agents.
Here, we present detailed protocols we have developed or optimized for studying spotted fever rickettsiosis (SFR) in the guinea pig model, focusing primarily on tick-borne transmission with relevant alternate methods as appropriate. For details on establishing and maintaining colonies of naturally infected and uninfected ticks, we refer readers to Levin and Schumacher (2016). First, we describe the procedure for tick transmission of spotted fever group Rickettsia spp. (SFGR; Basic Protocol 1), and a supporting method for experimentally infecting ticks for animal studies using microinjection (Support Protocol 1). If natural transmission is not indicated for the study, we offer a method for needle inoculation of guinea pigs with SFGR (Alternate Protocol 1). Next, we cover how to passively monitor the course of infection by observing clinical signs (Basic Protocol 2) and actively monitor the course of infection by collecting biological specimens for later analysis (Basic Protocol 3), including a protocol for inhalant anesthesia of guinea pigs (Support Protocol 2). Subsequent protocols describe methods for monitoring the rickettsial burden (Basic Protocol 4) and the guinea pig's immune response to infection (Basic Protocols 5-7, Support Protocol 4 and Alternate Protocol 2). Basic Protocol 5, which introduces polychromatic flow cytometry for use in guinea pigs, includes Support Protocol 3 for harvesting and freezing guinea pig splenocytes for use as reference controls.
STRATEGIC PLANNING
Biosafety and IACUC Considerations for Working with Spotted Fever Group Rickettsia spp
Working with SFGR always entails addressing biosafety concerns. Although the Varela-Stokes laboratory and others developed or use most of the methods presented here with Rickettsia parkeri and Rickettsia amblyommatis working under BSL-2 and ABSL-2 conditions, some SFGR, e.g., Rickettsia rickettsii and Rickettsia conorii , require BSL-3 and ABSL-3 facilities, practices, and procedures. Additionally, the Centers for Disease Control and Prevention (CDC) manual Biosafety in Microbiological and Biomedical Laboratories , 6th edition (Meechan, Hatcher, & Potts, 2020) includes R. parkeri with these other SFGR, generalizing recommendations for using cultivated isolates of pathogenic Rickettsia species at BSL-3. We advise working with your Institutional Biosafety Committee (IBC) to determine whether BSL-2 practices or BSL-2 with enhanced precautions are safe and acceptable when working with species such as R. parkeri that are less virulent than R. rickettsii (Londono, Mendell, Walker, & Bouyer, 2019). Before beginning work, ensure that you have obtained all the necessary protocol and facilities approvals from your Institutional Animal Care and Use Committee (IACUC) and IBC. Once work begins, ensure that all personnel remain vigilant, consistently use the engineering controls, follow proper work practices, and wear the personal protective equipment (PPE) described in the approved protocols.
Selection of Guinea Pigs
Both outbred and inbred guinea pig strains are used in rickettsial research. The selection of the guinea pig strain is influenced by the availability of the strain, space, time, and resources for breeding, and, of course, the research question. The most common strain for rickettsial work is the outbred Dunkin Hartley (or Hartley) guinea pig—the two names are considered analogous as a biomedical model, with the Dunkin Hartley originating in The Netherlands from Hartley colonies in England; we will use the Hartley terminology in this article. Hartley is the only laboratory strain of guinea pigs currently available from commercial sources in the United States. As a versatile, easy-to-handle, and hardy biomedical model, we recommend Hartley guinea pigs for research questions focused on tick-borne transmission, virulence, and pathogenicity of the rickettsial organism, as well as for pilot or proof-of-concept studies on rickettsial disease and host response to rickettsial infection (examples include Blanton, Mendell, Walker, & Bouyer, 2014; Goddard, 2003; Levin et al., 2020; Snellgrove, Krapiunaya, Scott, & Levin, 2021; Walker & Henderson, 1978; Walker, Harrison, Henderson, & Murphy, 1977). Hairless outbred euthymic guinea pigs, including the IAF strain (named for the Institute Armand Frappier, which identified the spontaneous mutation in 1978), were a popular research model for decades, often employed in dermatologic studies. Despite the appeal of the hairless strain for quickly visualizing tick attachment, hairless guinea pigs are no longer commercially available in the United States, and their utility in tick or tick-borne disease research was evidently never assessed based on available scientific literature.
Inbred strains offer the principal advantage of lower biological variability and thus better data reproducibility—ideal for immunological studies. However, consider using outbred guinea pigs for translational studies, as the variability resembles that of a human population. Among the many inbred strains developed in the mid to late 20th century, only strains 2 and 13 are extant. Neither strain is available commercially, nor can you acquire them quickly in numbers greater than what may be acceptable for a pilot study. Thus, starting with breeding pairs and developing a colony for research projects is highly advisable if transitioning to the guinea pig model for disease studies. Knowing that the gestation of guinea pigs is approximately three times the murine gestation period and that inbred litters may have 2-4 pups keeps expectations realistic.
When selecting the guinea pig strain for rickettsial research, consider the short- and long-term research goals. The age and sex of the guinea pigs are usually a consideration. Guinea pigs are typically at least 12 weeks old at sexual maturity and should be at least this age for evaluation of clinical signs. Early studies relied on intraperitoneal infection with rickettsiae to assess clinical signs, noting a scrotal reaction as a classic and consistent result of infection, which led to a reliance on male guinea pigs for most studies. In our hands, daily visual comparisons to pre-study photographs using guinea pigs exposed to three to five ticks infected with pathogenic rickettsiae have indicated that they do not always develop scrotal edema, and we recommend utilizing both males and females when possible or, if using only males, acknowledging sex as a potential biological variable, as more data are available on males and additional research is needed to evaluate sex as a variable. Further reading on laboratory guinea pigs and their utility as a biomedical model is available and recommended for a more in-depth understanding of the model (Shomer, 2015).
Selection of Ticks and Rickettsia ssp.
In studies involving tick-borne transmission of pathogens, it is always preferable to choose tick species that are natural vectors for the agent(s) under investigation. Even when studying novel tick-Rickettsia associations (e.g., vector competence of an invasive tick), it is necessary to have a known natural vector for a positive control. The source of ticks is a critical question when selecting ticks: i.e., laboratory reared or wild caught. These are also not mutually exclusive because laboratory-reared tick colonies require periodic additions of ticks from wild populations to maintain genetic diversity and prevent adverse effects of inbreeding, which may affect tick vitality and fecundity as early as in the fourth or fifth generation (Troughton & Levin, 2007). Although regular supplementation with wild-caught ticks maintains gene flow and high genetic variation among laboratory-reared ticks, laboratory colonies may be genetically distinct from their wild counterparts; the implications of these genetic differences for rickettsial biology and transmission are unknown (Araya-Anchetta, Busch, Scoles, & Wagner, 2015; Monzon, Atkinson, Henn, & Benach, 2016). Thus, we suggest that you interpret data from studies using laboratory colonies within the context of the study, with limitations acknowledged and caution applied when imputing broader implications for host-tick-Rickettsia systems in nature.
When starting a laboratory-reared tick colony for rickettsial research, there are several factors to consider, including the expense, labor, time, and facilities required. Researchers may also purchase colony-reared ticks from other tick-rearing laboratories (e.g., Oklahoma State University Tick Rearing Laboratory) or request them through BEI Resources (specific-pathogen-free [SPF] colonies established and maintained at the CDC). We encourage researchers to plan well in advance, regardless of source, as availability may be limited. Depending on the tick species, you may collect unfed wild-caught ticks by methods commonly used for surveillance, including flagging/dragging or using carbon dioxide traps (Newman et al., 2019; Salomon, Hamer, & Swei, 2020). Before the study, screen a subset of ticks for tick-borne pathogens or other tick-associated microbes that may affect data. At a minimum, screen ticks for Rickettsia spp., and be aware that endosymbionts, including those in the Rickettsia genus, are common; acknowledge their presence in colonies regardless of whether they are known to affect results.
When selecting the Rickettsia species, it is essential to understand different species’ virulence and the appropriate biosafety levels required for working with the organisms in a laboratory and animal setting. Selecting the Rickettsia species to use in a guinea pig study depends on the research objectives and study hypothesis. For studies on spotted fever rickettsioses in the Americas, there are three confirmed causative agents: the most virulent species, Rickettsia rickettsii (agent of Rocky Mountain spotted fever), Rickettsia parkeri , and Rickettsia sp. 364D. The latter two agents cause a milder disease typically distinguishable from Rocky Mountain spotted fever, with one or more eschars—red, erythematous lesion with necrotic center on the skin—rather than a maculopapular rash. Of the three species, R. parkeri and R. rickettsii have been studied using the guinea pig model, and tick-borne transmission has been extensively investigated for R. rickettsii and R. parkeri (Alugubelly et al., 2021; Goddard, 2003; Levin et al., 2020; Philip, Lane, & Casper, 1981). One may also employ guinea pigs to assess the pathogenicity of other SFGR and evaluate putative nonpathogenic endosymbionts (Snellgrove et al., 2021). We will not cover the cultivation of Rickettsia spp. here but refer you to the protocol for R. rickettsii by Ammerman et al. (Ammerman, Beier-Sexton, & Azad, 2008) with necessary modifications for the appropriate BSL level.
Disease Progression and Timing of Experiments
Spotted fever group rickettsiae are often present in salivary glands of unfed ticks; contrary to popular belief, they may not require a reactivation period after tick attachment for successful transmission, though more extended attachment periods lead to a larger inoculation dose (Levin et al., 2020). Guinea pigs typically develop clinical signs that begin with a fever within 1 week of exposure; other clinical signs may include scrotal edema, lividity, discoloration of ears, lethargy, and development of an eschar, depending on SFGR virulence and species. In our hands, where we allowed adult Amblyomma maculatum (Gulf Coast ticks) infected with R. parkeri to feed to repletion in a feeding chamber, we noted an eschar between 11 and 13 days after initial tick exposure (Cross et al., 2022). Expect injection of SFGR, even intradermal injection that attempts to mimic tick transmission, to result in faster development of clinical signs if the inoculation dose is higher than that delivered by an infected tick.
To our knowledge, no published studies have followed disease progression in guinea pigs infected with SFGR from tick bite to recovery despite the advantage of the guinea pig's size, which allows the collection of multiple samples over multiple time points without the need to euthanize animals at each timepoint. To assess disease progression, we suggest a study period of at least 14 days for most SFGR, allowing ticks to feed to repletion if necessary for the study question. This study period also allows the collection of serological data (i.e., rickettsial IgG titers) 2 weeks after the earliest exposure (day of tick placement), as well as whole blood, with the expectation that additional assay development over time will be necessary to expand on the previously published assay for immunophenotyping peripheral leukocytes (Stokes et al., 2020). Testing of whole blood samples may not be sensitive enough for diagnosis of rickettsial infection, considering the rarity of circulating rickettsiae in the early stages of infection; however, skin biopsies and ear notches are useful samples for detection of rickettsial dissemination at multiple time points (Levin, Snellgrove, & Zemtsova, 2016).
Basic Protocol 1: TICK TRANSMISSION OF SFGR TO GUINEA PIGS
Spotted fever group Rickettsia spp. (SFGR) are naturally transmitted to a vertebrate host in tick saliva during blood feeding, as are most tick-borne pathogens. Tick saliva serves as a medium for pathogen transfer and, significantly, also modulates host immune responses, creating a favorable environment for promoting pathogen transmission and infection. This phenomenon of enhanced pathogen transmission—called “saliva-assisted transmission”—has been documented for several tick-borne pathogens. Needle inoculation of bacteria into model animals does not replicate the environment and conditions of saliva-assisted transmission. In addition, the mode and route of inoculation can influence the development of infection in a vertebrate host along with its physiological and immunological responses. Thus, the natural tick-borne mode of infection is preferred in studies of pathogen-vector-host relationships. On the other hand, feeding uninfected ticks on animals is essential for xenodiagnosis of occult infections.
Here, we provide step-by-step instructions on the feeding of ixodid ticks on guinea pigs for introducing tick-borne Rickettsia spp. into the guinea pig via infected tick bite and the acquisition by ticks of pathogens from infected animals.
One can achieve the preliminary introduction of the Rickettsia species of interest into ticks by hemocoel microinjection (Support Protocol 1), capillary tube feeding, or immersion in cell culture or by feeding of ticks on infected animals (xenodiagnosis) with the subsequent maintenance in a natural transmission cycle where infected and uninfected colony-reared ticks are sequentially fed on susceptible laboratory animals.
Procedures described in this protocol involve ticks purposely infected with SFGR. The biological risk of working with live ticks is associated with their obligatory hematophagy in all life stages. Hazards include their ability to crawl under personal protective equipment (PPE) and personal clothing and remain hidden or attached to the host, as well as to survive on or under furniture (e.g., on a counter, in an elevator, on a door handle, on a telephone receiver) for extended periods. The Arthropod Containment Guidelines of the American Society of Tropical Medicine and Hygiene/American Committee of Medical Entomology contains general recommendations for safe arthropod handling practices, safety equipment, and facilities (American Committee of Medical Entomology and American Society of Tropical & Hygiene, 2019). Although airlocks impede escape and dispersal by flying insects, ticks can and will crawl through doorways. Therefore, we recommend surrounding the doorframes to laboratories holding ticks with a lining of petroleum jelly or carpet tape to prevent ticks from walking out of the designated facility. Replace the petroleum jelly or tape at least once a month. If using petroleum jelly is not practical, a similar method should be in place to prevent ticks from leaving the laboratory in the event of a spill or escape.
CAUTION : Appropriate PPE for personnel entering the designated facility includes a white gown or coverall, a hairnet or a cap (hair must not be touching the gown), and properly fitting gloves (BMBL, 2020). When putting on the gloves, ensure that you cover your wrists by pulling them over the sleeves of a gown or coverall so that ticks cannot crawl under the sleeves. Thoroughly inspect your PPE for ticks and appropriately remove it when exiting the facility—gloves, gowns, and coveralls worn inside the tick laboratory must not be allowed outside the area designated for tick work because ticks can hitch a ride on clothing and packaging.
Materials
- Guinea pigs (desired strain, aged 4 weeks)
- Biatane Non-Adhesive Foam, DuoDerm, or equivalent dressings
- Glue or adhesive, skin compatible (Ostobond Skin Bond, Kamar Adhesive, or equivalent)
- Anesthetics (appropriate injectables such as ketamine/xylazine, or inhalants, i.e., isoflurane—see Support Protocol 2)
- Detergent (e.g., dish soap)
- Petroleum jelly (Vaseline or equivalent)
- 10% (w/v) chlorine bleach solution
- Noncorrosive disinfectant (e.g., Lysol Disinfectant Spray or equivalent)
- 70% (v/v) ethanol
- PPE: White gown or coverall and disposable gloves (latex or nitryl with tight-fitting cuffs)
- Masking tape (for securing glove-cuff to the sleeve and capturing stray ticks)
- Cages to house guinea pigs individually
- Moats (water trays; of dimensions larger than the cage footprint, to place under cages and fill with water)
- LeFlap (Monarch Labs)
- Hypafix tape, optional
- Tubular cotton stockinette bandage (2.5-5 cm diameter tight-woven–for tick feeding bags/chambers)
- Nylon stocking (tight-woven pantyhose tights or equivalent–for tick feeding bags/chambers)
- Scissors
- 5- to 6-ml and 10- to 20-ml plastic syringes
- Hair clipper with clipper blades, no. 40 or 50 (Oster, Wahl or equivalent)
- Plastic bottle or beaker (3- to 6-cm diameter)
- Elizabethan collars (e-collars), optional
- Guinea pig jackets (Lomir)
- Pipe cutter
- Nylon mesh
- Bench-top vacuum pump
- Vacuum trap flask
- 50- to 100-ml side-arm flasks fitted with a rubber stopper and a glass tube bent at an obtuse angle
- Tubing (to connect collection flasks to the vacuum pump)
- Plastic container (Ziploc or equivalent) for restraining guinea pig during tick placement and collection
- 1- to 2-gallon waste bucket with a lid, for 10% (w/v) chlorine bleach solution (should be filled one-half to two-thirds full)
- Fine-tipped forceps
- Rubber bands
- White sorting tray
- Paintbrushes (e.g., Loew-Cornell 795 size 2 and 6 or equivalent)
- Permanent marker (fine-tip Sharpie or equivalent)
- Zip-top closable plastic bags (1/2-1 gallon, e.g., Ziplock or equivalent)
- Cotton balls
- Additional reagents and equipment for guinea pig anesthesia with isoflurane (Support Protocol 1; optional)
Preparing guinea pigs for tick infestation
1.Acclimate the guinea pigs in individual cages situated over water moats (if used) in the facility designated for tick feeding.
2.Prepare containment chambers, as well as double-layered feeding bags or plastic feeding chambers.
- a.
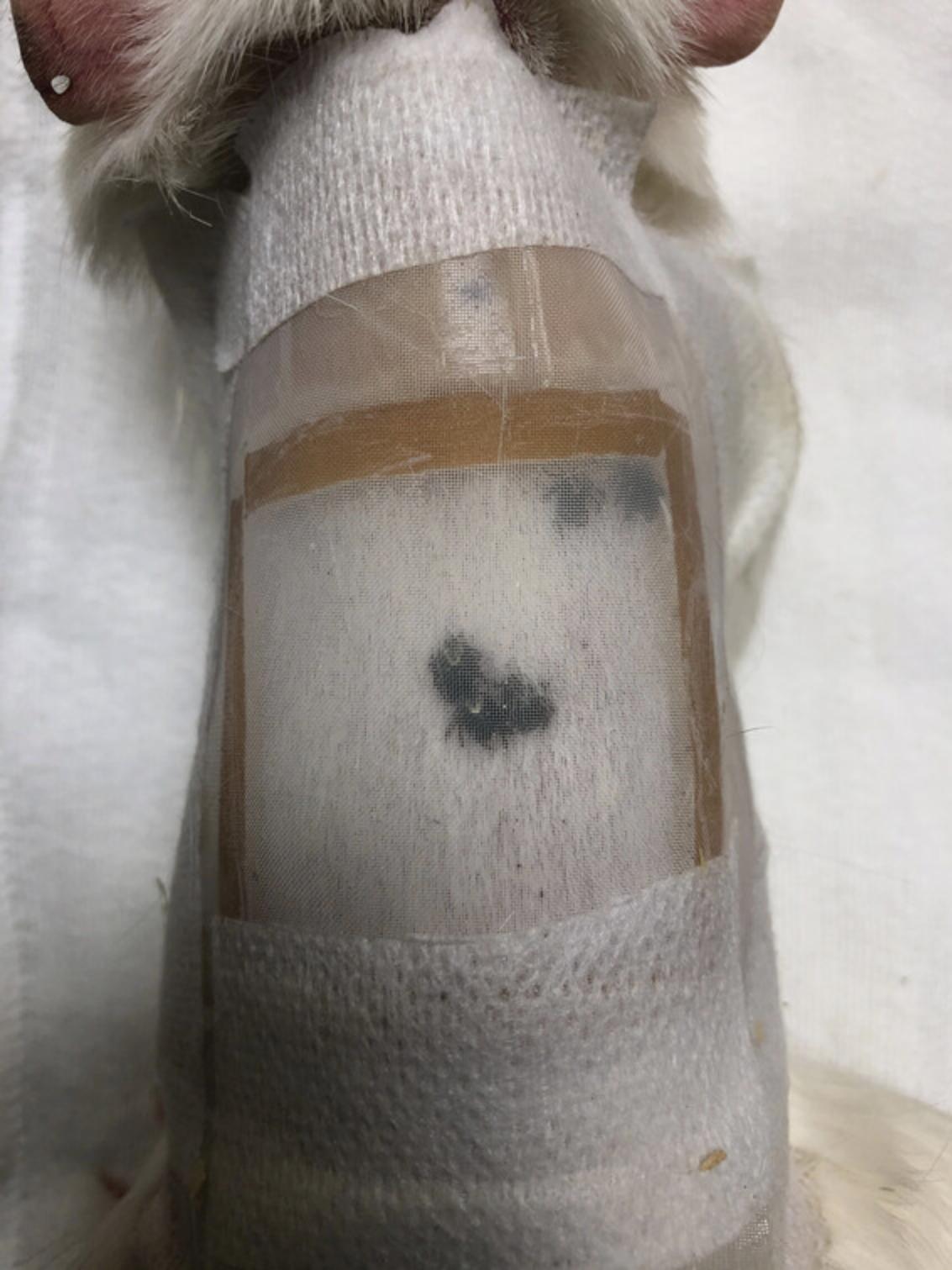
Containment chambers : Use chambers made of LeFlap (∼8-10 cm square, depending on the size of the dorsum) as the primary chamber, with reinforcement from other materials including DuoDERM and Biatane Non-Adhesive Foam to elevate the center area where ticks feed, and adhered to the skin using Ostobond Skin Bond, which may be supplemented with Hypafix tape (Fig. 1).
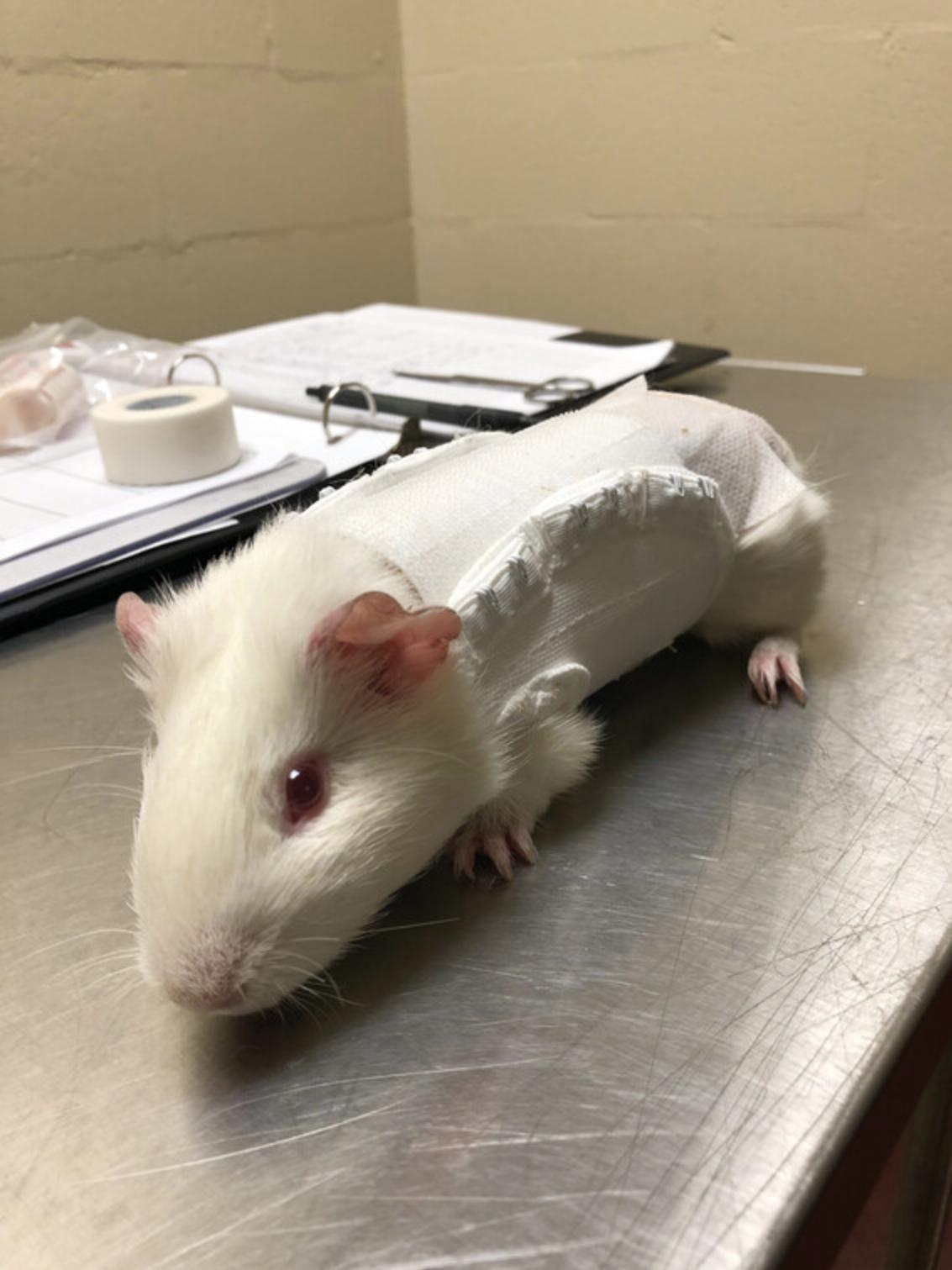
This method was modified from Embers et al. (Embers, Grasperge, Jacobs, & Philipp, 2013). We have also found this type of chamber to be successful with a Lomir guinea pig jack secured over the chamber, which removes the need for an e-collar and is well-tolerated by guinea pigs (Fig. 2).
- b.
Feeding bags : Cut the desired number of stockinette and nylon sleeves for the tick feeding bags (Fig. 3). Each feeding bag holding adult ticks requires two 10-cm segments of 2.5- to 5.0-cm-diameter cotton stockinette; each feeding bag containing larval or nymphal ticks requires one 10-cm segment of cotton stockinette for the outer layer and one 10-cm segment of a nylon stocking (pantyhose) for the inner layer.
The back of a guinea pig usually provides enough space for only a single feeding bag.
- c.
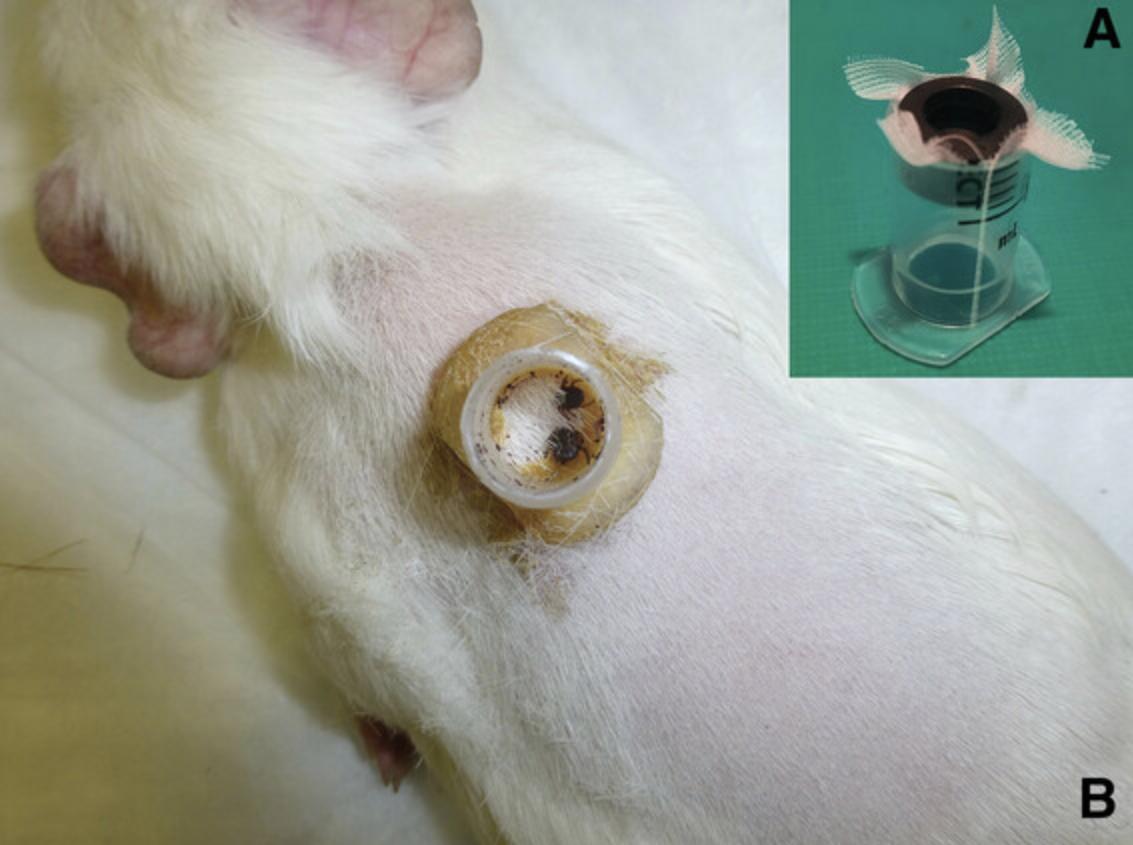
Feeding chambers : Alternatively, cut a 15- to 20-mm length of the syringe barrel (5-10 ml; 15-20 mm diameter) with the flange; remove the rubber seal from the syringe plunger and use it as a stopper to plug the capsule, preventing tick escape (Fig. 4).
Up to two plastic feeding capsules may fit on the shaven dorsum of a large guinea pig, between the shoulders and the bottom of the ribcage, if necessary.

3.Anesthetize the guinea pig using sedatives and dosage appropriate for body weight.
4.Clip the hair on the guinea pig's dorsum from neck to midriff as close to the skin as possible using no. 40 or 50 surgical clipper blades.
5.If using feeding bags, prepare them using a plastic bottle or a beaker 3-6 cm in diameter.
6.Affix containment chamber, feeding bags, or plastic feeding chambers to the shaven area on the animal's dorsum using Ostobond Skin Bond, Kamar Adhesive, or equivalent skin-compatible glue.
7.Following your approved IACUC protocol, keep the guinea pig warm and monitor heart rate and respiratory rate until complete recovery, then return the guinea pig to its cage.
Preparing ticks for infestation
8.Assemble a “tick syringe” (Fig. 5).
-
Remove the plunger from a 5- to 6-ml syringe (without needle) and remove the rubber stopper from the plunger. Cut a 4- to 6-mm airhole in the center of the rubber stopper.
-
Use a pipe cutter to cut off the needle-port part of the syringe barrel, making sure not to crack the barrel; keep the flanged part.
-
Place the rubber stopper with an airhole into a piece of precut nylon mesh, and push the nylon mesh and stopper into the flanged end of the syringe barrel (2 cm deep).
-
Remove the plunger with a seal from another syringe of the same size and use it to plug the open end of the cut barrel, creating a closed chamber.

9.Determine the number of ticks from the infected cohort that you need to place on each guinea pig to ensure that at least one Rickettsia- infected tick feeds to repletion based on (a) the prevalence of infection with the infected cohort and (b) the expected feeding success.
10.Aspirate ticks designated for each feeding bag or chamber into a “tick syringe” using a vacuum pump.
-
Remove the plunger from the tick-syringe assembly and connect the flanged end of the barrel to a vacuum trap flask and then to the pump using Tygon tubing. Keep the plunger nearby.
-
Count the designated number of ticks into a small beaker with water containing a trace of detergent (dish soap).
-
Aspirate the water with the ticks from the beaker into a tick syringe. Water will pass through the nylon mesh and the airhole in the stopper and collect in the trap flask while leaving ticks in the tick syringe.
-
Immediately plug the open end of the tick syringe with the plunger.
Placement of ticks on guinea pigs
11.Fill the moat under the guinea pig cage with water to the depth of 1.0-2.5 cm; cover the edges of the moat with petroleum jelly.
12.Prepare a waste container filled with 10% (w/v) bleach.
13.Remove the guinea pig from the cage and place it in a restraining plastic container.
14.Open the containment chamber, feeding bag, or plastic chamber.
15.Holding the prepared tick syringe flange down, tap it several times on the counter to knock all ticks to the mesh-covered rubber stopper.
16.Quickly remove the plunger, insert it into the flange end of the tick syringe, and push (inject) ticks with the mesh and rubber stopper into the feeding bag or chamber.
17.Place the emptied tick syringe into the waste container.
18.Close the containment chamber, feeding bag, or plastic chamber with a mesh cover reinforced with Hypafix tape, rubber bands, or a stopper.
19.Return the guinea pig to its cage.
Post-infestation monitoring
20.The day after tick placement, remove nylon mesh and rubber stopper from the feeding bag or chamber without disturbing the attached ticks.
21.Check the condition of the containment chamber, feeding bags, or plastic chambers and the engorgement status of ticks daily.
22.Collect, clean, and house the engorged ticks.
23.Keep a daily log of the number of ticks placed on animals and those recovered.
24.Conduct and record clinical assessments daily. These assessments may include rectal temperature (or alternatively, temperature by microchip and reader), observations of body posture, scrotal edema, and ear auricle discoloration as described in Basic Protocol 3, steps 7-12.
Support Protocol 1: LABORATORY INFECTION OF TICKS BY INJECTION
Studying SFR in guinea pigs requires a source of infected ticks to transmit the SFGR via tick transmission—the route of naturally acquired SFR in humans. Although generating infected ticks by feeding them on infected animals is time-consuming, costly, and typically reserved for pathogens that reach high circulating blood levels in the host, the use of infected hosts to infect nymphal and adult ticks is well documented (Levin, Zemtsova, Killmaster, Snellgrove, & Schumacher, 2017; Matsumoto, Brouqui, Raoult, & Parola, 2005; Schumacher, Snellgrove, & Levin, 2016; Stanley et al., 2020; Zemtsova, Killmaster, Mumcuoglu, & Levin, 2010). You may acquire infected ticks from wild populations if SFGR infection rates are high in the region and take advantage of transovarial transmission of SFGR from engorged female ticks. Larval offspring from engorged females are likely to harbor SFGR and can serve as a source of infected larval stages. One might also consider immersion of immature stages—larvae and nymphs—if using these stages in guinea pig-tick-Rickettsia studies or capillary feeding (Baldridge et al., 2007; Matsumoto et al., 2005; Ye et al., 2014).
Here, we present a protocol to generate SFGR-infected adult ticks for SFGR transmission to guinea pigs by hemocoel microinjection of engorged nymphs. By allowing engorged nymphs to undergo ecdysis, one can use adult males and females for SFGR transmission. We focus on adult ticks because they are easier to visualize and, thus, easier to contain on animals and keep track of for biosafety purposes. Note that injection into the unfed adult tick anal pore or hemocoel has been used for other organisms besides SFGR (Levin et al., 2009; Yang et al., 2022). We began using injection of engorged nymphal stages based on the publication by Goddard (2003) and through our collaborations with Dr. Goddard. The protocol presented here is a modified version of what Dr. Goddard used in his study with R. parkeri and guinea pigs.
CAUTION : Perform all activities involving handling infectious materials at the proper biosafety level conditions for the bacteria cultured (see Basic Protocol 1).
Materials
- 70% (v/v) ethanol
- Live co-culture of Rickettsia sp. (protocol not presented here) in Vero or tick embryonic cells harvested when 75%-90% of the cells are infected, with 1 ml placed in sterile tube, such as a 1.5-ml tube or 2-ml cryovial
- Engorged nymphal ixodid ticks (preferably within 2 days of detachment from host, ideally ≤24 hr after dropping), wiped gently with 70% (w/v) ethanol before use
- Biosafety cabinet
- Sterile petri dishes with lid (Fisher FB0875713A or FB0875713, or equivalent)
- Small sharps container (Uline S-15307 or equivalent)
- 1-ml syringes, with slip tip (Fisher 14-823-434 or equivalent)
- 21- or 23-G, 5/8- or 1-inch single-use needle, for drawing up Rickettsia sp. culture (Fisher 14-826C, 14-826-6C, or equivalent)
- 30-G, 1/2-inch sterile needles (Fisher 14-826F or equivalent)
- Forceps, stainless steel serrated, two pairs (Fisher 12-000-169 or equivalent; other lengths are okay)
- Kimwipes, 8.4 × 4.4 in. (Kimberly-Clark 34120 or 34155, or equivalent)
- Polystyrene containers, sterile
- Nylon mesh
- Rubber bands
- Small biohazard bag (Fisher 22-044561 or equivalent)
- Humidity chamber for housing ticks: Desiccator with porcelain plate (Fisher 08-615A) containing saturated salt solution on the bottom and a rack above the liquid on which to set tick vials
Prepare biosafety cabinet
CAUTION : Turn on the biosafety cabinet at least 5 min before starting. Then, perform SFGR injections within the biosafety cabinet with the sash lowered to proper level for safe use.
1.Clean the working surface of biosafety cabinet with 70% ethanol.
2.Place necessary supplies in the biosafety cabinet. Include two sterile petri dishes for every ten ticks to be injected, a sharps container, syringes and needles, forceps, a vial of Rickettsia sp. culture in a rack, Kimwipes, polystyrene containers, mesh covers, and rubber bands. Once ticks are injected, you will place them in the container with a mesh covering secured by a rubber band. Keep a spray bottle of 70% ethanol and a small biohazard bag in the biosafety cabinet while working.
Tick injection
3.After gently inverting vials with cultured Rickettsia spp. (host cells from the co-culture are still present), draw up ∼0.5 ml of cultured material using a 21-G or 23-G needle, then replace the needle with 30-G needle for injection into the hemocoel.
4.Open the “stock” container of ticks and transfer up to 10 ticks to one sterile petri dish.
5.With a petri dish lid nearby, gently grasp the tick between the flat serrated edges of the forceps, with the tick's ventral side up and posterior end at the open end of the forceps.
6.Using a 1-ml syringe, squeeze out a small bleb of cultured rickettsiae, letting it remain at the tip of the needle, and then gently prick the posterior end of the engorged nymph body, preferably along the edge or at the anal pore if possible.
7.Place the injected tick in a fresh petri dish where the ticks can dry and recover before they are transferred to the polystyrene container.
8.Continue performing injections and transferring ticks to a new dish until finished with the first 10 ticks. Then examine the injected ticks and transfer all dry ticks to a polystyrene container with a top that has a 4- to 6-mm air hole cut in the center and a piece nylon mesh (2.5-4.0 cm square) under the lid.
9.Place the polystyrene container(s) in the humidity chamber, where the ticks will remain for ecdysis.
10.Dispse of the used Petri dishes and other contaminated non-sharps material in a biohazard bag; dispose of needles in the “sharps” container.
Housing ticks after injection
11.Check on the ticks periodically (≥3 times a week) for evidence of mold. Clean the ticks gently with Kimwipes and 70% ethanol, allowing them to dry before returning them to a fresh, sterile container.
Alternate Protocol 1: NEEDLE INOCULATION OF SFGR TO GUINEA PIGS
When infection of animals with SFGR via tick bite is impossible or impractical, one can introduce pathogens into guinea pigs by needle-inoculation of infected cell cultures, homogenates of tick tissues, or cryopreserved homogenates of tissues from previously infected guinea pigs. Often used for the initial introduction of a pathogen into the tick-animal transmission cycle, needle inoculation also allows standardization of the infectious dose between multiple animals, which is difficult when utilizing the tick-borne route. Intraperitoneal (IP) and subcutaneous (SC) are the primary routes for injection of 0.1-5.0 ml of infectious material. The IP route provides an additional benefit of allowing quick absorption of a larger volume of inoculum within the body cavity.
This protocol describes procedures for infecting guinea pigs with infectious cell culture containing Rickettsia spp. Via intraperitoneal (IP) and subcutaneous (SC) routes.
CAUTION : Perform all activities involving handling infectious materials at the proper biosafety level conditions for the bacteria cultured (see Basic Protocol 1). Pay special attention to the careful handling of Rickettsia- contaminated sharps (needles) and the possibility of aerosolizing the infectious material during the injection. When injecting infectious material, always use a Luer-lock (not slip-tip) syringe to secure the needle, ensuring no liquids leak. Perform SFGR injections within a biosafety cabinet with the sash lowered to protect the researcher against a potential spray of infectious material.
Additional Materials (also see Basic Protocol 1)
- Infectious material (i.e., SFGR cell culture)
- Antiseptic solution (rubbing alcohol or equivalent)
- 1- to 5-ml Luer-lock syringes (sized depending on the volume of the inoculum)
- 19- to 23-G. 1/2- to 5/8-inch Luer-lock needles
- Biosafety cabinet
- Disposable absorbent benchtop pads or paper
- Sharps container
- Gauze pads
Preparation of the inoculum
1.Determine the volume of infectious material to be injected.
2.Prefill syringes with the required volume of infectious material.
3.Sedate or anesthetize guinea pigs using sedatives and dosages appropriate for body weight; see Support Protocol 2 for anesthesia.
Intraperitoneal (IP) inoculation
4a. Place the guinea pig in the biosafety cabinet on its back with the head away from you.
5a. Locate the peritoneal cavity in the lower quadrant of the abdomen, lateral to the animal's midline.
6a. Swab the injection site with an antiseptic on a gauze pad.
7a. Tilt the animal's body downward with the head rolled back by lifting the right hind leg slightly so that its hind end is higher than its head.
8a. Insert the needle into the inguinal region (just above a line between the hip and the abdomen) at a 30-45° angle to the skin.
9a. Aspirate the syringe to ensure the intestines or urinary bladder were not penetrated accidentally.
10a. If nothing is drawn back into the syringe, inject the material and withdraw the needle.
11a. Discard the syringe with the needle into a sharps container without recapping.
Subcutaneous (SC) inoculation
4b. Place the sedated guinea pig in the biosafety cabinet on its abdomen.
5b. Pull the scruff upwards, creating a “tent” with the skin, and inject the inoculum here.
6b. Insert the needle under the skin along the spine with the needle pointing cranially.
7b. Aspirate the syringe.
8b. If nothing is drawn back into the syringe, inject the required amount of material and withdraw the needle.
9b. Discard the syringe with the needle in a “sharps” container without recapping. Use a new needle and syringe for each animal.
10b. Proceed to step 12.
Post-inoculation monitoring
12.Ensure that no blood or inoculum is present at the injection site and place the animal back in its cage. If needed, apply pressure until the bleeding stops and clean with gauze and 70% ethanol (or equivalent antiseptic).
13.Disinfect all work surfaces by spraying with 70% ethanol (or equivalent antiseptic).
14.Continue checking the recovery status of animals every 15 min until they completely recover from anesthesia and are moving with no unsteadiness.
Basic Protocol 2: MONITORING THE COURSE OF GUINEA PIG RICKETTSIAL INFECTION: CLINICAL SIGNS
Here, we describe steps for the daily monitoring and recording of clinical signs in guinea pigs exposed to SFGR through the course of infection. Because the normal body temperature of a guinea pig can fluctuate between 37.2°C and 38.8°C throughout the day, conduct daily observations at the same time of the day to minimize diurnal variability. It is important to schedule the first 1-2 time-point observations before the guinea pigs are exposed to rickettsial agents to collect baseline data.
In general, guinea pigs usually become febrile (>39.5°C) between the fourth and seventh day after infection, depending on the virulence of a rickettsial isolate, the inoculum, and the transmission route. In addition to abruptly rising body temperature, guinea pigs injected with pathogenic rickettsiae characteristically develop scrotal reactions including erythema and edema, brawny discoloration and lividity of ears due to necrotizing vasculitis, perivascular hemorrhage, and focal necrosis, as well as edema and dermatitis of footpads. In fulminant cases, animals become moribund on the 8-11th day after infection with dehydration and hypothermia.
Materials
- Clinical record forms
- Digital bench-top scale (Mettler Toledo or equivalent)
- Restraining box: plastic box or a cage for confining a guinea pig on the scale
- Digital thermometer, soft tip (or microchips and a reader)
- Thermometer probe covers (disposable)
- Lubricant (Vaseline or equivalent)
1.Each day, assess and note in the clinical record the level of activity, body posture, and food intake of each guinea pig in its cage before removal for clinical observation.
2.Weigh the guinea pig by placing it in a restraining box sitting on the tared bench-top scale.
3.Measure and record the core temperature:
- a.
Cover the tip of a thermometer with a disposable probe cover and lubricate.
Lubricating the probe helps alleviate irritation of the rectum due to repeated insertions.
- b.Restrain the guinea pig by hand.
- c.Insert ∼3 cm of thermometer probe into the rectum, slightly tilt it to the side so it can touch the rectal wall, and then hold in place until the digital thermometer beeps.
- d.Remove the thermometer, discard the probe cover, and record the temperature in the clinical observation form.
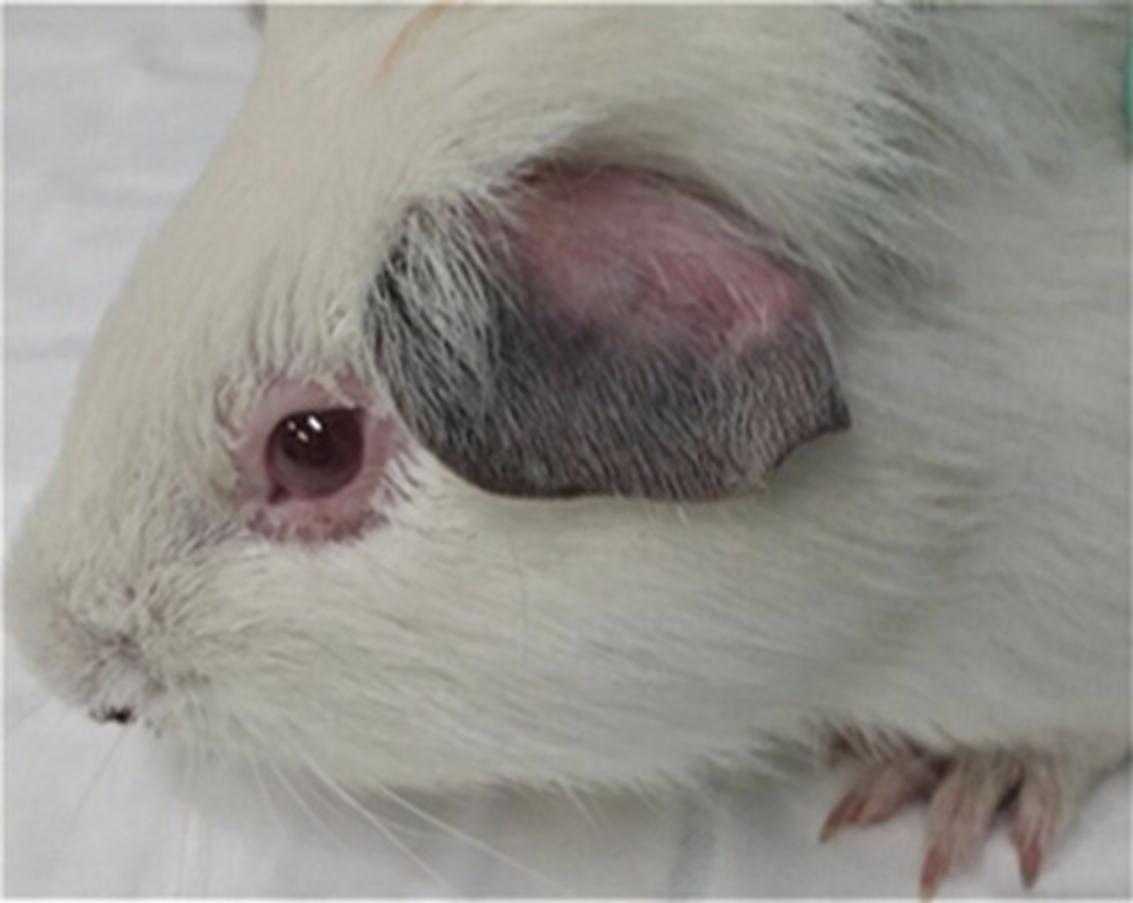
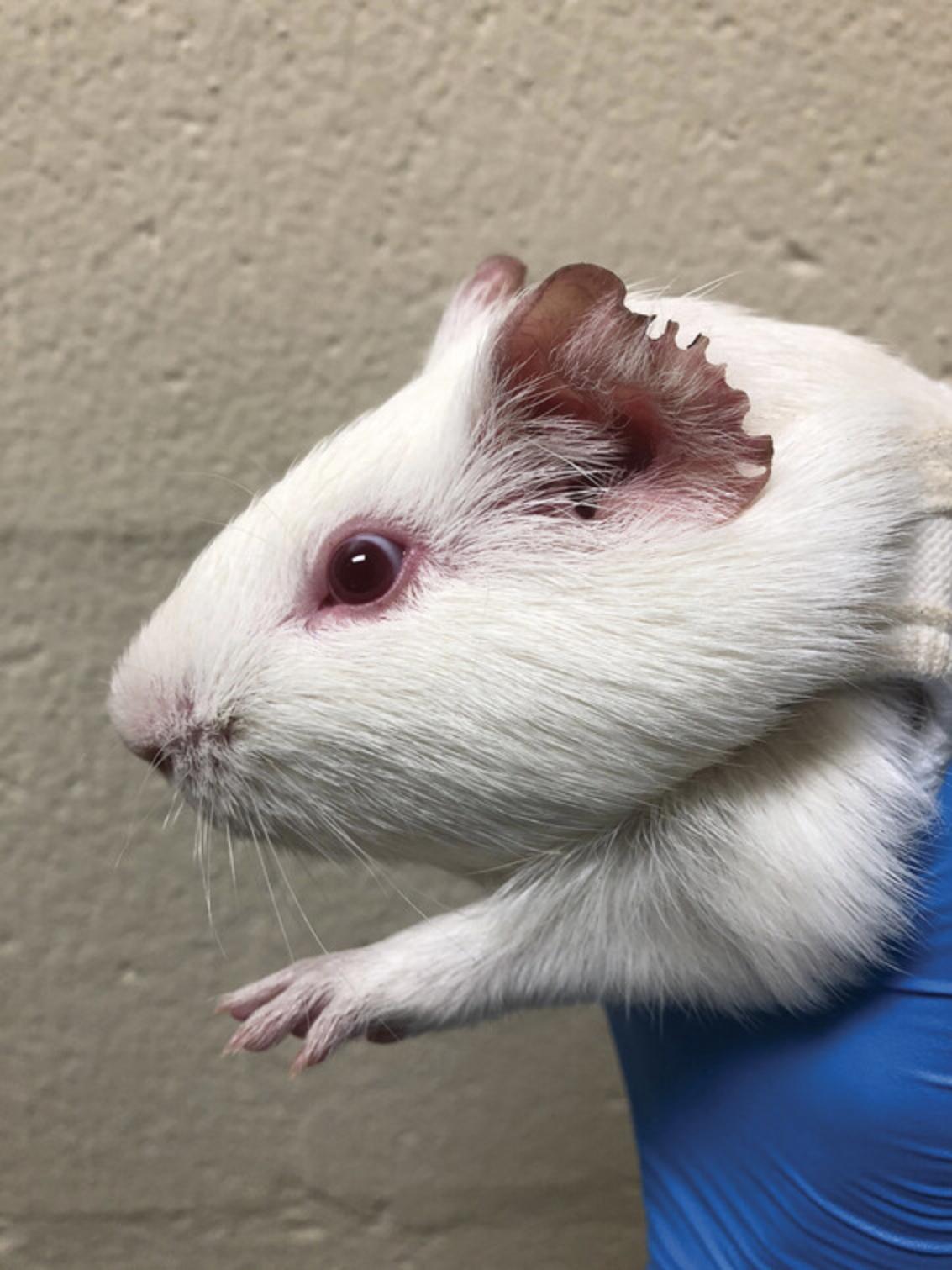
4.Examine ears, looking for lividity and purplish-brownish discoloration (Fig. 6)
5.Examine the genital area:
- a.
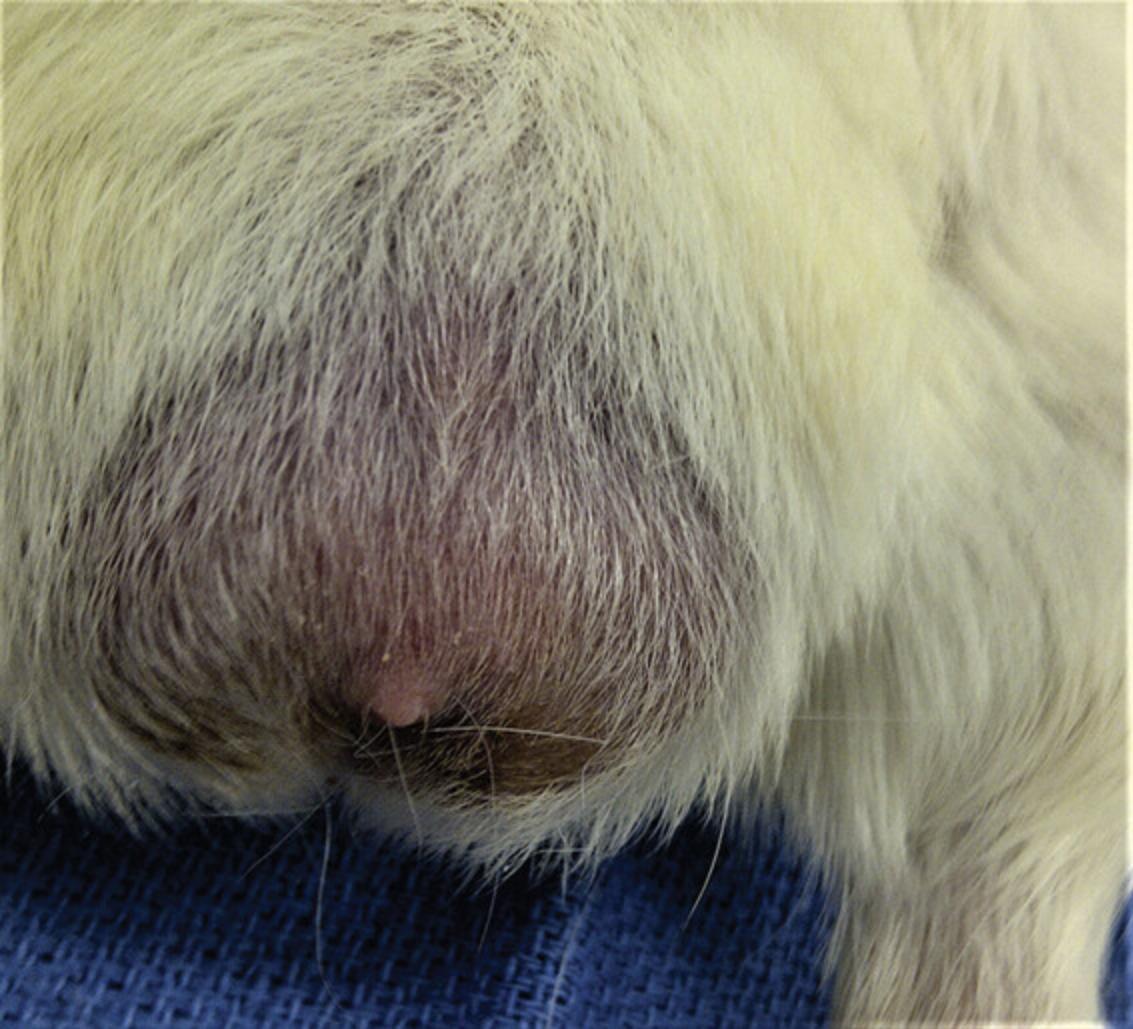
In the male guinea pig, observe the development of scrotal reactions, including erythema and edema (Fig. 7).
As the initial sign of a scrotal reaction, the skin of the scrotum becomes reddened and visible between hairs.
- b.In the female guinea pig, look for a swollen erythematous vulva.
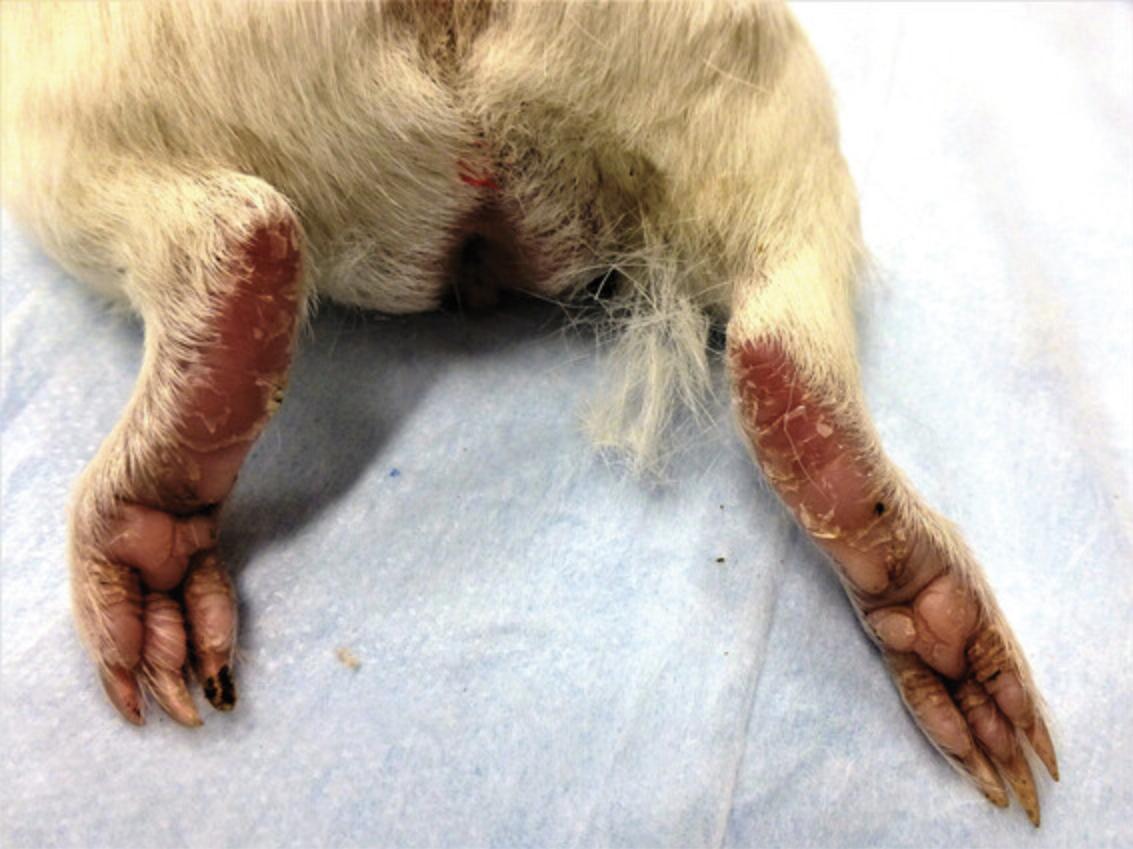
6.Examine footpads for the presence of erythema, edema, and desquamating dermatitis (Fig. 8).
7.Return the guinea pig to its cage.
8.Record all observations on the clinical observation form.
Basic Protocol 3: MONITORING THE COURSE OF GUINEA PIG RICKETTSIAL INFECTION: COLLECTION OF BIOLOGICAL SPECIMENS
The typical sexually mature guinea pig (3-4 months old), the age we recommend for most studies, will be at least 20 times the size of a sexually mature BALB/c mouse (6-8 weeks old). Thus, the larger size of the guinea pig expands the number of potential time points for antemortem sampling and the amount of the sample you can safely collect during infection (e.g., blood volume, skin biopsy size). Although the types of biological specimens obtained will depend on the study question and hypothesis, they will typically include blood and skin samples, at the tick bite site or peripheral sites, at various time points.
Blood collection sites will depend on the volume of blood needed for assays, with our locations of choice being the saphenous vein for volumes of ∼200 µl or less and the jugular vein for larger volumes. For larger volumes, do not exceed 5% of the guinea pig's total blood volume (75 ml/kg) weekly, or 7.5% blood volume biweekly (Williams & Kendall, 2015). For studies using a 550-g guinea pig, this would equal ∼2 ml total volume divided into multiple time points over the week. If needed, one may collect sufficient blood at three time points over a week for immunophenotyping using flow cytometry (see Basic Protocol 5), serology (see Basic Protocol 7), a complete blood count (CBC), and if required, PCR assays (see Basic Protocol 4). Attempts to detect rickettsiae in whole blood will rarely, if ever, bear fruit. We recommend using skin samples from the tick-bite site or ear pinna to assess rickettsial transmission and dissemination. We were guided by methods described by Birck et al. (Birck, Tveden-Nyborg, Lindblad, & Lykkesfeldt, 2014). Here we focus on blood collection techniques for the saphenous and jugular veins, adding tips or points for consideration, and techniques for collection of skin punch biopsies at tick bite sites and ear notches.
Materials
- Clippers (Wahl W9868)
- 70% (v/v) ethanol
- Petroleum jelly (Vaseline original 100% pure petroleum jelly or equivalent)
- 4 × 4-inch gauze sponges (Fisher 13-761-52 or equivalent)
- 25-G, 5/8-inch needle (Fisher 14-826AA or equivalent)
- 1- or 3-ml syringes (Fisher 14-823-30 or 14-823-30, or equivalent)
- Microtainer EDTA Blood Collection Tubes (Fisher 02-669-33 or equivalent) or microhematocrit capillary tube (Fisher22-362566 or 22-362574, or equivalent)
- Blood tube rocker
- 21-G needle (Fisher 14-826C or equivalent)
- Clipper blades, surgical #40 or #50 (Wahl or equivalent)
- 4-mm biopsy punch (Integra Miltex Standard Biopsy Punches, cat no. 3334)
- Fine, straight scissors (Fine Science Tools, multiple types available)
- Forceps (Fine Science Tools, multiple types available)
- 2.0-mm ear punch (World Precision Instruments, cat no. 500077)
- 1.5-ml Eppendorf Safe-Lock microcentrifuge tubes or equivalent, sterile (autoclaved while capped)
- Additional reagents and equipment for guinea pig anesthesia with isoflurane (Support Protocol 1; for jugular venipuncture procedure)
Blood collection by jugular venipuncture
1a. Place the guinea pig under anesthesia (see Support Protocol 2) and in dorsal recumbency.
2a. Adjust the position of the guinea pig's head while the mask is on so that the nose is pointing slightly toward the floor, with one person holding both forelegs caudally (Fig. 9).
3a. Shave the area for blood collection and disinfect the skin with 70% ethanol, wiping away hair and excess ethanol with 4 × 4-inch gauze sponge.
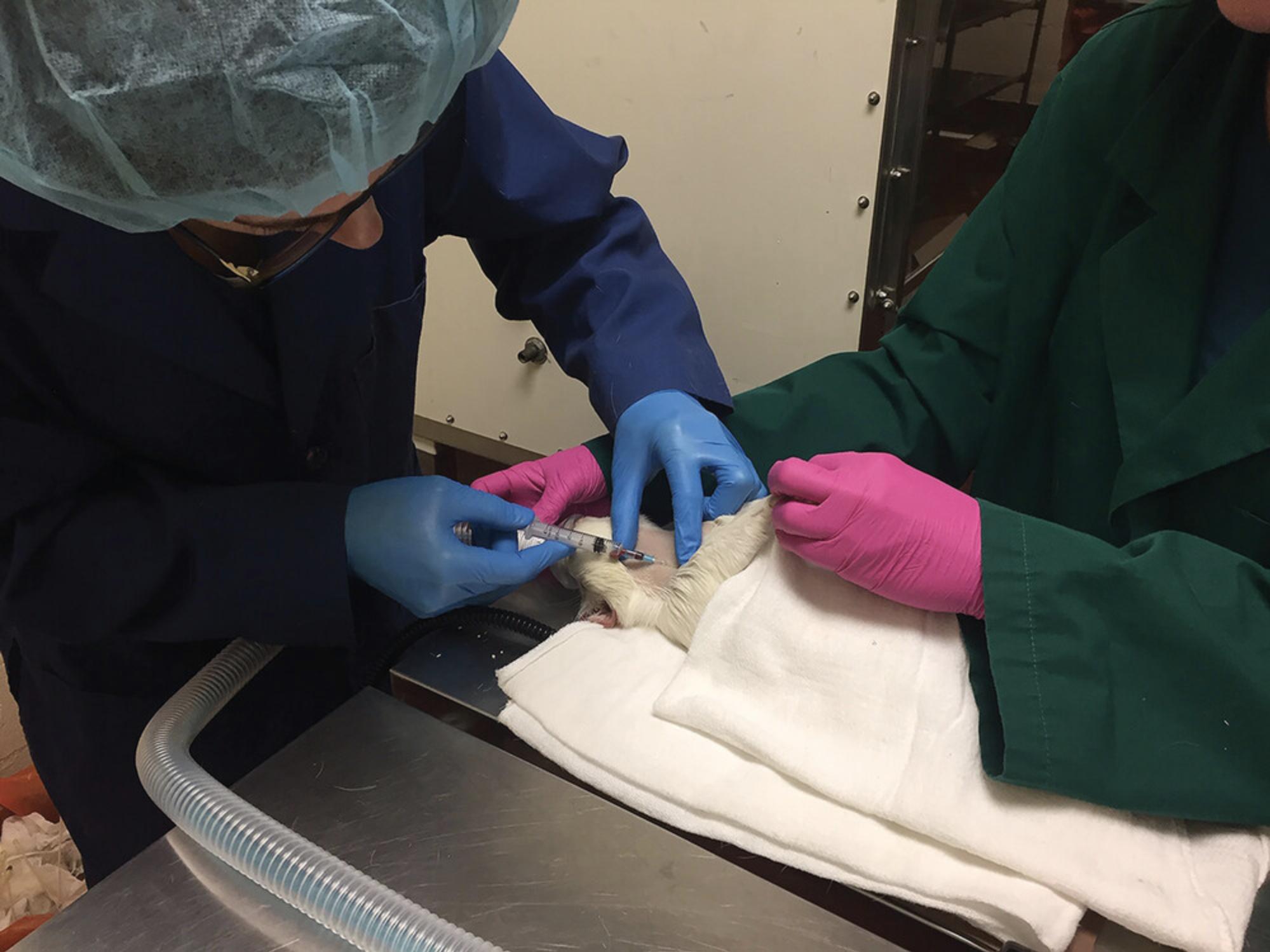
4a. With the index finger on the clavicle and distributing some pressure to hold the jugular vein cranially, and placing the thumb on the jawline, insert a 25-G, 5/8-inch needle attached to a syringe (1-ml or 3-ml depending on volume required), with needle bevel side up, into the skin at a slight ∼10-25° angle to the skin (Fig. 9).
5a. Keeping some negative pressure on the syringe and making slight adjustments to the position of the needle, watch for a flash of blood to enter the hub of the needle and then maintain negative pressure until a sufficient blood volume is obtained; after withdrawing the needle, release the forelegs, and apply slight pressure with gauze to the puncture site.
6a. Immediately dispense the blood into EDTA microtainer tubes, filling to no more than four-fifths of the maximum fill volume of each tube (i.e., 400 µl of a 0.5-ml tube) to avoid clotting. Cap each tube as it is filled and immediately invert 4-5 times, then place on a tube rocker until use.
Blood collection via saphenous venipuncture
1b. Starting with either an anesthetized or non-anesthetized guinea pig, shift it into a lateral recumbent position, and extend the opposite hind leg and down by holding out that foot—i.e., if the animal is in a left lateral recumbent position, extend the right hind leg.
2b. Shave the tarsal area of the leg to visualize vessels and clean with 70% ethanol and gauze.
3b. Apply a small amount of petroleum jelly to the skin in the region to better visualize the vessels and stimulate blood circulation.
4b. While firmly holding the foot to better expose the vein, puncture the vein using a 21-G needle.
5b. Place a microtainer tube immediately at the base of the site to collect blood before clotting.
6b. Proceed to collection of skin biopsies (steps 7-10) and/or ear notches (steps 11 and 12) if needed.
Collection of skin biopsies
7.With the guinea pig still under anesthesia, place it in a ventral recumbent position and shave the dorsum using surgical clipper blades if the area is not already shaved from the tick placement.
8.Wipe the area gently with gauze, using 70% ethanol on intact skin and ensuring the skin is dry before taking the biopsy.
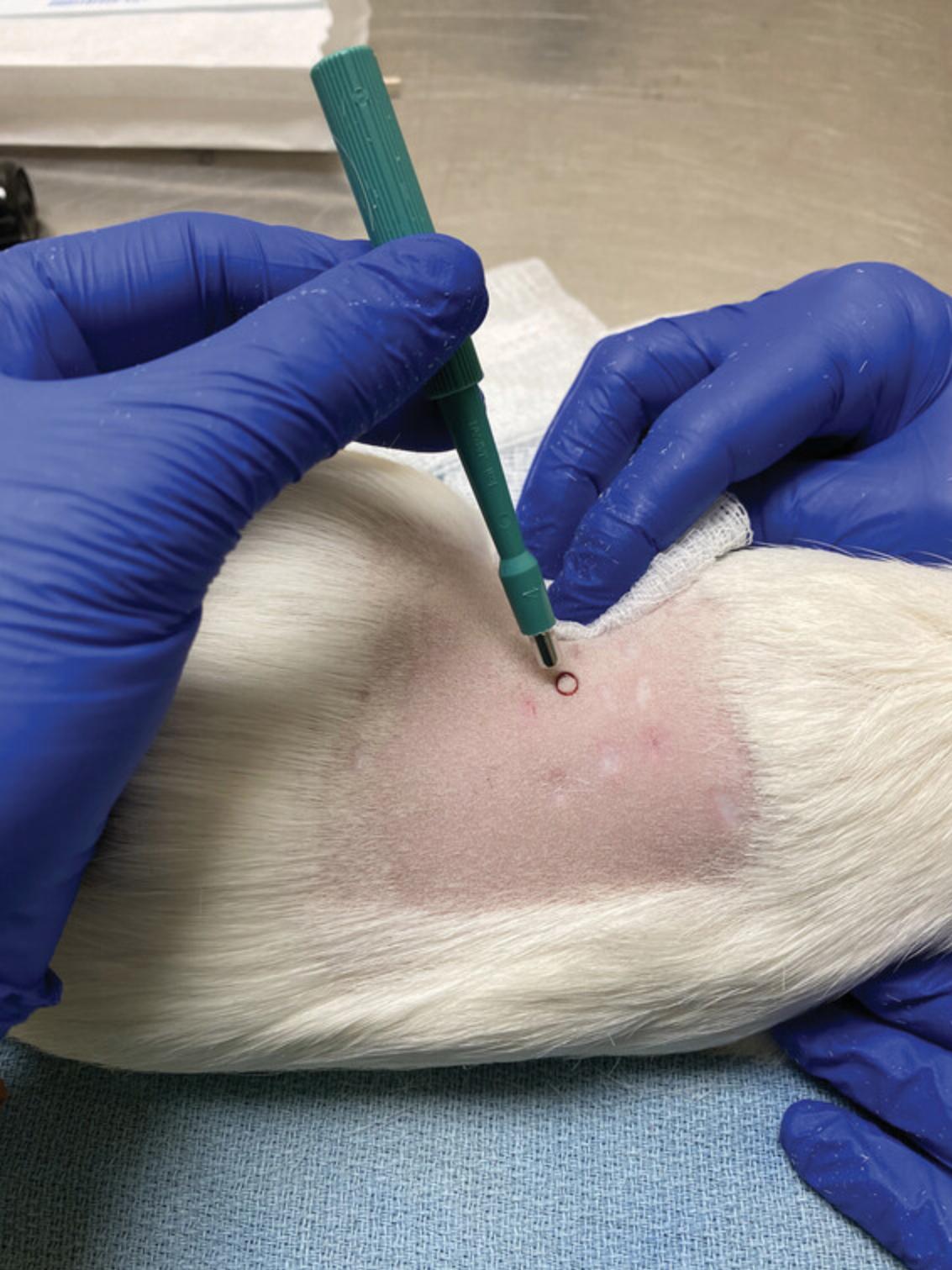
9.Place the 4-mm biopsy punch perpendicular to the skin surface and apply pressure, gently twisting it into the dermis until resistance is no longer apparent and you have reached subcutaneous fat. Draw the punch out (Fig. 10), remove the biopsy from the site or the punch itself, and apply pressure to the site to stop any bleeding.
10.Place the punch in a 1.5-ml microcentrifuge tube to transport to the laboratory.
Collection of ear skin sample
11.Starting with either an anesthetized or non-anesthetized guinea pig, take a 2.0-mm ear punch from the ear pinna margin.
12.Place the ear punch in a 1.5-ml microcentrifuge tube to transport it to the laboratory.
Support Protocol 2: GUINEA PIG ANESTHESIA
Anesthesia of guinea pigs is complicated, and you must handle animals carefully to reduce the chance of anesthetic complications. Animals must be acclimated to their surroundings, including handling, for ≥72 hr before any procedures are begun. Guinea pigs may hold feed in their oral cavity and hypersalivate under anesthesia, so passive suction or swabbing of the throat is necessary for short procedures, while glycopyrrolate is beneficial for more lengthy procedures under anesthesia. Give analgesics if there is a likelihood that the procedure will cause pain.
Materials
- Guinea pigs
- Puralube vet ointment (07-888-2572 Patterson Veterinary Supply) or equivalent sterile, non-medicated ophthalmic ointment
- Isoflurane (1182097 Henry Schein)
- Anesthesia induction box (75-2030 Harvard Apparatus)
- Rodent mask (07-8776926 Patterson Veterinary Supply)
- Oxygen tank (nexAir)
- Mobile anesthesia system (75-0238 Harvard Apparatus)
- Clean air cannister (07-893-7070 Patterson Veterinary Supply)
- Cotton-tipped swab (MDS202000Z Medline or equivalent)
- Infant suction device (Walmart or equivalent)
- Digital thermometer, soft tip (or microchips and reader)
- Towel or blanket
1.Remove hay from cage 2-3 hr before the anesthetic procedure to decrease the amount of food held in the animal's mouth. You can either leave pelleted feed in the cage or remove it 2 hr before anesthesia. Do not restrict water.
2.Apply a sterile non-medicated ophthalmic ointment to both eyes to prevent drying of the cornea and irritation due to isoflurane.
3.Place the guinea pig into the anesthesia induction box, close the lid, adjust the oxygen flow rate to 2 L/min and the isoflurane flow rate to 3%, and wait until the animal is laterally recumbent.
4.Remove the guinea pig from the induction chamber and place a mask on the animal set to provide 2 L/min oxygen and 2%-3% isoflurane. Use a non-rebreathing circuit.
5.Maintain the animal's body temperature with either a water-circulating heating pad or a heating pad compatible with use on animals.
6.Remove secretions from the mouth and throat area with a gentle suction device when performing short procedures such as blood collection. You may also use cotton-tipped swabs to remove feed and secretions.
7.Maintain the guinea pig on a mask until you complete the procedure. Monitor anesthetic depth by assessing jaw tone and palpebral reflex. Other parameters to monitor include respiratory rate and pattern, mucous membrane color, body temperature, and heart rate.
8.When finished with the procedure, turn off the isoflurane, remove the mask, and flush it with 100% oxygen; then replace the mask and maintain the animal on oxygen until it begins to awaken.
9.Remove the mask once the guinea pig has a palpebral reflex and has begun to move; keep the guinea pig wrapped in a towel or blanket to keep it warm until fully awake.
10.Provide food once the guinea pig is fully awake and ambulatory. The food will decrease the chance of post-anesthetic ileus.
Basic Protocol 4: MONITORING GUINEA PIG RICKETTSIAL BURDEN BY MULTIPLEX qPCR
This protocol describes how to simultaneously amplify multiple target sequences to monitor the rickettsial load down to 10 copies in a spotted fever group Rickettsia (SFGR) infection in host-tick pathogen studies through quantitative PCR (Ross, Stokes, Cross, Alugubelly, & Varela-Stokes, 2022). The three multiplex assays consist of (1) R. parkeri (Rp), R. amblyommatis (Ramb), and guinea pig (GP) targets; (2) R. parkeri , R. amblyommatis , and lone star tick (LST) targets; and (3) R. parkeri , R. amblyommatis , and Gulf Coast tick (GCT) targets.
Materials
- Primers and probes for R. parkeri ompB target (sequences 5′-3′):
- qOmpB_Rp_F (CGT GAC GGT GAT GTT GCT ATT A)
- qOmpB_Rp_R (CGG CAG CAT TTG TAG TTC TTG)
- qOmpB_Rp_p (/5HEX/AAC GGT GCA /ZEN/GTA CAA TTC GCT CAT /3IABkFQ/)
- Primers and probes for R. amblyommatis ompB target:
- qOmpB_Ramb_F (AAA GCA CCA CCG ACA ACA)
- qOmpB_Ramb_R (ACA TAC TGC CGA GTT ACG TTT AG)
- qOmpB_Ramb_p (/56-FAM/ACC GTT TAT /ZEN/ AAC TGT GCC GTC AGC A/ 3IABkFQ/)
- Primers and probes for guinea pig 12S rRNA target:
- Universal 12S-F (ACC GCG GTC ATA GCA TT)
- Universal 12S-R (GGG TAT CTA ATC CCA GTT TGG G)
- Cavia 12S-p (/5Cy5/AGT TAA TAA /TAO/ACC CCG GCG TAA AAA GTG /3IAbRQSp/)
- Primers and probes for lone star tick MIF target:
- LST-MIFf (CGA ATC GTC TCT GCG TCT TT)
- LST-MIFr (TTT GCA GCG TTG AGA AAG TAT G)
- LST-MIFp (/5Cy5/TGA GTG CGA /TAO/TTT CCG TAC AGA GCA /3IAbRQSp/)
- Primers and probes for Gulf Coast tick MIF target (Lee et al., 2017):
- AmacMIF.18F (CCA GGG CCT TCT CGA TGT
- AmacMIF.99R (CCA TGC GCA ATT GCA AAC C
- AmacMIF.63 (TGT TCT CCT TTG GAC TCA GGC AGC
- Water, molecular biology grade (Fisher Scientific BP2819-1)
- Brilliant Multiplex QPCR Master Mix (Agilent Technologies, Inc. 600553)
- 1.5-ml Eppendorf Safe-Lock microcentrifuge tubes (or equivalent)
- TempAssure 0.2 ml PCR 8-Tube Strips, Att. Optical Caps (USA Scientific 1402-3900)
- Optical Cap, 8× Strip (Agilent Technologies, Inc. 401425)
- AriaMx 96 Well Optical Plates (Agilent Technologies, Inc. 401494)
- Fisherbrand™ microplate centrifuge (or equivalent)
- Agilent AriaMx Real-Time PCR (or equivalent)
In the day(s) before starting
1.Order primers and probes and prepare stock solutions.
2.Produce plasmids and adjust the stock concentration to 108 copies/µl for each qPCR target.
Start experimental preparation
3.Calculate the volume of the qPCR master mix needed based on the total number of samples. Include standard curve, unknowns, no-template control (NTC) samples, and two extras to ensure enough volume is present. You will run duplicates for each sample.
4.Prepare the appropriate qPCR master mix(es), depending on the SFGR species and host target being measured and according to the recipes in Tables 1-3; add all components into a 1.5-ml microcentrifuge tube in each case (Tables 1-3).
| Component | n = 1 (µl) |
|---|---|
| H2O | 0.937 |
| Multiplex Master Mix (Brilliant) 2× | 12.5 |
| Rp probe (HEX) qOmpB_Rp_p (300 nM) | 0.75 |
| Primer qOmpB_Rp_F (300 nM) | 0.75 |
| Primer qOmpB_Rp_R (600 nM) | 1.5 |
| Probe (CY5) LST-MIF p (400 nM) | 1.0 |
| Primer LST-MIF f (75 nM) | 0.188 |
| Primer LST-MIF r (300 nM) | 0.75 |
| Probe (FAM) qOmpB_Ramb_p (50 nM) | 0.125 |
| Primer qOmpB_Ramb_F (150 nM) | 0.375 |
| Primer qOmpB_Ramb_R (300 nM) | 0.75 |
| ROX (1:500 dilution) | 0.375 |
| Total volume | 20.0 |
-
Working solutions for primers and probes are 10 µM.
| Component | n = 1 (µl) |
|---|---|
| H2O | 0.625 |
| Multiplex Master Mix (Brilliant) 2× | 12.5 |
| Rp probe (HEX) qOmpB_Rp_p (400 nM) | 1.0 |
| Primer qOmpB_Rp_F (300 nM) | 0.75 |
| Primer qOmpB_Rp_R (600 nM) | 1.5 |
| Probe (CY5) Cavia 12S-p (200 nM) | 0.5 |
| Primer Universal 12S-F (150 nM) | 0.375 |
| Primer Universal 12S-R (300 nM) | 0.75 |
| Probe (FAM) qOmpB_Ramb_p (200 nM) | 0.5 |
| Primer qOmpB_Ramb_F (150 nM) | 0.375 |
| Primer qOmpB_Ramb_R (300 nM) | 0.75 |
| ROX (1:500 dilution) | 0.375 |
| Total volume | 20.0 |
-
Working solutions for primers and probes are 10 µM.
| Component | n = 1 (µl) |
|---|---|
| H2O | 0.625 |
| Multiplex Master Mix (Brilliant) 2× | 12.5 |
| Rp probe (HEX) qOmpB_Rp_p (400 nM) | 1.0 |
| Primer qOmpB_Rp_F (300 nM) | 0.75 |
| Primer qOmpB_Rp_R (600 nM) | 1.5 |
| Probe (CY5) Amac MIF.63 (200 nM) | 0.5 |
| Primer Amac MIF.18F (150 nM) | 0.375 |
| Primer Amac MIF.99R (300 nM) | 0.75 |
| Probe (FAM) qOmpB_Ramb_p (200 nM) | 0.5 |
| Primer qOmpB_Ramb_F (150 nM) | 0.375 |
| Primer qOmpB_Ramb_R (300 nM) | 0.75 |
| ROX (1:500 dilution) | 0.375 |
| Total volume | 20.0 |
-
Working solutions for primers and probes are 10 µM.
5.Aliquot 40 μl master mix for each sample into PCR 8-Tube Strips.
6.Aliquot 10 μl of unknown template, or 10 μl of molecular-grade H2O for the NTC, into the correct strips and cap the tubes before proceeding.
7.Perform a 1:10 serial dilution from 107-101 copies using the 108 (per target) stock plasmid mix and diluting with molecular-grade H2O.
8.Add 10 μl of each standard dilution to its corresponding sample in the 8-Tube strip containing the master mix. Cap the tubes between each one.
9.Load 22 μl of each sample into two duplicate wells of the 96-well optical plate.
10.Ensure that the caps are tight and pulse-centrifuge the plate.
Multiplex qPCR and analysis
11.Run qPCR plate on Agilent AriaMx (or equivalent) using the following thermal profile:
| 1 cycle: | 10 min | 95°C | (initial denaturation) |
| 40 cycles | 5 s | 95°C | (denaturation) |
| 1 min | 60°C | (annealing/extension). |
12.Analyze results on AriaMx software (or equivalent).
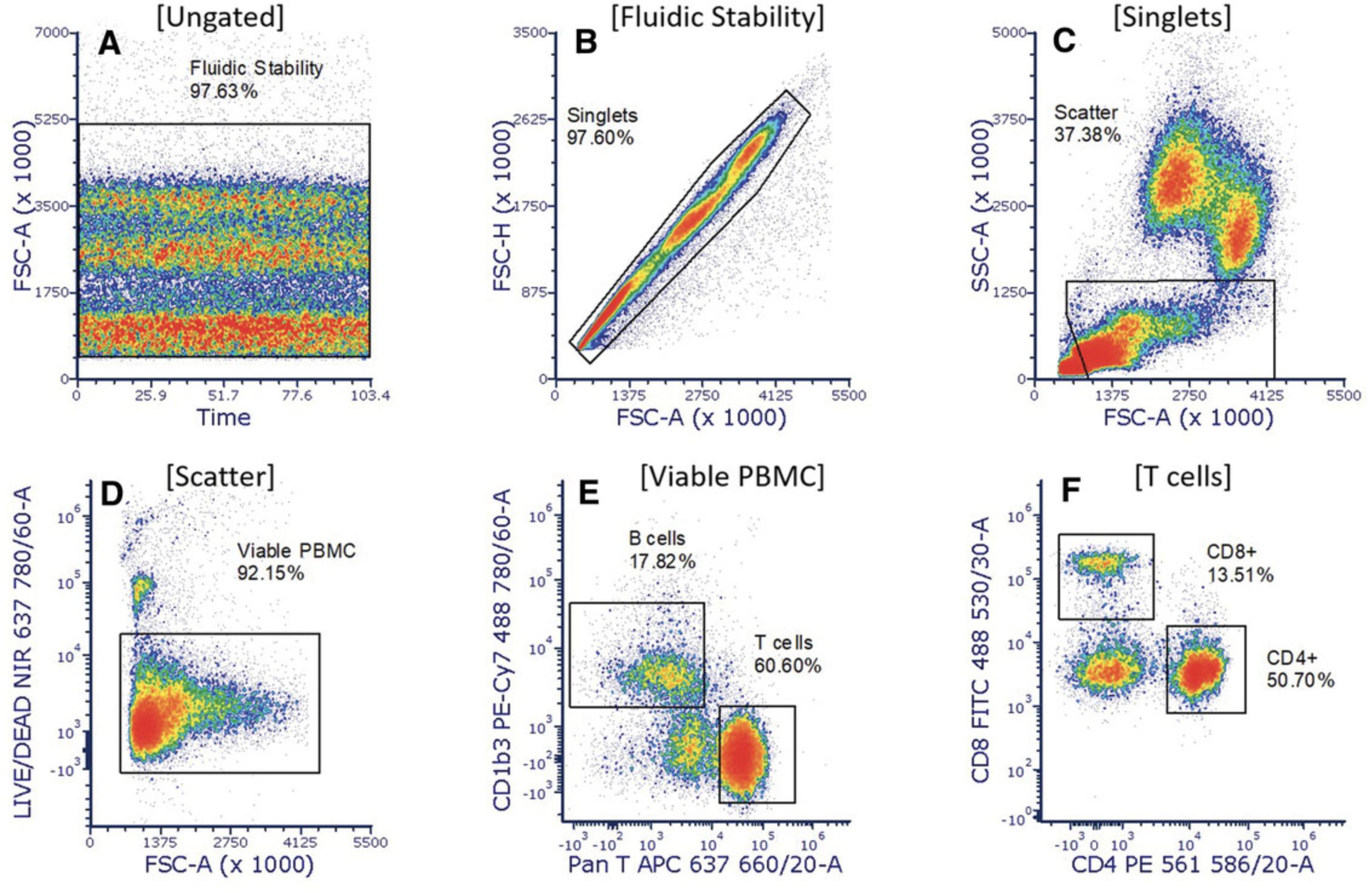
Basic Protocol 5: MONITORING GUINEA PIG IMMUNE RESPONSE TO RICKETTSIAL INFECTION: BLOOD LEUKOCYTES BY FLOW CYTOMETRY
Here, we describe a method for monitoring the immune response to rickettsial infection by immunophenotyping blood leukocytes via polychromatic flow cytometry at predetermined time points (Stokes et al., 2020). Rather than sacrificing a group of mice at each time point, you follow individual guinea pigs over the course of the study. Schedule the first time point to occur before you place the ticks on the guinea pigs to collect baseline data. Also, it is essential to maintain a rapid and consistent tempo at all time points when performing this protocol to achieve the highest reproducibility. This consistency is best accomplished by working as a team of at least two people. Finally, for clarity in the steps that require calculations, we will assume that there will be ten experimental (unknown) samples, five Fluorescent Minus One (FMO) controls, five compensation controls, one reference control, one unstained control, and one or two “extras” to ensure adequate volume.
Materials
- Lightning-Link PE-Cy7 Conjugate (Expedeon 762-0005)
- Mouse Anti Guinea Pig CD1b3 (Bio-Rad MCA566GA)
- Ca2+- and Mg2+-free phosphate-buffered saline (PBS), pH 7.4 (Gibco 10-010-049)
- Bovine serum albumin (BSA; Sigma A3059-100G)
- Guinea pig serum (Sigma G9774-5ML)
- Fetal bovine serum (FBS), heat inactivated (Atlanta Biologicals S11510H)
- Gibco RPMI 1640 medium without phenol red (Fisher 11-835-030)
- Near-IR LIVE/DEAD Fixable Dead Cell Stain (Invitrogen L34976)
- DMSO, anhydrous (Invitrogen D12345)
- Frozen guinea pig blood (Basic Protocol 3; 100 μl/per sample for each control and experimental group)
- Propidium iodide staining solution (Invitrogen 00-6990-50)
- BD Pharm Lyse Lysing Buffer (BD Biosciences 555899)
- Whatman™ pH Indicators Integral Comparison Strips (Fisher 09-876-20)
- Cell culture water (Sigma W3500)
- True-Stain Monocyte Blocker (BioLegend 426102)
- Mouse Anti Guinea Pig Anti T-Lymphocytes (PAN):APC (Bio-Rad MCA751APC)
- Mouse Anti Guinea Pig Anti CD4:PE (Bio-Rad MCA749PE)
- Mouse Anti Guinea Pig Anti CD8:FITC (Bio-Rad MCA752F)
- 16% formaldehyde solution, methanol-free (Thermo Scientific 28906)
- Viability Dye Compensation Standard (Bangs Laboratories 451)
- OneComp eBeads Compensation Beads (Invitrogen 01-1111-42)
- 0.1-µm filters (PALL 4481)
- Beckman Coulter Allegra X-14R tabletop centrifuge with canister kit, tube racks (13 mm), and Biocert covers (or equivalent)
- 15-ml polypropylene centrifuge tubes (Fisher 05-538-53D)
- 12 × 75-mm flow tubes (Fisher 14-959-6)
- 1.5-ml Eppendorf LoBind (Protein) microcentrifuge tubes (Fisher 13-698-794)
- Countess Cell Counter Chamber Slides (Invitrogen C10228)
- Thermo Fisher Countess II FL Automated Cell Counter (or equivalent)
- Tube Revolver/Rotator (Thermo Scientific 88881001) or equivalent
- 1-ml BD syringes with attached 25-G, 5/8-inch needle (Fisher 14-826-88 or equivalent)
- 250- to 500-μl BD Microtainer, EDTA (Fisher 02-669-33)
- 13-mm Nalgene Unwire™ test tube racks (Fisher 14-809-45) or equivalent
- NovoCyte Quanteon flow cytometer (or equivalent)
- Additional reagents and equipment for the preparation of splenocytes (Support Protocol 3)
In the day(s) before starting sample preparation
1.Conjugate PE-Cy7 to Mouse Anti Guinea Pig CD1b3.
2.Prepare 1.0% (v/v) BSA in Ca2+- and Mg2+-free PBS (FCM-PBS), filter through a 0.1-µm filter, and store at 4°C.
3.Determine the optimal antibody concentrations through titration.
4.Prepare aliquots of splenocytes to use as reference controls (see Support Protocol 3).
5.Prepare and freeze 275-µl aliquots of guinea pig serum for blocking Fc receptors.
6.Prepare and freeze 15-ml aliquots of 5% (v/v) FBS in RPMI 1640 without phenol red (cRPMI).
7.Thaw a 15-ml aliquot of cRPMI for thawing the reference control.
Immediately before starting sample preparation
8.Prepare Near-IR LIVE/DEAD Fixable Dead Cell stain.
- a.Bring one vial of the fluorescent reactive dye (Component A) and the vial of anhydrous DMSO (Component B; or Invitrogen D12345) to room temperature (they should be thawed before the caps are removed).
- b.
Add 150 μl DMSO to the vial. Mix well and visually confirm that all the dye has dissolved. Protect from light.
The manufacturer suggests using 50 μl DMSO. However, the best practice is to empirically determine the lowest concentration of stain that gives consistent separation of viable vs. dead cells and with which the dead cells do not stain brighter than the Viability Dye Compensation Standard. Also, amine-reactive stains are not stable for long after being suspended in DMSO. You will obtain the most consistent results if you prepare the stain fresh on the day of use.
9.Bring the FCM-PBS (from step 2) to room temperature.
10.Thaw splenocytes for reference control (5 × 106 cells).
- a.Prewarm cRPMI to 37°C.
- b.Thaw cryovial at 37°C for 1.5-2 min; flick vial with a finger every few seconds.
- c.Dropwise (over 30 s), add ∼1 ml prewarmed cRPMI to the cryovial and then transfer contents to a 15-ml centrifuge tube containing 8 ml prewarmed cRPMI.
- d.Centrifuge 5 min at 300 × g. Decant the supernatant.
- e.Add 4.8 ml (to ∼5 ml total volume) of prewarmed cRPMI and gently resuspend the pellet. Then transfer 1 ml (∼1 × 106 cells) to a flow tube (keep the other 4 ml in a separate flow tube as extra).
- f.
Remove cells for counting and keep the rest at 37°C until needed (step 19).
All centrifugation steps are performed at room temperature.
Mix splenocytes in the flow tubes gently by pipetting up and down. Take 100 μl of splenocytes from the center of suspension from the extra tube and place them into a 1.5-ml microcentrifuge tube.
- g.Add 5 μl propidium iodide staining solution to the 100 μl of splenocytes and mix gently. Incubate at room temperature in the dark for 7 min.
- h.Load a Cell Counter Chamber Slide with 10 μl (each side) of the stained splenocytes using a 20-μl pipet. Let settle for 1 min before reading.
- i.
Take two readings (one from each side of the chamber slide) on the Countess II FL Automated Cell Counter. Take an average of the two readings and record the cell count and viability.
Set the light source on the cell counter to “RFP.”
The reference control comes from one animal at a single time point. Therefore, it should generate consistent results between time points and thus provide assay-specific quality control (QC) by controlling for run-to-run variation in sample processing and staining. Although often left out of longitudinal studies, this control gives confidence that results in the experimental samples are due to the “treatment” rather than instrument, technical, or analysis variability between runs.
11.Prepare 36 ml of 1× lysing buffer (2.0 ml/sample × 18, for 10 experimental samples + 5 FMO controls + 1 unstained control + 1 reference control + 1 extra) from 10× BD Pharm Lyse Lysing Buffer stock solution, as follows.
- a.Dilute 3.6 ml of 10× solution to 36 ml with 32.4 ml cell culture water.
- b.Warm the 1× solution to room temperature.
- c.
Check the pH of the 1× solution with pH paper strips. The pH should be 7.1-7.4.
Deionized water made in the laboratory (e.g., “Millipore water”) will usually work; however, in our hands we found that cell culture water consistently has an acceptable pH.
12.Prepare the surface antigen antibody cocktail by adding in the order listed:
- a.66.0 μl True-Stain Monocyte Blocker = 13.2 samples × 5 μl block/sample
- b.88.4 μl CD4 antibody:PE = 13.2 samples × 6.7 μl antibody/sample
- c.33.0 μl Pan T antibody:APC = 13.2 samples × 2.5 μl antibody/sample
- d.13.2 μl CD8 antibody:FITC = 13.2 samples × 1 μl (1:4) antibody/sample
- e.
17.2 μl CD1b3 antibody:PE-Cy7 (from step 1) = 13.2 samples × 1.3 μl antibody/sample.
Store on ice in the dark.
CD8 is used at 1:400. To enhance pipetting accuracy and precision, make 24 μl of a 1:4 dilution by adding 6 μl of antibody to 18 μl FCM-PBS. Save the leftover at 4°C in the dark to use in step 26.
The multiplier “13.2” is derived from counting all the samples that will get the entire surface antigen cocktail plus ten percent. There are 10 experimental samples, one reference control, one FMO for the viability stain, plus ten percent in this example. The total volume of the cocktail is 217.8 μl. Each sample gets 16.5 μl for surface staining, for a total volume of 198.0 μl. Thus, 217.8 μl leaves 19.8 μl extra to account for the “angel's share,” i.e., the amount lost to evaporation.
The dilutions (volumes) of the antibodies were based on our antibody titrations of the lot numbers used in our study. You will need to perform your own titrations (see step 3 above) before starting the experiment.
We included monocyte blocker in the cocktail to prevent nonspecific binding of PE-Cy7 to monocytes. The volume is based on the manufacturer's recommendation.
13.Prepare 24 ml of methanol-free 1% formaldehyde (1.0 ml/sample × 24 samples: 10 experimental + 5 FMO controls + 5 compensation controls + 1 unstained control + 1 reference control + 2 extra) by diluting 1.5 ml of 16% formaldehyde with 22.5 ml PBS (protein-free). Keep at 4°C until ready to use.
14.Thaw an aliquot of guinea pig serum and bring to room temperature. You will need 240 μl of serum (10 μl/sample × 24 samples: 10 experimental + 5 FMO controls + 5 compensation controls + 1 unstained control + 1 reference control + 2 extra).
15.Prepare Viability Dye Compensation Standard (Bangs Beads).
- a.
Bring the vial of beads to room temperature while end-over-end mixing on the Tube Revolver/Rotator.
Do not leave the vial of beads at room temperature for >25-35 min before preparation.
- b.Place one drop of beads in a flow tube designated for Near-IR LIVE/DEAD and another drop in another tube to use for a negative population.
- c.Wash both tubes of beads by adding 0.5 ml PBS (protein-free), vortexing lightly, and centrifuging 5 min at 300 × g. Decant the supernatant.
- d.Wash the beads a second time just as in step 15c (above).
- e.Resuspend the beads for the Near-IR LIVE/DEAD in 50 μl PBS (protein-free) and the beads for the negative population in 100 μl PBS (protein-free). Place a cap on the tubes to reduce evaporation.
Start sample preparation
16.Draw ∼250 μl blood (100 μl for experimental FCM sample plus 150 μl “extra” for make up the minimum volume of the Microtainer) from the jugular vein of each of the ten study guinea pigs using 1-ml syringes with 25-G, 5/8-inch needles. From one guinea pig, draw ∼800-900 μl blood (100 μl for the experimental sample + 500 μl for the FMO controls + 100 μl for the unstained control + 100 to 200 μl “extra”). Dispense each ∼250-μl blood sample into a BD Microtainer. For controls, dispense ∼800-900 μl of blood from the syringe into two or three BD Microtainers (total volume ∼800-900 μl). Invert the Microtainers several times to mix the EDTA with the blood. Transport the sample preparation area.
17.Dispense 100 μl blood (∼106 WBC) into each of 17 flow tubes. For example (here GP refers to individual guinea pigs, i.e., biological replicates):
-
Unstained control
-
FMO-L/D NIR
-
FMO-FITC
-
FMO-PE
-
FMO-PE-Cy7
-
FMO-APC
-
GP-1
-
GP-2
-
GP-3
-
GP-4
-
GP-5
-
GP-6
-
GP-7
-
GP-7
-
GP-8
-
GP-9
-
GP-10
18.Add 2 ml of 1× lysing buffer to each flow tube and lightly vortex.
19.Incubate the tubes at room temperature for 5 min. Add the tube from step 10f (reference control) to the test tube rack (no lysis buffer) with the blood during the incubation.
20.Centrifuge 5 min at 300 × g. Decant the supernatant and break the pellets (with two strikes).
21.Wash by adding 3 ml PBS (protein-free) and centrifuge 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
22.Resuspend the cells in 1 ml PBS (protein-free) and lightly vortex.
23.Prepare single-stained beads for compensation and apply viability stain to the cells.
-
a.Prepare the Viability Dye Compensation Standard beads:
-
i.Add 9 μl of the reconstituted Near-IR LIVE/DEAD fixable dead cell stain directly onto the bead suspension and mix by pipetting up and down.
-
ii.Incubate at room temperature in the dark for 30 min.
-
iii.Add 3 ml FCM-PBS to the sample tube.
-
iv.Centrifuge 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
-
b.Apply viability stain to the cells:
-
i.Add 1 μl of the reconstituted Near-IR LIVE/DEAD Fixable Dead Cell Stain to all cell suspensions from step 22 (except for the FMO-L/D NIR) and mix by light vortexing.
-
ii.Incubate for 30 min at 4°C in the dark.
-
iii.Centrifuge 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
-
iv.Add 3 ml FCM-PBS to each tube.
-
v.Centrifuge 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
-
c.Prepare the OneComp Beads.
-
i.Label a flow tube for each of the four colors represented by the four antibodies (i.e., FITC, PE, PE-Cy7, and APC).
-
ii.Lightly vortex the OneComp/UltraComp beads for 10-15 s to completely resuspend.
-
iii.Add 1 drop of OneComp Beads to each tube.
-
iv.Add 1 μl anti-CD8:FITC, 1 μl anti-CD4:PE, 1 μl anti-CD1b3:PE-Cy7, and 1 μl anti-PAN T:APC to the appropriately marked tubes and mix by pipetting up and down. Deposit the antibody directly onto the bead suspension.
-
v.Incubate for 30 min at 4°C in the dark.
-
vi.Add 3 ml FCM-PBS to each tube.
-
vii.Centrifuge 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
24.Add 10 μl of 100% guinea pig serum to each tube to block Fc receptor and nonspecific binding of the antibody to the cells and mix by pipetting up and down.
25.Incubate 30 min at 4°C in the dark.
26.While the incubation proceeds, prepare FMO cocktails for step 27 as follows.
-
a.FMO-L/D NIR: From assay cocktail (step 12).
-
b.FMO-No CD8:FITC
-
5.0 μl Monocyte Blocker
-
1.0 μl FCM-PBS
-
2.5 μl PAN T:APC
-
6.7 μl CD4:PE
-
1.3 μl CD1b3:PE-Cy7.
-
c.FMO-No CD4:PE
-
5 μl Monocyte Blocker
-
6.7 μl FCM-PBS
-
2.5 μl PAN T:APC
-
1.0 μl CD8:FITC (1:4 dilution)
-
1.3 μl CD1b3:PE-Cy7.
-
d.FMO-No CD1b3:PE-Cy7
-
5 μl Monocyte Blocker
-
1.3 μl FCM-PBS
-
2.5 μl PAN T:APC
-
6.7 μl CD4:PE
-
1.0 μl CD8:FITC (1:4 dilution).
-
e.FMO-No PAN T:APC
-
5 μl Monocyte Blocker
-
2.5 μl FCM-PBS
-
6.7 μl CD4:PE
-
1.0 μl CD8:FITC (1:4 dilution)
-
1.3 μl CD1b3:PE-Cy7.
The FCM-PBS in the FMOs is to replace the antibody that is missing. Again, the final staining volume is critical and must be the same for the FMO controls as for the experimental samples and the reference control.
27.Label surface antigens. Add the appropriate antibody directly to each sample and mix by pipetting up and down.
-
For each experimental sample (GP 1-10), the reference control, and the L/D NIR FMO, add 16.5 μl of the surface antigen antibody cocktail from step 12.
-
For each FMO, add the appropriate cocktail from step 26.
28.Incubate for 30 min at 4°C in the dark.
29.Wash all the samples by adding 3 ml FCM-PBS and centrifuging 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
30.Wash all samples by adding 3 ml PBS (protein-free) and centrifuging 5 min at 300 × g. Decant the supernatant and break the pellet (two strikes).
31.Add the suspension prepared for the negative population (in step 15) to the Near-IR LIVE/DEAD compensation control.
32.Pulse vortex all samples (cells and beads) and then immediately add 1 ml of 1% formaldehyde solution. Vortex lightly again and incubate at 4°C in the dark for 30 min.
33.Centrifuge 5 min at 800 × g. Decant the supernatant and break the pellet (2 strikes).
34.Resuspend pellets in 225 μl FACS-PBS.
35.Pellet cells by centrifuging 5 min at 300 × g.
36.Cap tubes and keep at 4°C, protected from light. Analyze within 24 hr for best reproducibility.
37.Analyze on a NovoCyte Quanteon (or equivalent) using the following lasers/filters:
-
488-nm laser with 530/30 bandpass filter (CD8:FITC).
-
637-nm laser with 660/20 bandpass filter (PAN T:APC) and 780/60 bandpass filter (LIVE/DEAD Near-IR)
-
561-nm laser with 586/20 bandpass filter (CD4:PE) and 780/60 bandpass filter (CD1b3:PE-Cy7).
38.Analyze using third-party software (Fig. 12).
Support Protocol 3: HARVESTING AND FREEZING GUINEA PIG SPLENOCYTES
This support protocol describes the collection and freezing of splenocytes for use as reference controls in longitudinal studies involving flow cytometry. Process the spleen within 30 min of euthanizing the guinea pig and before removing other organs for fixation or freezing.
NOTE : Follow the American Veterinary Medicine Association (AVMA) approved guidelines for CO2 anesthesia of rodents (https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.584/attachments/Guidelines-on-Euthanasia-2020.pdf).
Materials
- Guinea pig
- Ca2+- and Mg2+-free phosphate-buffered saline (PBS), pH 7.4 (Gibco 10-010-049), sterile, 4°C
- Fetal bovine serum (FBS), heat inactivated (Atlanta Biologicals S11510H)
- Gibco RPMI 1640 medium without phenol red (Fisher 11-835-030)
- DMSO, Hybri-Max grade (Sigma D2650-5 × 5ML)
- Biosafety cabinet
- CO2 tank and appropriate container for euthanizing animal, as approved by the Institutional Animal Care and Use Committee
- 150 × 15-mm petri dishes (Fisher FB0875714 or equivalent)
- Ice bucket or Tupperware container filled with ice
- Scalpel, sterile
- gentleMACS 70-μm SmartStrainers (Miltenyi Biotec 130098462 or equivalent)
- 50-ml polypropylene centrifuge tubes (Fisher 07-201-332)
- 5-ml syringes (Fisher 14-823-16D or equivalent)
- Beckman Coulter Allegra X-14R tabletop centrifuge with canister kit (or equivalent), 4°C
- 2-ml cryogenic tubes (Fisher 03-337-7D or equivalent)
- Mr. Frosty™ freezing container (Fisher 15-350-50)
1.Euthanize the guinea pig by CO2 inhalation.
2.Working in a biosafety cabinet, open the body cavity and remove the spleen under sterile conditions.
3.Place the spleen in a sterile 150 × 15-mm petri dish that is sitting in Tupperware container (or equivalent) filled with ice.
4.Add enough cold, sterile PBS to keep moist and slice the spleen into several small pieces using a sterile scalpel blade.
5.Place the pieces of spleen into a 70-μm strainer attached to a 50-ml polypropylene centrifuge tube.
6.Gently press the pieces of spleen through the strainer using the rubber side of the plunger from a 5-ml syringe.
7.Centrifuge in precooled centrifuge 5 min at 300 × g , 4°C.
8.Remove the supernatant, resuspend the pellet in 10 ml cRPMI, and place the tube on ice.
9.Perform a cell count and adjust the concentration of cells in the tube to 5 × 106 cells/ml by pelleting and then diluting in freshly prepared cold freezing medium.
10.Aliquot into cryogenic tubes.
11.Freeze slowly, by placing the cryogenic tubes in a Mr. Frosty freezing container and leaving at –80°C for at least 4 hr before transferring to an ultra-low-temperature freezer (–150°C).
Basic Protocol 6: MONITORING GUINEA PIG IMMUNE RESPONSE TO RICKETTSIAL INFECTION: LEUKOCYTE INFILTRATION OF SKIN AT THE TICK BITE SITE BY FLOW CYTOMETRY
In this protocol, we present a method to monitor leukocyte infiltration in guinea pig skin following the bite of a Rickettsia-infected tick, as previously described (Cross et al., 2022). Infected ticks are placed on the guinea pig for an appropriate time (determined by pilot studies or previous research) and then skin biopsies are collected. The skin biopsies undergo enzymatic and mechanical dissociation followed by immunofluorescence staining and flow cytometric analysis. As with any light-sensitive experiment, it is important to process samples quickly and efficiently. We recommend working in a three-person team for this protocol to achieve the best results. For the reagent preparation calculations described below, we assume eight samples for the tissue dissociation steps (five experimental tissue samples, two blood samples for the FMO controls, and one reference control). After tissue dissociation, there are then 22 samples during the remaining sample preparation steps (five experimental, six FMO controls, six compensation controls, one unstained control, one unstained sample for the CD45 FMO preparation, one reference control, and two extras).
Materials
- Lightning-Link PE-Cy7 Conjugate (Abcam ab102903)
- Lightning-Link Rapid DyLight 405 Conjugate (D:405; Abcam ab201798)
- Mix-n-Stain (CF594) Antibody Labeling Kit (Biotium 92236)
- Mouse anti-Guinea Pig CD1b3 Antibody (Bio-Rad MCA566GA)
- Mouse anti-Guinea Pig CD45 Antibody (Bio-Rad MCA1130)
- Mouse anti-Human L1 Antibody (Bio-Rad MCA387)
- Bovine serum albumin (BSA; Sigma A3059-100G)
- Ca2+- and Mg2+-free phosphate-buffered saline (PBS), pH 7.4 (Gibco 10-010-049)
- Gibco RPMI 1640 medium without phenol red (Fisher 11-835-030)
- Fetal bovine serum (FBS), heat inactivated (Atlanta Biologicals S11510H)
- Guinea pig serum (Sigma G9774-5ML)
- Multi Tissue Dissociation Kit 1 (Miltenyi 130-110-201)
- DNase I (ThermoFisher Scientific 18047-019)
- Magnesium chloride solution (Sigma 63069-100ML)
- Guinea pig splenocytes for use as a reference control (see Support Protocol 3)
- Gentamicin solution (Sigma G1397-10ML)
- MACS Tissue Storage Solution (Miltenyi 130-100-008)
- Near-IR LIVE/DEAD Fixable Dead Cell Stain (Invitrogen L34976)
- DMSO, anhydrous (Invitrogen D12345)
- BD Pharm Lyse Lysing Buffer (BD Biosciences 555899)
- Cell culture water (Sigma w3500)
- 16% formaldehyde solution, methanol-free (Thermo Scientific 28906)
- 10× Intracellular Staining Permeabilization Wash Buffer (BioLegend 421002)
- Millipore purified water (0.1 µm filtered)
- Propidium iodide staining solution (Invitrogen 00-6990-50)
- Mouse anti-Guinea Pig Anti CD8:FITC Antibody (Bio-Rad MCA752F)
- Guinea pig
- Viability Dye Compensation Standard (Bangs Laboratories 451)
- 0.4% trypan blue stain (ThermoFisher Scientific T10282)
- Simply Cellular anti-Mouse Beads for Violet Laser (Bangs Laboratories 835)
- OneComp eBeads Compensation Beads (Invitrogen 01-1111-42)
- Mouse anti-Guinea Pig Anti CD4:PE Antibody (Bio-Rad MCA749PE)
- True-Stain Monocyte Blocker (BioLegend 426102)
- Novocyte Quality Control and Calibration Particles (Agilent 8000004)
- 12 × 75-mm Falcon round-bottom polypropylene flow tubes (Fisher 14-959-11A)
- 12 × 75-mm Falcon round-bottom polystyrene flow tubes (Fisher 14-959-1A)
- 12 × 75-mm Falcon round-bottom polystyrene flow tubes with 35-µm Cell Strainer Cap (Fisher 08-771-23)
- 13-mm Nalgene Unwire™ test tube racks (Fisher 14-809-45 or equivalent)
- 15- and 50-ml polypropylene centrifuge tubes (Fisher 05-538-53D and Fisher 05-538-55A)
- Beckman Coulter Allegra X-14R Tabletop Centrifuge with Canister Kit, tube racks (13 mm), and Biocert covers (or equivalent)
- 60-ml syringe with Luer-Lok Tip (BD Biosciences 301035)
- gentleMACS C Tubes (Miltenyi 130-096-334)
- 0.2-µm syringe filter (PALL 4652)
- 1-ml syringe with attached 25-G, 5/8-inch needle (Fisher 14-826-88 or equivalent)
- pH paper, H 6.0-8.1, 0.3-unit increments (Cytiva Whatman 2629-990)
- Countess Cell Counter chamber slides (ThermoFisher Scientific C10228)
- Countess II FL Automated Cell Counter (Life Technologies) with EVOS LED RFP Light Cube (Invitrogen AMEP4952) or equivalent
- 250- to 500-µl EDTA-coated BD Microtainers (Fisher 02-669-33)
- Tube Revolver/Rotator (Thermo Scientific 88881001) or equivalent
- gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec) or equivalent
- gentleMACS 70 µm SmartStrainers (Miltenyi 130-098-462)
- NovoCyte Quanteon Flow Cytometer (or equivalent)
- 0.1-µm cap filters (PALL 4481)
In the day(s) before starting sample preparation
1.Conjugate PE-Cy7 to Mouse anti-Guinea Pig CD1b3, DL405 to Mouse anti-Guinea Pig CD45, and CF594 to Mouse anti-Human L1.
2.Titrate all the antibodies to determine the optimal antibody concentrations, as described in Basic Protocol 6, step 3.
3.Prepare 1.0% (v/v) BSA in Ca2+- and Mg2+-free PBS (FMC-PBS), filter through a 0.1-µm filter, and store at 4°C.
4.Prepare RPMI 1640 without phenol red + 5% FBS (cRPMI), filter through a 0.1-µm filter, and divide into 40-ml aliquots. Store at –20°C.
5.Prepare and freeze 250-µl aliquots of guinea pig serum for blocking Fc receptors.
6.Reconstitute enzymes in Multi Tissue Dissociation Kit 1 and divide into appropriate-sized aliquots for your experiment. Store at –20°C.
7.Prepare dilutions of supplemental DNase I. Store at –20°C.
8.Prepare 72-µl aliquots of MgCl2. Store at –20°C.
9.Prepare aliquots of guinea pig splenocytes to use as reference controls.
10.Thaw 110 ml cRPMI (step 4).
11.Add 0.1% (w/v) gentamicin to the MACS Tissue Storage Solution and prepare a 250-µl aliquot in separate microcentrifuge tubes for each sample.
Immediately before starting sample preparation
12.Bring one vial of Near-IR LIVE/DEAD Fixable Dead Cell stain (Component A) and the vial of anhydrous DMSO (Component B; or Invitrogen D12345) to room temperature before removing the caps.
13.Bring aliquots of cRPMI, Enzyme D, Enzyme R, Enzyme A, DNase I, MgCl2, and FCM-PBS to room temperature.
14.Bring the guinea pig serum to room temperature. You will need 220 µl of serum (5 µl/sample × 22 samples: 5 experimental + 6 FMO controls + 2 unstained controls + 1 reference control + 6 compensation controls + 2 extra) × 2 (for surface and intracellular staining).
15.Prepare 6.0 ml lysing buffer (2 ml/sample × 3 samples: 2 experimental + 1 extra) from 10× stock solution of BD Pharm Lyse Lysing Buffer.
-
Dilute 0.6 ml of 10× solution to 6.0 ml with 5.4 ml cell culture water.
-
Warm the 1× solution to room temperature.
-
Check the pH of the 1× solution. The pH should fall within the range of 7.1-7.4.
16.Prepare 10 ml of 2% formaldehyde solution (1.0 ml/sample × 19 samples: 5 experimental + 6 FMO controls + 1 unstained control + 1 reference control + 5 compensation controls + 1 extra) by diluting 2.4 ml methanol-free 16% formaldehyde solution with 16.6 ml PBS (protein-free). Keep at 4°C until ready to use.
17.Prepare 152 ml of 1× permeabilization wash buffer (2.0 ml/sample × 4 washes × 19 samples (5 experimental + 6 FMO controls + 1 unstained control + 1 reference control + 5 compensation controls + 1 extra) by diluting 15.2 ml of 10× Intracellular Staining Permeabilization Wash Buffer with 136.8 ml Millipore water.
18.Thaw splenocytes for reference control (5 × 106 cells).
- a.Prewarm cRPMI to 37°C.
- b.Thaw cryovial at 37°C for 1.5-2 min; flick vial with a finger every few seconds.
- c.
Dropwise (over 30 s), add ∼1 ml prewarmed cRPMI to the cryovial and then transfer contents to a 15-ml centrifuge tube containing 8 ml of prewarmed cRPMI.
All centrifugation steps are performed at room temperature.
- d.Centrifuge 5 min at 300 × g. Decant the supernatant.
- e.Add 4.8 ml prewarmed cRPMI (gives ∼5 ml total volume) and gently resuspend the pellet. Then transfer 1 ml (∼1 × 106 cells) to a flow tube (keep the other 4 ml in a separate flow tube as extra).
- f.
Remove cells for counting and keep the rest at 37°C until needed (step 19).
Mix splenocytes in the flow tubes gently by pipetting up and down. Take 100 μl of splenocytes from the center of suspension from the extra tube and place them into a 1.50ml microcentrifuge tube.
- g.Add 5 μl propidium iodide staining solution to the 100 μl of splenocytes and mix gently. Incubate at room temperature in the dark for 7 min.
- h.Load a Cell Counter Chamber Slide with 10 μl (each side) of the stained splenocytes using a 20-μl pipet. Let settle for 1 min before reading.
- i.
Take two readings (one from each side of the chamber slide) on the Countess II FL Automated Cell Counter. Take an average of the two readings and record the cell count and viability.
Set the light source on the cell counter to “RFP.”
The Reference Control comes from one animal at a single time point. Therefore, it should generate consistent results between time points and thus provide assay-specific quality control (QC) by controlling for run-to-run variation in sample processing and staining. Although often left out of longitudinal studies, this control gives confidence that results in the experimental samples are due to the “treatment” rather than instrument, technical, or analysis variability between runs.
19.Once at room temperature, filter the cRPMI through a 0.2-µm filter using a 60-ml syringe.
20.Prepare the following enzyme cocktail in a 50-ml tube and mix by gently inverting the tube 10-12 times. The final cocktail should contain 0.25% Enzyme A, 0.375% Enzyme D, 0.920% Enzyme R, and an additional 50 U/ml DNase I.
- a.25,918.2 µl cRPMI (2945.25 µl × 8.8 samples)
- b.66 µl MgCl2 (7.50 µl × 8.8 samples)
- c.66 µl Enzyme A (7.50 µl × 8.8 samples)
- d.99 µl Enzyme D (11.25 µl × 8.8 samples)
- e.242.88 µl Enzyme R (27.60 µl × 8.8 samples)
- f.
7.83 µl DNase I (0.89 µl × 8.8 samples).
Adding an extra 10% ensures adequate volume.
21.Aliquot 3 ml of the enzyme cocktail into each of five C Tubes.
22.Warm the remaining cRPMI to 37°C (for use in step 43).
23.Prepare antibody dilutions and store them on ice in the dark.
-
Anti-CD8:FITC is used at 1:1600. Hence, for accurate pipetting, make 64 μl of 1:16 dilution by adding 4 μl of antibody to 60 μl FCM-PBS. You will dilute this further in the antibody cocktail and FMOs.
-
Anti-CD45:DL405 is used at 1:160. Hence, for accurate pipetting, make 16 μl of 1:1.6 dilution by adding 10 μl of antibody to 6 μl FCM-PBS. You will dilute this further in the cocktail and FMOs.
-
Anti-CF594:L1 is used at 1:800. Hence, for accurate pipetting, make 64 μl of 1:16 dilution by adding 4 μl of antibody to 60 μl FCM-PBS; 2 μl of 1:16 in 100 μl = 1:800. You will dilute this further during intracellular staining.
Start sample preparation
24.Draw ∼500 µl of blood from the jugular vein of one guinea pig using a 1-ml syringe with a 25-G, 5/8-inch needle. Dispense ∼350 µl of blood into each of two BD Microtainers, cap the tubes, and invert several times to mix the EDTA with the blood. Place EDTA tube with blood on a tube revolver/rotator at room temperature while you collect tissue samples.
25.Take a 4-mm skin-punch biopsy from each guinea pig at the site of a tick bite. Place each biopsy in 250 µl of MACS Tissue Storage Solution in a 1.5-ml microcentrifuge tube. Transport blood and tissue to the sample preparation area and store at room temperature.
26.Transfer 100 µl blood (∼106 leukocytes) into each of two flow tubes.
27.Add 2 ml of 1× lysing buffer (from step 15) to each flow tube and lightly vortex.
28.Incubate the tubes 5 min at room temperature.
29.Centrifuge for 5 min at 350 × g. Decant the supernatant and break the pellet (two strikes).
30.Wash the lysed cells by adding 3 ml PBS (protein-free) and centrifuging for 5 min at 350 × g. Decant the supernatant with a “bump” and break the pellet (2 strikes).
31.Bring the volume of each tube of lysed cells to exactly 100 µl with PBS (protein-free) and pool the two samples.
32.Bring the vial of Viability Dye Compensation Standard beads to room temperature while end-over-end mixing on the tube revolver/rotator.
33.Mince each 4-mm punch biopsy into ∼15 small pieces using a scalpel (Fig. 13).
34.Transfer each tissue sample into a separate C Tube containing the enzyme cocktail simultaneously.
35.Transfer 100 µl lysed blood from step 31 to each of two C Tubes.
36.Transfer the reference control cells into an eighth C Tube.
37.Tightly close each C Tube until it clicks into place. Attach each C Tube upside down onto the sleeve of the gentleMACS Dissociator, and then attach a heating unit over each tube.
38.Run the gentleMACS pre-loaded program titled “37C_Multi_A_01” for the full 41 min. Continue to the next step while the program is running.
39.Immediately after beginning the program, prepare the Near-IR LIVE/DEAD Fixable Dead Cell stain (see step 12) by adding 100 µl of DMSO to the vial of reactive dye. Mix well and visually confirm that all the dye has dissolved. Protect from light.
40.After 20 min of the dissociation program has elapsed, prepare the Viability Dye Compensation Standard beads as follows.
-
Place two drops of beads in a flow tube designated for Near-IR LIVE/DEAD and another three drops in a tube for a negative population.
-
Wash both tubes by adding 0.5 ml PBS (protein-free), vortexing lightly, and centrifuging 5 min at 350 ×g. Decant the supernatant.
-
Wash the beads a second time as in step 40.b. (above).
-
Re-suspend the Near-IR LIVE/DEAD beads in 50 µl PBS (protein-free) and the negative population beads in 100 µl PBS (protein-free). Place a cap on the tubes to reduce evaporation.
41.Detach the C Tubes from the gentleMACS Dissociator and place them in the centrifuge. Pulse to 100 × g to collect the samples at the bottom of the tube. Do NOT decant.
42.Use a 1000-µl pipet to apply each cell suspension to a separate 70-µm MACS Smart Strainer, placed in 15-ml tubes.
43.Wash each MACS Smart Strainer with 10 ml of prewarmed (37°C) cRPMI.
44.Centrifuge the samples 5 min at 350 × g. Decant the supernatant and break the pellet (two strikes).
45.Wash samples by adding 3 ml PBS (protein-free) and centrifuging 5 min at 350 × g. Decant the supernatant with a “bump” and break the pellet (two strikes).
46.Resuspend in PBS (protein-free):
-
Tissue samples: Resuspend each tube in 870 µl PBS (protein-free) and vortex lightly. Assume ∼130 µl residual volume after decanting in step 45. After adding PBS, the volume will be ∼1000 µl.
-
Blood samples: Resuspend each tube in 885 µl PBS (protein-free) and vortex lightly. Assume ∼130 µl residual volume after decanting in step 45. After adding PBS and removing 15 µl for staining in step 48, the volume will be ∼1000 µl in each of the two tubes, so ∼2000 µl total.
-
Reference control: Resuspend in 885 µl PBS (protein-free) and vortex lightly. Assume ∼130 µl residual volume after decanting in step 45. After adding PBS and removing 15 µl for staining in step 48, the volume will be ∼1000 µl.
47.Use a 1000-µl pipet to pass each cell suspension through a separate 35-µm cell-strainer cap placed on separate flow tubes.
48.From the reference control and blood tubes, remove cells for trypan blue viability staining.
-
a.Mix the cell suspensions in the flow tubes by gently pipetting up and down; then, transfer 15 µl from the middle of the flow tube to a microcentrifuge tube containing 15 µl trypan blue.
-
b.Mix by pipetting up and down; then, load 10 µl of stained cell suspension into each side of a disposable Cell Counter Chamber Slide.
-
c.Let the samples settle for 1 min.
-
d.Take brightfield readings with the Countess II FL Automated Cell Counter.
-
i.Use a gate that excludes any particle <6 µm.
-
ii.Record the cell count, viability, and average size of live cells.
49.Transfer the samples to flow tubes:
- a.Tissue samples : They should already be in flow tubes. Ensure that there is exactly 1 ml in each tube.
- b.Blood samples : Aliquot 200 µl (equivalent to ∼20 µl whole blood) into eight separate flow tubes (6 FMO controls, 1 unstained control, and 1 unstained population for the CD45 FMO). Add 800 µl PBS to each tube to bring the volume to 1 ml.
- c.
Reference control : It should already be in a flow tube. Ensure there is exactly 1 ml in each tube.
The staining volume is critical for the next step.
50.Prepare the single-stained compensation beads and add viability stain to the cells.
- a. Prepare the Viability Dye Compensation Standard:
- i. Add 18 µl of the reconstituted Near-IR LIVE/DEAD fixable dead cell stain directly onto the bead suspension prepared in step 40.Mix by pipetting up and down.
- ii. Incubate at room temperature in the dark for 30 min. Immediately move on to step 50b.
- b. Prepare the Simply Cellular for Violet Laser Beads:
- i. Manually shake the Simply Cellular Beads for 10-15 s to completely resuspend.
- ii. Add 1 drop of Simply Cellular Beads to one tube.
- iii. Add 1 µl anti-CD45:DL405 to the tube. Deposit the antibody directly onto the bead suspension and mix by pipetting up and down.
- iv. Incubate at room temperature in the dark for 30 min. Immediately move on to step 50.c.
- c. Prepare the OneComp Beads:
- i. Lightly vortex the OneComp Beads for 10-15 s to completely resuspend.
- ii. Add 1 drop of OneComp Beads to each of four tubes.
- iii. Add 1 µl anti-CD8:FITC (step 23) to one tube, 1 µl anti-CD4:PE to one tube, 1 µl anti-CD1b3:PE-Cy7 to one tube, and 1 µl anti-L1:CF594 one tube. Deposit the antibody directly onto the bead suspension and mix by pipetting up and down.
- iv. Incubate for 30 min at 4°C in the dark. Immediately move on to step 50.d.
- d. Add viability stain to the cells:
- i. Add 1 µl of the reconstituted Near-IR LIVE/DEAD Fixable Dead Cell Stain to the cell suspensions from step 39 (except for the LIVE/DEAD FMO). Mix by vortexing lightly.
- ii. Incubate for 30 min at 4°C in the dark.
- iii. Centrifuge 5 min at 350 × g. Decant the supernatant and break the pellet (two strikes).
All experimental samples, FMOs, and compensation controls will emerge around the same time, and you should process them together moving forward.
51.Add 3 ml FCM-PBS to each tube.
52.Centrifuge 5 min at 350 × g. Decant the supernatant and break the pellet (two strikes).
53.Add 5 µl of 100% guinea pig serum to each tube to block the Fc receptors and nonspecific binding of antibody to the cells.
54.Add 5 μl Monocyte Blocker to each tube and mix by pipetting up and down.
55.Incubate for 30 min at 4°C in the dark.
56.Prepare the surface antigen-antibody cocktail and FMOs during the 30-min blocking incubation above (using the 1:16 CD8 and 1:1.6 CD45 dilutions that you made in step 23).
- a. For the FMOs : Prepare as follows and store on ice in the dark:
- i. FMO-No LIVE/DEAD: Comes from the assay cocktail.
- ii. FMO-No CD8:FITC:
- 5.0 µl FCM-PBS
- 6.7 µl CD4:PE
- 1.0 µl CD1b3:PE-Cy7.
- 1.0 µl CD45:DL405 (use the 1:1.6 dilution).
- iii. FMO-No CD4:PE:
- 10.7 µl FCM-PBS
- 1.0 µl CD8:FITC (use the 1:16 dilution)
- 1.0 µl CD1b3:PE-Cy7
- 1.0 µl CD45:DL405 (use the 1:1.6 dilution).
- iv. MO-No CD1b3:PE-Cy7:
- 5.0 µl FCM-PBS
- 1.0 µl CD8:FITC (use the 1:16 dilution)
- 6.7 µl CD4:PE
- 1.0 µl CD45:DL405 (use the 1:1.6 dilution).
- v. FMO-No CD45:DL405:
- 5.0 µl FCM-PBS
- 1.0 µl CD8:FITC (use the 1:16 dilution)
- 6.7 µl CD4:PE
- 1.0 µl CD1b3:PE-Cy7.
- vi. FMO-No L1:CF594:
- 4.0 µl FCM-PBS
- 1.0 µl CD8:FITC (use the 1:16 dilution)
- 6.7 µl CD4:PE
- 1.0 µl CD1b3:PE-Cy7
- 1.0 µl CD45:DL405 (use the 1:1.6 dilution).
- b. For the surface antigen-antibody cocktail : Prepare as follows and store on ice in the dark:
- i. 30.8 μl FCM-PBS (7.7 samples × 4.0 μl FCM-PBS/sample).
- ii. 7.7 μl CD8:FITC (7.7 samples × 1.0 μl (1:16) Ab/sample).
- iii. 51.6 μl CD4:PE (7.7 samples × 6.7 μl Ab/sample).
- iv. 7.7 μl CD1b3:PE-Cy7 (7.7 samples × 1.0 μl Ab/sample).
- v. 7.7 μl CD45:DL405 (7.7 samples × 1.0 μl [1:1.6] Ab/sample).
Seven samples will receive the surface antigen-antibody cocktail (5 experimental, 1 reference control, 1 LIVE/DEAD FMO); we add an extra 10% in case of pipetting error, so the total volume of the cocktail will be 105.5 μl. Each sample gets 13.7 μl for surface staining, for a total volume of 95.9 μl. This volume leaves 9.6 μl extra in case of pipetting error.
We included 4.0 μl FCM-PBS per sample to replace the volume of another antibody we originally had in the panel. You could remove this volume of FCM-PBS from the surface antigen-antibody cocktail and FMO cocktails so long as you perform antibody titrations using the same surface staining volume.
57.Label surface antigens.
-
For each FMO: Add the appropriate cocktail (13.7 μl) from step 56a directly onto each sample and mix by pipetting up and down.
-
For each experimental sample, the reference control, and the LIVE/DEAD FMO: Add 13.7 μl of the surface antigen-antibody cocktail from step 56.b. directly onto the sample and mix by pipetting up and down.
58.Incubate for 30 min at 4°C in the dark.
59.Wash all the samples by adding 3 ml of FCM-PBS. Centrifuge 5 min at 350 × g. Decant the supernatant and break the pellet (two strikes).
60.Wash all samples by adding 3 ml PBS (protein-free). Centrifuge 5 min at 350 × g. Decant the supernatant and break the pellet (two strikes).
61.Add the negative population suspension prepared in step 40d to the LIVE/DEAD Viability Dye compensation control.
62.To the LIVE/DEAD Viability Dye compensation control only:
- a.Add 50 µl FCM-PBS to bring the volume up to 200 µl.
- b.Pellet cells by centrifuging 5 min at 350 × g.
- c.
Cap tube and store at 4°C, protected from light. Analyze within 24 hr for best reproducibility.
The LIVE/DEAD Viability Dye compensation control will not perform well if it is subjected to the intracellular staining steps below.
63.Add one drop of the blank population (Tube B) to the Simply Cellular for Violet Laser Beads.
64.Add one tube of unstained cells to the CD45 DL405 FMO tube.
65.Pulse vortex all samples (cells and beads) and then immediately add 1 ml of 2% formaldehyde solution. Vortex lightly again and incubate at 4°C in the dark for 30 min.
66.Centrifuge 5 min at 800 × g. Decant the supernatant and break the pellet (two strikes).
67.Wash by adding 2 ml Permeabilization Wash Buffer, vortexing lightly, and incubating for 5 min at room temperature in the dark. Centrifuge 5 min at 800 × g. Decant the supernatant and break the pellet (two strikes).
68.Repeat step 67.
69.Add 5 µl of 100% guinea pig serum to each tube to block the Fc receptor and nonspecific binding of the antibody to the cells and mix by pipetting up and down.
70.Incubate for 30 min at 4°C in the dark.
71.Label intracellular antigen (use the 1:16 L1 dilution that you made in step 23).
-
For the L1:CF594 FMO: Add 15.0 μl FCM-PBS and mix by pipetting up and down.
-
For each experimental sample, the reference control, and the other FMOs: Add 13.0 μl FCM-PBS and 2 μl L1 CF594 (use the 1:16 dilution) and mix by pipetting up and down.
72.Incubate for 30 min at 4°C in the dark.
73.Wash by adding 2 ml Permeabilization Wash Buffer. Centrifuge 5 min at 800 × g. Decant the supernatant and break the pellet (two strikes).
74.Repeat step 73.
75.Resuspend pellets in 200 µl FCM-PBS.
76.Pellet cells by centrifuging 5 min at 350 × g.
77.Cap tubes and keep at 4°C, protected from light. Analyze within 24 hr for best reproducibility.
78.Analyze by flow cytometry on NovoCyte Quanteon (or equivalent) using the following lasers/filters:
-
405-nm laser with the 445/45 bandpass filter (CD45: DL405).
-
488-nm laser with the 530/30 bandpass filter (CD8:FITC).
-
637-nm laser with the 780/60 bandpass filter (LIVE/DEAD Near-IR).
-
561-nm laser using the 586/20 bandpass filter (CD4:PE), the 615/20 bandpass filter (L1:CF594), and the 780/60 bandpass filter (CD1b3:PE-Cy7).
79.Analyze using third-party software (Fig. 14).
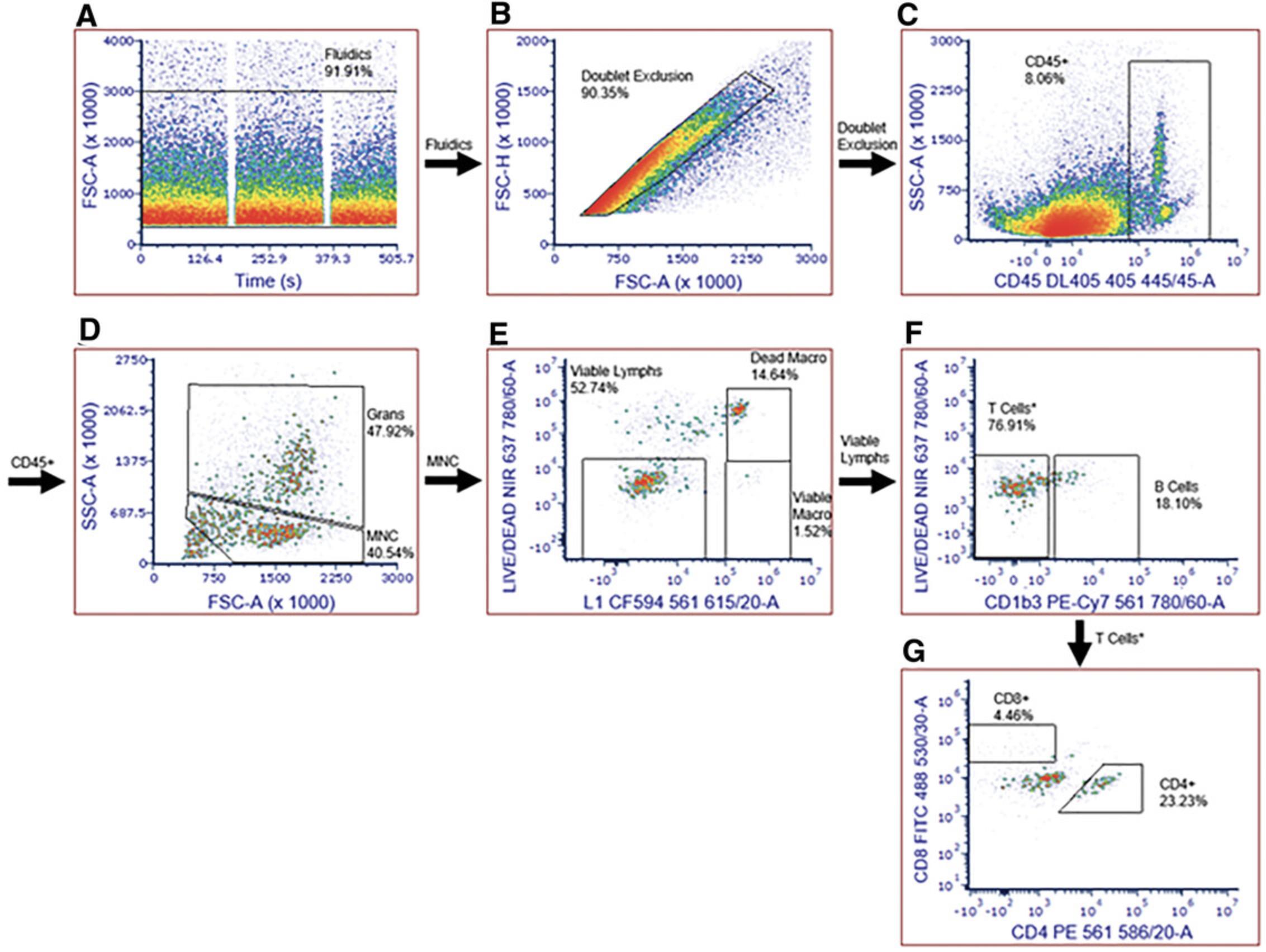
Basic Protocol 7: MONITORING GUINEA PIG IMMUNE RESPONSE TO RICKETTSIAL INFECTION: ANTIBODY TITER BY ELISA
This protocol describes an enzyme-linked immunosorbent assay (ELISA) for monitoring the immune response in guinea pigs to rickettsial infection (Alugubelly et al., 2021). The principal advantages of this assay over the immunofluorescence assay (IFA) are that it is quantitative rather than semi-quantitative, more objective, and more suitable for high-throughput studies or diagnostics. With regard to objectivity, where an IFA titer might be reported at 128, an ELISA readout will provide an exact titer; the titer of 128 from the IFA—presumably negative at 256—might be reported as 225 by ELISA, i.e., much closer to 256 than the 128 reported by IFA. Additionally, the ELISA reliably quantifies positives down to a titer of 75, which would be considered negative by IFA. However, in our hands, a titer of 75 is always greater than the optical density (OD) of the negative + 5 SD, which suggests approximately one chance in a million of a false-positive of a sample that falls within the standard curve (75-1200). Suppose a clinician needs a simple determination of positivity; in that case, this can be obtained with high confidence, albeit non-quantitatively, from a result falling outside the standard curve (<75) by observing the OD negative + 3 SD rule. Considering objectivity, we know that a plate reader is more objective than a human, who may or may not be well-trained and highly experienced and is potentially more prone to error. Finally, using 96-well plates and an automated reader will provide higher throughput.
One person can perform most of the experiment. To aid in understanding the calculations we use, we are taking a 96-well plate ELISA as our template for this protocol and adding a few “extra” wells to ensure that we have an adequate volume of reagents needed to perform the experiment.
Materials
- ELISA Strips, slightly hydrophilic (Thermo Fisher Scientific 467120)
- Water, molecular biology grade (Fisher BP28191)
- Goat anti-Guinea Pig IgG (H+L) Secondary Antibody, HRP conjugated (Thermo Fisher A18775)
- 10× ELISA Wash Buffer (Bio-Rad BUF031A)
- FCM-PBS: PBS + 1.0% (v/v) BSA (filtered through a PALL 4481 0.1-µm filter; see Basic Protocol 5, step 2)
- Millipore purified water (filtered through a 0.1-µm filter; Millipore-Sigma)
- TMB substrate solution (Thermo Fisher Scientific N301)
- Stop solution for TMB substrates (Thermo Fisher Scientific N600)
- Reagent reservoirs (Fisher 07-200-127 or equivalent)
- Tube Revolver/Rotator (Thermo Scientific 88881001) or equivalent
- 0.1-µm filters (PALL 4481)
- Thermo Scientific™ Nunc™ Sealing Tapes (Thermo Fisher Scientific 232698)
- Protein (Eppendorf 022431064)
- Fisherbrand™ accuWash™ microplate washer (or equivalent)
- Microplate shaker (Fisher 88-861-023 or equivalent)
- Synergy/H1 microplate reader (BioTek) or equivalent
- Microplate analysis software (Gen5 version 3.09, BioTek, or equivalent
- Additional reagents and equipment for coating ELISA plates (Support Protocol 4)
In the week(s) before starting
1.Coat the ELISA plates (strips).
2.Obtain seropositive and seronegative control plasma and determine the titer of the seropositive control by IFA.
In the day(s) before starting
3.Reconstitute the goat anti-guinea pig IgG (H+L) secondary antibody.
-
Add 1.1 ml molecular-biology-grade (protease-free) water per 1 mg antibody.
-
Mix gently by pipetting up and down.
-
Place on tube revolver/rotator for 30-45 min at room temperature or end-over-end mixing.
-
Centrifuge to remove aggregates.
Immediately before starting
4.Bring the coated ELISA strips to room temperature at least 1 hr before the start of the experiment.
5.Bring 10× ELISA Wash Buffer, guinea pig plasma (step 2), and 65 ml FCM-PBS to room temperature at least 1 hr before the start of the experiment.
6.Place 10× Wash Buffer in the cell culture incubator and occasionally swirl until in solution.
7.Prepare ∼320 ml of 1× wash buffer (96 wells per full ELISA plate + 10 extra wells [∼10% overage] = 106 wells; 106 wells × 0.3 ml/wash × 8 washes = 254.4 ml for washes + 60 ml [for dead space at bottom of plate washer] = 314.4 ml).
- a.Dilute 32.0 ml of 10× ELISA Wash Buffer to 320.0 ml with 288.0 ml Millipore water.
- b.Place in the plate washer reservoir.
- c.
Prime the plate washer (use the “prime” button on the plate washer).
The plate washer only needs priming before the first wash.
8.Prepare a stock positive control plasma with a titer of 1200.
- a.In this example, the antibody titer of seropositive plasma after 59 days of infection was ≈7000 (as determined by IFA; see step 2).
- b.
Take 10 µl of plasma with titer 7000 and dilute to 58.3 µl with 48.3 µl FCM-PBS ≡ plasma with a titer of 1200.
The amount of antibody in 1 µl seropositive plasma with a titer of 7000 ≡ (is equivalent to) 5.83 µl of seropositive plasma with a titer of 1200. Note that we achieved a titer of 7000 through repeated inoculations of our donor guinea pig.
Perform assay protocol
9.Prepare 1:7000 dilution of stock seropositive plasma (titer 1200) for the standard curve (Fig. 15): Make 3.6 ml diluted plasma at a 1:1200 dilution by taking 3.0 µl from the titer 1200 stock (step 8) and mixing it with 3597.00 µl of FCM-PBS in a 5-ml tube. Mix well by gently pipetting 5-7 times with the pipet set to 1000 µl. This dilution yields plasma with a final dilution of 1:7000 from the original seropositive plasma.
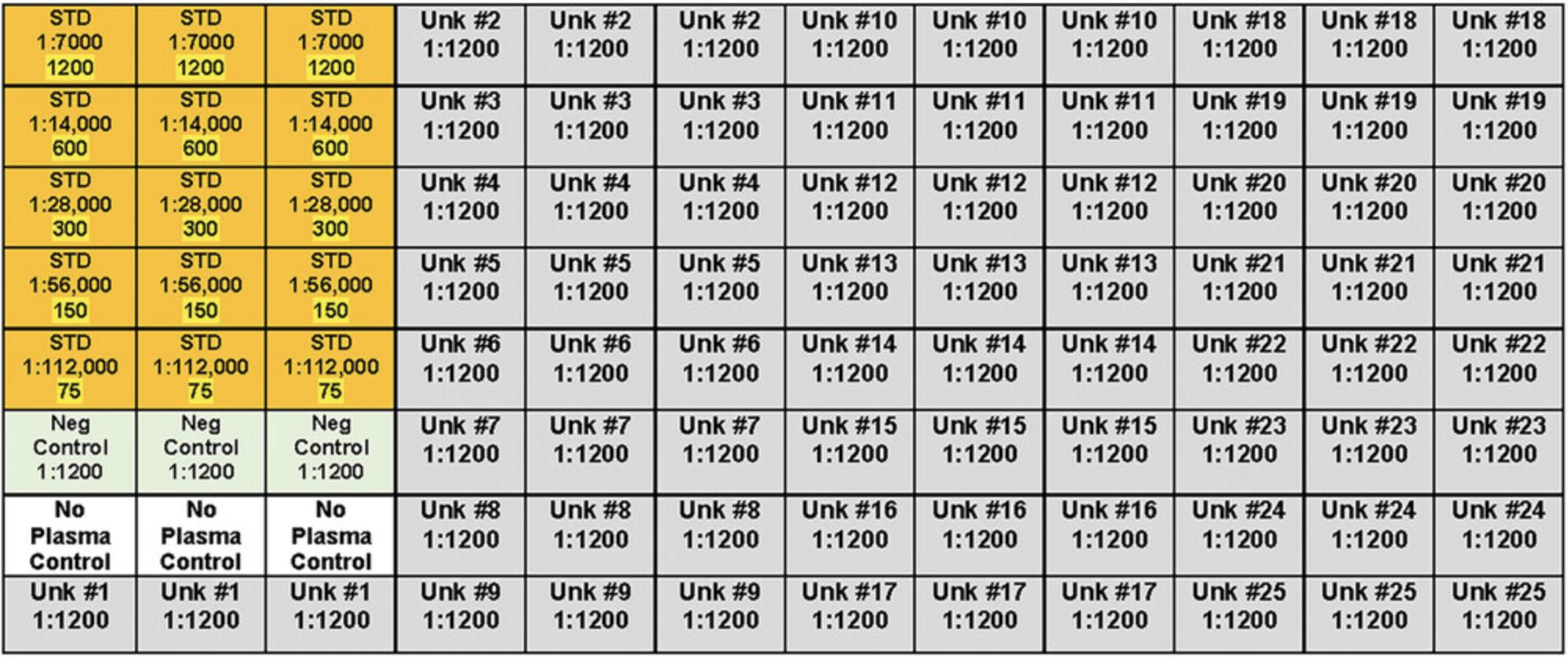
10.Prepare samples for the standard curve.
- a.1:7000 (titer 1200);
- b.1:14,000 (titer 600);
- c.1:28,000 (titer 300);
- d.1:56,000 (titer 150);
- e.
1:112,000 (titer 75).
Start with 1100 µl of 1:7000 (titer 1200) from step 10a (above) and then serially dilute by carrying 700 µl forward into 700 µl FCM-PBS for each dilution listed below. For each dilution, mix well by gently pipetting 5-7 times with the pipet set to 1000 µl before moving to the next dilution.
11.Prepare negative plasma controls (Fig. 15): Make 3.6 ml of diluted seronegative plasma at a 1:1200 dilution by taking 3.0 μl seronegative plasma and mixing it with 3597.0 μl of FCM-PBS in a 5-ml tube. Mix well by gently pipetting 5-7 times with the pipet set to 1000 μl.
12.Prepare “Unknowns” (dilution 1:1200). For each unknown (Fig. 15), make 3.6 ml diluted plasma at a 1:1200 dilution by taking 3.0 µl from an “Archive” stock and mixing it with 3597.0 µl of FCM-PBS in a 5-ml tube. Mix well by gently pipetting 5-7 times with the pipet set to 1000 µl.
13.Add 100 µl of the appropriately diluted plasma samples corresponding to the plate setup shown in Fig. 15.
14.Cover the plates (strips) with sealing tape.
15.Incubate at room temperature for 2 hr on the microplate shaker at 250 rpm.
16.Wash the plates (strips) four times with 300 µl of 1× wash buffer using a plate washer. During the fourth wash, let the plate soak for 5 min before removing the buffer.
17.Add 100 µl of the HRP-conjugated secondary antibody (diluted 1:12,000 in FCM-PBS) to each well.
18.Cover plates (strips) with sealing tape.
19.Incubate at room temperature for 1 hr with gentle (250 rpm) shaking
20.Wash the plates (strips) four times with 300 µl of 1× wash buffer using a plate washer. During the fourth wash, let the plate soak for 5 min before removing the buffer.
21.Add 100 μl TMB substrate to each well and incubate for 30 min.
- a.Place the plate on the shaker.
- b.Set the timer to the desired and predetermined substrate incubation time (30 min).
- c.Start the timer with the addition of substrate to the first well or set of wells.
- d.Use a consistent pipetting pattern and rate to add substrate to all the wells (e.g., always move from the first row to the last row).
- e.Cover with aluminum foil and start the shaker.
- f.
When the timer signals the end of the incubation period, stop the reaction (step 24).
It is essential that the timing of the reaction in every well in every plate be controlled as precisely as possible to have an endpoint assay that provides reliable and consistent results. Because enzyme-substrate reactions are kinetic, timing from the start to the end of the reaction will affect the final concentration of the product developed. To ensure precise timing, follow this procedure for every assay that you perform regardless—even when only a few wells are involved.
Turn on the plate reader when there is 15-20 min left in incubation (as it needs a 20-min warmup).
22.Stop the reaction by adding 100 μl stop solution to each well. Tap the side of the plate with your fingertips until no blue is visible.
23.Read the plate at 450 nm.
24.Analyze the results.
Support Protocol 4: COATING ELISA PLATES WITH RICKETTSIAL ANTIGEN
Here we describe the preparation of rickettsial antigen for, and antigen coating of, ELISA plates for Basic Protocol 8.The protocol is written for use with the BSL-2 SFGR, R. parkeri , and should be modified appropriately for a BSL-3 SFGR. We use whole-cell rickettsiae for ELISA for plating, grown in Vero cells, and collected by mechanical disruption. We recommend following protocols as in Ammerman et al. to grow SFGR (Ammerman et al., 2008).
CAUTION : Perform all activities involving handling infectious materials at the proper biosafety level conditions for the bacteria cultured (see Basic Protocol 1).
Materials
- R. parkeri or other BSL-2 SFGR
- Vero cells
- Tissue-culture-treated 75-ml vented flasks (Fisher 13-680-65 or equivalent)
- Millipore water (0.2 µm filtered)
- 5× ELISA Coating Buffer (Bio-Rad BUF030A)
- ELISA Ultrablock (Bio-Rad BUF033A)
- Bacterial Viability Kit (Invitrogen L7012)
- Tube Revolver/Rotator (Thermo Scientific 88881001) or equivalent
- Class II biosafety cabinet
- Sharps container
- Cell scraper, sterile (Fisher 08-771-1A or equivalent)
- 1-, 5-, 10-, 25-, and 50-ml serological pipets
- 50-ml Luer-Lok syringes without needle (Fisher 13-689-8 or equivalent)
- 27-G, 1/2-inch needles (Fisher 14-826-48 or equivalent)
- 25-G, 5/8-inch needle (Fisher 14-826AA or equivalent)
- 18-G, 3-inch needles (Fisher 14-821-16R or equivalent)
- Whatman Swin-Lock Plastic Filter Holder, autoclaved (Fisher 09-927-168)
- Nucleopore Track-etched Membrane Filters, 3.0 µm, 47 mm (Fisher 09-800-917)
- Petroff-Hausser counting chamber (Fisher 02-671-51B) or equivalent
- ELISA Reagent Reservoir (Fisher PI15075) or equivalent
- ELISA Strips (plates)
- Slightly Hydrophilic Immuno Clear Standard Modules (Thermo Scientific, 467120)
- Sealing tape for 96-well plates (Thermo Scientific 15036)
In the week(s) before starting
1.Cultivate R. parkeri or other BSL-2 SFGR in 75-cm2 (T75) tissue culture flasks with Vero cells until 75%-90% of cells are infected.
2.Determine the total number of bacteria needed to seed wells with 5 × 107 rickettsiae per well and thus the total number of T75 flasks needed to achieve that goal.
Immediately before starting
3.Prepare 1× coating buffer, making 33.0 ml for each set of eight strips, or 96 wells (96 coated wells + 14 extra wells = 110 wells × 0.1 ml = 11.0 ml total × 3 plates = 33.0 ml total), by diluting 6.6 ml of 5× ELISA Coating Buffer to 33.0 ml with 26.4 ml Millipore water.
4.Leave mixing on the end-over-end mixer until use.
Harvesting rickettsiae
5.Harvest SFGR-infected Vero cells in a Class II biosafety cabinet that contains the required materials, including a small sharps container. Use a cell scraper or serological pipet to scrape and blow off, respectively, SFGR-infected Vero cells from the T75 flask(s) into culture medium.
6.Transfer suspension(s) to appropriately sized tubes (e.g., 50 ml)
7.Set up a 50-ml syringe (plunger removed) with a 25-G, 5/8-inch needle over a fresh 50-ml tube and transfer suspension to the syringe. Replace the plunger and pass suspension through to mechanically rupture host cells.
8.Draw up the suspension with an 18-G, 3-inch needle on a 50-ml syringe, carefully replace the needle with a 27-G, 1/2 inch needle, and repeat the host cell rupture.
9.Centrifuge the 50-ml tube(s) with the suspension for 10 min at 1000 × g to pellet the cellular debris.
10.Transfer the suspension to a 50-ml syringe (with the plunger removed) attached to a holder with a Nucleopore 3-µm filter sitting over a fresh 50-ml tube and filter passively into the tube.
11.Aliquot 1-1.2 ml of suspension into 1.5-ml microcentrifuge tubes while continually mixing the solution by pipetting up and down.
12.Centrifuge 10 min at 12,000 × g and combine pellets into one tube. Repeat centrifugation, and then wash pellet three times with PBS by resuspending and then centrifuging 10 min at 12,000 × g each time.
13.Make 1 ml each of 1:100 and 1:1000 dilutions of enriched rickettsial suspension in 1.5-ml microcentrifuge tubes for Live/Dead staining and counting using the Bacterial Viability Kit. Place the remainder of the rickettsial suspension in a 56°C water bath for 60 min to heat-kill the bacteria.
14.Load a Petroff-Hauser counting chamber for total bacterial count under epifluorescence.
15.For seeding the ELISA plate, use the volume of rickettsiae needed (determined before starting the protocol) to achieve a number for seeding and centrifuge that volume for 10 min at 12,000 × g to pellet the rickettsiae.
16.Remove the supernatant and resuspend the pellet in freshly prepared 1× coating buffer to achieve the desired concentration. Mix on end-over-end mixer for 5-15 min.
17.Transfer the suspension to an ELISA reagent reservoir and use a multichannel pipet to aliquot 100 µl into each well of Slightly Hydrophilic Immuno Clear Standard Modules.
18.Incubate the plate(s) at 4°C overnight; protect the plate from light and cover with sealing tape to prevent evaporation.
Day 2: Washing antigen-coated plates
19.Before starting, prepare 275.0 ml of 1× wash buffer for each set of eight strips, or 96 wells (96 coated + 20 extra = 116 wells × 3 plates × 0.3 ml/wash × 2 washes = 208.8 ml total + 60 ml [for dead space at bottom of automatic plate washer] = 268.8 ml), by diluting 27.5 ml of 10× ELISA Wash Buffer to 275.0 ml with 247.5 ml Millipore water.
20.Dump the coating buffer in the plate(s) from step 18 by flicking the plate(s) over a sink and then tapping the plate(s) upside down on absorbent paper.
21.Wash plate(s) twice with the plate washer with 300 µl of 1× wash buffer.
22.Examine a strip under an inverted microscope to visualize antigen (rickettsiae) on wells.
23.Block nonspecific binding by adding 200 µl Ultrablock to each well. Cover the plate(s) with sealing tape, place foil over it to protect from light, and incubate at room temperature overnight (24-26 hr)
Day 3: Final plate processing
24.Dump the Ultrablock from step 23 by flicking the plate(s) into the sink and tapping the plate(s) upside down on absorbent paper.
25.Dry the plate(s) in the biosafety cabinet (fan on, lights off) for 2-3 hr.
26.Place the strips in a sealed plastic bag or storage cabinet with desiccant and protected from light; store dried for up to 1 year at 4°C.
Alternate Protocol 2: MONITORING GUINEA PIG IMMUNE RESPONSE TO RICKETTSIAL INFECTION: ANTIBODY TITER BY IMMUNOFLUORESCENCE ASSAY
Indirect immunofluorescence assays (IFA) are used to monitor the guinea pig's immune response to rickettsial infection by detecting antibodies against Rickettsia spp. Although we typically assess multiple serum samples over a longitudinal study to monitor the development of an immune response, it is crucial to include a timepoint before placing ticks on the guinea pigs or before inoculation with infectious agents to collect baseline data. Following the U.S. Centers for Disease Control and Prevention (CDC) criteria for human diagnostics, we recommended waiting at least 14 days after exposure or infestation to evaluate seroconversion in guinea pigs.
First, you process samples by applying pre-diluted sera from test animals onto antigen-coated slides. Next, add a fluorescence-labeled secondary antibody against the guinea pig IgG in the sera. Then, evaluate the results through fluorescence microscopy to quantify the immune response. Finally, results are reported as the final titer (dilution) at which you detect specific fluorescence for a sample. Titers corresponding to dilutions are reported as the reciprocal of the dilution: e.g., if the highest dilution with a positive result is 1/128, then the titer is reported as 128.Alternatively, some people define the titer to be 1:128; but we will use 128 for this protocol.
CAUTION : Perform all activities involving handling infectious materials at the proper biosafety level conditions for the bacteria cultured.
Materials
- Rickettsia spp.
- Ca2+- and Mg2+-free phosphate-buffered saline (PBS), pH 7.4 (Gibco 10-010-049)
- Sodium azide (CAS 26628-22-8)
- Bovine serum albumin (BSA), gamma-globulin free (Sigma A7030)
- Eriochrome black T powder (CAS 1787-61-7)
- Deionized water (in house system or commercially available)
- Acetone (CAS 67-64-1)
- Anti-Guinea Pig IgG (H+L) Antibody, FITC-Labeled (Sera Care 5230-0303)
- Goat Serum Donor Herd (Sigma G6767)
- Glycerol (CAS 56-81-5)
- DABCO (CAS 280-57-9)
- Guinea pig serum (at least 40 μl per sample)
- Coverslip sealant (Biotium 23005 or equivalent)
- Biosafety cabinet class 2 type A2 (NuAire NU-543 or equivalent)
- 5-mm sterile glass beads (Thomas Scientific 1177Q81 or equivalent)
- Polypropylene Falcon tubes (size dependent on antigen production volume)
- Serological pipet (Cole Parmer EW-25200 or equivalent)
- Disposable serological pipets (size dependent on antigen production volume)
- Centrifuge (Thomas Scientific 75377406 or equivalent)
- Glass or Teflon-coated slides, well number based on user preference (Immuno-Cell 61.100.32 or equivalent)
- Microfiber cloth or Kim wipes (KimTech 34705 or equivalent)
- Capillary tube (Flinn Scientific GP7046 or equivalent)
- Calibrated pipets or piston-type pipettors with appropriate filtered disposable tips
- Inverted scope (Zeiss Axio Vert.A1 or equivalent)
- Coplin jar or slide staining chamber (Fisher Scientific 08-813A or equivalent)
- Slide storage boxes (Electron Microscopy Science 71550 or equivalent)
- Desiccator (Fisher Scientific 08-647-26 or equivalent)
- Desiccant pack (Fisher Scientific 09-928-142 or equivalent)
- Vortex (Scientific Industries SI-P236 or equivalent)
- 1.5-ml polypropylene screw-cap tubes (Thermo Scientific 3467TS or equivalent)
- Buffer reservoirs (VWR 82031-548 or equivalent)
- U- or V-bottom microplates (Corning 3799 or equivalent)
- Lab tape (Thomas Scientific LT-18WH or equivalent)
- Humidity chamber (in-house or Fisher Scientific 23-769-522)
- Incubator (Thermo Forma 3110 or equivalent)
- Magnetic stir bar (VWR 58948-138 or equivalent)
- Magnetic stirrer (Fisher Scientific S88857200 or equivalent)
- Plastic wash bottle (Thermo Scientific 2407-1000 or equivalent)
- Light-safe microcentrifuge tubes (Cole Parmer UX-06333-80 or equivalent)
- Amber glass oval dropper bottle (Spectrum Chemical 550-81013-CS or equivalent)
- 24 × 60-mm coverslips (Corning 2975-246 or equivalent)
- Refrigerator, 2-8° C
- UV fluorescence microscope (ZEISS Axio Lab.A1 or equivalent)
- Slide folder (Mortech Manufacturing BH014 or equivalent)
Preparation for IFA
In-house production of IFA slides
1.Inoculate a cell culture with the appropriate Rickettsia species to coat IFA slides.
2.Propagate Rickettsiae to a suitable infection level.
3.Add 5-mm-diameter sterile glass beads to the culture flask to remove the monolayer.
4.Transfer the lifted monolayer to an appropriately sized polypropylene Falcon tube using a serological pipet.
5.Pipet mix material to disperse any large clumps.
6.Centrifuge samples for 30 min at 10,000 × g , 20°C.
7.Remove the supernatant with an appropriately sized serological pipet and discard.
8.Resuspend the pellet in antigen diluent using one-third the volume of the reference value noted in step 7.
9.Inactivate antigen stocks.
10.Clean blank glass or Teflon-coated slides with a microfiber cloth or Kimwipes.
11.Apply 3 µl of antigen (from step 9) with a micropipet or capillary tube to a single well.
12.Observe the concentration of cells under an inverted microscope before the well dries.
- a.If there are too many cells, dilute the antigen with antigen diluent and reassess the number of cells until you determine an appropriate concentration is present.
- b.
If there are too few cells, centrifuge the antigen for 10 min at 1000 × g , room temperature, remove supernatant, and resuspend in a smaller volume of antigen diluent.
The well should not be overpopulated with cells (clustered heavily throughout the well and difficult to differentiate individual cells) or underpopulated (difficult to find cells close together).
13.Aliquot 3 µl of appropriately diluted antigen to each well on slides.
14.Allow the slides to dry for at least 2 hr.
15.Place slides into a slide staining chamber or Coplin jar and fix in acetone for 15 min.
16.Remove slides and allow to air dry.
17.Resuspend the Anti-Guinea Pig IgG (H+L) Antibody, FITC-Labeled conjugate, with sterile water.
Preparation of secondary antibody conjugate
18.Determine the optimal dilution of the anti-guinea pig IgG-FITC conjugate using a checkerboard titration.
19.Prepare IFA buffer, 1.65% Eriochrome counterstain, and mounting medium solutions (see recipes in Reagents and Solutions) before beginning the remainder of the protocol.
Indirect immunofluorescence assay
20.Warm the IFA slides to room temperature before use.
21.Thaw test and control sera and vortex briefly.
22.Aliquot an appropriate amount of IFA buffer into buffer reservoir for use in steps 23 and 24.
23.Prepare initial working dilutions of test and control sera in U- or V-bottom microplates.
- a.Pre-record sample identifiers and testing dilutions for all slides.
- b.Tape off microplates in the same setup as the slide you are using to avoid later loading errors.
- c.
Load appropriate volumes of IFA buffer into all wells designated for starting 1/16 dilutions before loading samples.
Commonly used dilution systems are 1/10 and 1/16.We use the 1/16 system throughout this protocol.
The necessary starting volume will depend on how many assays or replicates you plan to use for assessment. For a single assay run, a standard dilution setup for a 1/16 dilution is 5 µl sera + 75 µl IFA buffer (see recipe).
- d.
Load serum samples into appropriate wells to make 1/16 dilutions of all samples.
Make the 1/16 dilutions in a 96-well U- or V-bottom plate for immediate use. Keep the plate refrigerated and covered (for example, with aluminum foil). Use within 2 hr, as the antibody will bind to the plate if left too long.
Although you should avoid this for longer-term storage, it is possible to make the 1/16 dilutions in polypropylene screw-cap tubes. The diluted serum will be stable at 4°C for at least 1 month. You can also freeze these dilutions for long-term storage. Higher dilutions of antibodies are not stable, so we do not recommend further dilution until the day of use.
24.Prepare the remaining dilutions of test and control sera, pipet mixing all samples throughout step 24.
- a.For the positive control: Dilute to the known endpoint titer; see step 24c for suggested mixing volumes. Always assess positive controls using twofold serial dilutions to the endpoint titer.
- b.
If screening serum: Make 1/256 dilutions of test sera in the same 96-well plate by aliquoting 5 µl of the 1/16 dilution into a new well with 75 µl of IFA buffer.
Use this process to assess where it is logical to start the endpoint titration. If a sample is positive at 1/16 and not 1/256 in the screen, you only need to evaluate dilutions 1/16 through 1/128 in a subsequent run. If a sample is positive at 1/256, then the subsequent endpoint titration should begin at 1/256 to save time and slide space.
- c.
If titrating to the endpoint: Dilute the test sera samples using twofold dilutions by aliquoting 35 µl of the previous dilution with 35 µl of IFA Buffer into a fresh well.
Plate templates with suggested volumes to generate for both the screening and endpoint titer setups described in step 23 are provided in Table 4.
Endpoint titers will vary; we recommended running samples out to 1/2048 or 1/4096 for the end-dilution value on the first assessment.
| For each setup, read from left to right starting in row 1. | |||||||
|---|---|---|---|---|---|---|---|
| Screening slide | |||||||
| Positive control | 1/32 | 1/64 | 1/128 | 1/256 | 1/512 | 1/1024 | 1/2048 |
| 75 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent |
| 5 µl serum | 35 µl 1/16 | 35 µl 1/32 | 35 µl 1/64 | 35 µl 1/128 | 35 µl 1/256 | 35 µl 1/512 | 35 µl 1/1024 |
| Sample 1 1/16 | 1/256 | IFA buffer only | IFA buffer only | Sample 2 1/16 | 1/256 | IFA buffer only | IFA buffer only |
| 75 µl diluent | 75 µl diluent | 75 µl diluent | 75 µl diluent | ||||
| 5 µl serum | 5 µl 1/16 | 5 µl serum | 5 µl 1/16 | ||||
| IFA buffer only | IFA buffer only | Sample 3 1/16 | 1/256 | IFA buffer only | IFA buffer only | IFA buffer only | IFA buffer only |
| 75 µl diluent | 75 µl diluent | ||||||
| 5 µl serum | 5 µl 1/16 | ||||||
| IFA buffer only | IFA buffer only | IFA buffer only | IFA buffer only | Negative control | |||
| 75 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | ||||
| 5 µl serum | 35 µl 1/16 | 35 µl 1/32 | 35 µl 1/64 | ||||
| End-point titer slide | |||||||
| Positive control | 1/32 | 1/64 | 1/128 | 1/256 | 1/512 | 1/1024 | 1/2048 |
| 75 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent |
| 5 µl serum | 35 µl 1/16 | 35 µl 1/32 | 35 µl 1/64 | 35 µl 1/128 | 35 µl 1/256 | 35 µl 1/512 | 35 µl 1/1024 |
| Sample 1 1/16 | 1/32 | 1/64 | 1/128 | 1/256 | 1/512 | 1/1024 | 1/2048 |
| 75 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent |
| 5 µl serum | 35 µl 1/16 | 35 µl 1/32 | 35 µl 1/64 | 35 µl 1/128 | 35 µl 1/256 | 35 µl 1/512 | 35 µl 1/1024 |
| Sample 2 1/16 | 1/32 | 1/64 | 1/128 | 1/256 | 1/512 | 1/1024 | 1/2048 |
| 75 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent |
| 5 µl serum | 35 µl 1/16 | 35 µl 1/32 | 35 µl 1/64 | 35 µl 1/128 | 35 µl 1/256 | 35 µl 1/512 | 35 µl 1/1024 |
| IFA buffer only | IFA buffer only | IFA buffer only | IFA buffer only | Negative control | |||
| 75 µl diluent | 35 µl diluent | 35 µl diluent | 35 µl diluent | ||||
| 5 µl serum | 35 µl 1/16 | 35 µl 1/32 | 35 µl 1/64 | ||||
25.Remove dry, room-temperature slides from the desiccator and apply 10 µl of each working dilution sample created in steps 23 and 24 from the microplates to slide wells.
26.Place the slides into a humidified chamber with a closed top and incubate slides and chamber in an incubator at 37°C, 0% CO2 for 30 min.
27.Remove the humidified chamber containing slides from the incubator.
28.Remove the slides from the humidified container.
29.Spray the slides with PBS from a wash bottle to remove sera.
30.Place slides into a slide staining dish filled with PBS and containing a magnetic stir bar.
31.Place the slides onto a magnetic stirrer and wash the slides with the stir bar at 60 rpm for 5 min.
32.Gently decant the PBS.
33.Repeat steps 31 and 32 for a second wash.
34.Dilute the anti-guinea pig-FITC conjugate to the predetermined working concentration using IFA Buffer.
35.Centrifuge the diluted conjugate for 5 min at 10,000 × g , room temperature.
36.Repeat steps 31 and 32 for a third wash.
37.Remove slides from the staining dish after the third wash and tap the edges against a paper towel to remove excess PBS.
38.Allow the Teflon of the slides to air dry.
39.Apply 10 µl of the conjugate to each well of all slides.
40.Place the slides into the humidified chamber.
41.Place humidified chamber into an incubator at 37°C, 0% CO2 for 30 min.
42.Remove the slides from the incubator.
43.Remove the slides from the humidified container.
44.Spray the slides with PBS from a wash bottle to remove sera.
45.Place the slides into a slide staining dish containing a magnetic stir bar and filled with PBS.
46.Place slides onto a stirrer and wash the slides with the stir bar set at 60 rpm for 5 min.
47.Gently decant the PBS.
48.Fill slide staining dish with PBS for a second wash.
49.Add 600 µl of 1.65% Eriochrome T counterstain per 150 ml of PBS to the center of the dish.
50.Place the slides onto a stirrer and wash the slides with the stir bar on at 60 rpm for 5 min.
51.Decant wash buffer.
52.Fill the staining dish with PBS and perform a third wash at 60 rpm for 5 min.
53.Decant wash buffer.
54.Remove the slides and tap the edges of the slides against a paper towel to remove excess PBS.
55.Allow slides to air dry.
56.Add a small drop (covering approximately one-third of the well) of mounting medium to each well of every slide.
57.Apply a coverslip to each slide, ensuring that all the wells are covered.
58.Place slides into a light-proof container (such as a slide folder) and allow them to settle for at least 15 min at 2-8°C.
59.Examine the slides under a UV fluorescent light source at 40× magnification.
60.Confirm that the negative control and any wells containing only conjugate are negative, and the positive control is within the expected range.
61.Read the results for experimental wells after ensuring that controls have expected outcomes.
REAGENTS AND SOLUTIONS
Antigen diluent
- 100 ml PBS, pH 7.38
- 0.1 g (0.1% w/v) sodium azide
- 1 g (1.0% w/v) gamma-globulin-free BSA
- Mix using gentle stirring. Filter sterilize the diluent and store up to 6 months at 4°C.
Eriochrome T counterstain, 1.65%
- 1.65 g Eriochrome Black T Powder
- 100 ml deionized water
- Mix and store up to 6 months at room temperature in a light-proof bottle.
IFA buffer
- 100 ml PBS, pH 7.38
- 0.1 g (0.1% w/v) sodium azide
- 1 g (1.0% w/v) gamma-globulin-free BSA
- 1 ml (1% v/v) heat-inactivated, normal goat (or other) serum
- Mix using gentle stirring
- Filter sterilize and store up to 6 months at 4°C.
Match serum to the species in which the secondary antibody was raised (here, goat anti-guinea pig).
Mounting medium (anti-fade)
- 90 ml glycerol
- 10 ml PBS, pH 7.38
- 3.37 g (0.3 M) DABCO
- Mix and store up to 6 months at 4°C in a light-proof bottle.
COMMENTARY
Background Information
H.T. Ricketts “arrived in Missoula, Montana, April 21, 1906, equipped for the bacteriologic and hematologic study of the so-called Rocky Mountain spotted fever and for the study of the infectious agent by means of animal inoculations.” After unsatisfactory preliminary experimentation with rabbits, he found the guinea pig to be an excellent model for his study, reporting that the clinical signs in guinea pigs reflected many of the symptoms seen in human infections (Ricketts, 1906). More recently, the advantages of using a guinea pig as a model for infectious diseases have been reviewed several times; they include the marked similarity of the immune system to that of a human and their relatively large size for a rodent model, which allows for multiple samples for multiple assays in longitudinal studies (Broad_Institute, 2022; Mestas & Hughes, 2004; Padilla-Carlin et al., 2008; Stokes et al., 2020). Given the advantages of the guinea pig model, why do many researchers continue to espouse the mouse?
Compared to the guinea pig model, the principal advantages of the murine model are the abundance of immunological reagents, the availability of strains with specific gene knockouts and mutations, and the comparatively modest maintenance cost. However, when conducting research on infectious diseases affecting humans, one must carefully weigh these advantages against the shortcomings of the murine model if one desires relevant and robust data. The principal disadvantages of a murine model are the dissimilarity of a mouse's immune system from that of a human and its small size, which limits blood collection. Advocates of the murine model point to the development of humanized mice as a better mouse model for the study of human disease (Pearson, Greiner, & Shultz, 2008; Tsuji & Akkina, 2019). However, the initial cost of developing humanized strain combined with the potential need to spend several hundred dollars or more per mouse only to sacrifice them in groups at every time point of a study may alter the relative cost dynamic between the two models. Additionally, the size limitation imposed by the mouse forces the investigator onto the proverbial horns of a dilemma by forcing them to choose between following the same individual over a study, but with limited material for assays, and sacrificing groups of mice at each timepoint. Even if the cost of humanized mice falls in the future, the inability to obtain an adequate sample for many assays at a single timepoint still limits the utility of humanized mice for longitudinal studies. In contrast, the guinea pig does not appear to have intractable limitations.
The guinea pig genome sequence, at full (7×) coverage, and a comparative chromosome map to humans are available (Broad_Institute, 2022; Romanenko et al., 2015). CRISPR/Cas9 gene-edited guinea pigs with knockouts such as RAG2 and interferon-lambda receptor have been produced as proof of concept (personal communication, Dr. Zhongde Wang, Utah State University). Thus, the primary disadvantage of the guinea pig as a model organism remains the paucity of immunological reagents. Researchers can address this scarcity of reagents by generating increased demand for guinea pig-specific antibodies through by a shift in the paradigm that has promoted overdependence on, and overinvestment in, the fundamentally flawed murine model for infectious disease research.
We present this compendium of protocols to help address the need for established methods for leveraging the guinea pig as a model for spotted fever rickettsiosis. Our additional objective is to stimulate interest in the guinea pig as a general model for infectious disease and thereby inspire the development of a more comprehensive suite of immunological reagents for this species. We recognize that researchers can adapt most of these protocols for other infectious disease studies when they desire results that reflect a more relevant immune system than that of a mouse.
Critical Parameters
Tick feeding and colony development
Pay special attention to preventing inbreeding in tick colonies, which can cause teratogenic effects leading to colony collapse. Early indications of inbreeding in ixodid ticks include decreased longevity of unfed ticks, low feeding success with increased feeding duration, and smaller size of engorged female ticks. Periodic introduction of wild-caught ticks into a colony helps establish gene flow and avert inbreeding. Clean and sterilize humidity chambers and incubators at least monthly to prevent entomopathogenic fungi and mite infestation (see Troubleshooting).
Tick transmission of SFGR to guinea pigs
Guinea pigs infected with SFGR are usually most infectious to ticks at 7-9 days after infection; ticks completing their engorgement during this period tend to have the highest prevalence of infection.
Monitoring the course of infection
Pay special attention to the animal's core temperature; an abrupt drop in temperature to <36.2°C indicates organ failure and calls for immediate termination.
Monitoring rickettsial burden by multiplex qPCR
Keep pipetting steps to a minimum to reduce human error and variation. As you make the serial dilutions for the standard curve, mix each sample well before proceeding with the subsequent dilution to help ensure acceptable R 2 and efficiency values.
Monitoring blood leukocytes by flow cytometry
In Basic Protocol 5, when performing the titrations in step 3, it is important to use the saturating titer (highest SI) for the Pan T antibody and not a separating (sub-saturating) titer such as one might use for the other antibodies. Saturation is called for because, in our hands, the Pan T antibody appears to be somewhat cross-reactive with B cells. Thus, the concentration must be almost perfect to achieve a clean separation. This issue has been brought to the manufacturer's attention, and they are now suggesting the use of clone MsGp7 (also a Pan T), which has no such reported cross-reactivity. We have not yet tested the MsGp7 clone but hope to adopt it in future studies.
For most of the wash and incubation steps, using FCM-PBS is essential for providing protein support for the cells to maintain viability. However, for step 21 (before viability staining) and step 30 (before addition of formaldehyde), it is important to use PBS (protein-free) because the BSA in the FCM-PBS will reduce the effectiveness of the viability staining and formaldehyde fixation.
When labeling the surface antigens in step 27, maintaining the same total staining volume (i.e., antibody concentration) within and between assays is crucial for accurate placement of gates based on FMOs and inter-assay reproducibility. At the same time, the number of cells in the staining reaction is more forgiving. For example, if you titrate your antibody using 106 cells per tube, the titration is still valid if the experimental samples have anywhere from 5 × 105 to 2 × 106.
Monitoring leukocyte infiltration of skin at the tick bite site by flow cytometry
The most crucial parameters associated with Basic Protocol 6 are the dissociation conditions, the antibody concentrations, and the staining volumes. The dissociation enzyme concentrations were empirically determined to balance cell yield and viability, i.e., to maximize the number of live cells recovered from the dissociation of a punch biopsy. We recommend optimizing the concentration of Enzyme R before Enzyme D if you make an adjustment because Enzyme R is used at a higher concentration. Do not alter the concentration of Enzyme A/DNase I.
Determine appropriate antibody concentrations through titrations before using the panel in a study. Always calculate and plot the Stain Index when choosing the optimal concentration. You must perform a new titration with a new lot of antibodies or with a new conjugation.
Again, the staining volumes for both surface and intracellular staining are critical. Therefore, specific steps call for a “bump” to the tube to bring the volume to a reproducible amount. We recommend practicing this bump until you can reproducibly recover the same volume. And remember that adding this bump at other decanting steps adds no value and will lead to lower cell recovery.
Monitoring the antibody titer by ELISA
In Basic Protocol 7, when preparing secondary antibody dilution for step 17, measuring out the 36 ml FCM-PBS during the primary antibody incubation is acceptable. However, do not add the secondary antibody until immediately before addition to the wells. Proteins (e.g., antibodies) will adsorb to many plastics and glass, which can significantly change the concentration of a dilute solution; the use of FCM-PBS as the diluent will reduce this adsorption.
TMB substrate is sensitive to light and contamination from various oxidizing agents. To avoid contamination and premature expiration, avoid contacting TMB solution with any potential source of contamination. Never pipet directly from the bottle; pour the required amount into a tube and pipet from the tube; and do not return excess TMB to the primary storage container.
In step 25, although you are adding a stop solution to stop the reaction, it is important to read the plate immediately after adding the stop solution to keep consistency in absorbance readings between assays. Setting up a required endpoint assay on the instrument the day before or after step 5 will reduce the time between the end of the kinetic reaction and the acquisition of data.
Monitoring the antibody titer by IFA
An IFA depends on many factors to produce a reliable result. Making and using superior-quality IFA slides is crucial to the outcome of this procedure. The appropriate dilution of samples and the IFA conjugate are the most important steps during the protocol. Lastly, although performing an IFA is relatively straightforward, evaluating the results of an IFA run requires a skilled microscopist to obtain accurate, reproducible data between assays.
Troubleshooting
For lists of the categories of problems that may arise with this procedure along with their possible causes and solutions, see Table 5 (Basic Protocol 4), Table 6 (Basic Protocol 5), Table 7 (Basic Protocol 6), Table 8 (Basic Protocol 7), and Table 9 (Alternate Protocol 2).
| Problem | Possible cause | Solutions |
|---|---|---|
| Cross-contamination of samples | Poor technique | Avoid creating aerosols |
| Only open cap of one sample at a time | ||
| Use filter tips |
| Problem | Possible cause | Solution |
|---|---|---|
| Nonspecific staining | Fc receptors not blocked | Make sure guinea pig serum is used to block Fc receptors |
| Insufficient washing | Ensure all washing steps are observed | |
| Failure to titrate antibodies | Nonspecific staining may be reduced or eliminated by using an empirically determined antibody concentration—often lower than manufacturer's suggestion | |
| Weak staining | Failure to observe optimized incubation times and temperatures | Find and consistently observe the optimal time and temperature for antibody incubations |
| Incorrectly diluted antibodies | Ensure that antibodies are used at the correct concentration as determined by titration |
| Problem | Possible cause | Solution |
|---|---|---|
| Low cell yield | Insufficient dissociation | Increase enzyme concentrations and/or length of dissociation |
| Overaggressive dissociation | Decrease enzyme concentrations and/or length of dissociation | |
| Unsuccessful intracellular staining | Failed permeabilization | Optimize steps 67-74: try adjusting permeabilization wash incubation time in steps 67-68 or adding/removing a permeabilization wash step before intracellular staining |
| Unsuccessful lysis of red blood cells | Temperature of the room at the time of lysis | Optimize the lysis conditions at one temperature, and ensure that all future experiments take place at the same temperature |
| Problem | Possible cause | Solution |
|---|---|---|
| Weak or no signal | Reagents not at room temperature | All reagents should be at room temperature at the start of assay |
| Expired reagents | Do not use reagents past their expiration dates | |
| Reagents prepared or added incorrectly | Review the protocol | |
| Incorrect dilutions | Check your calculations | |
| Plate read at wrong wavelength | Read plate at 450 nm | |
| Too much signal | Insufficient washing | Follow washing procedure in the protocol |
| Incorrect dilutions | Check your calculations | |
| Incorrect incubation time | Adhere to the protocol | |
| High background | Insufficient washing | Follow washing procedure in the protocol |
| Substrate exposed to light before use | Store substrate in the dark, and limit exposure to light when performing assay | |
| Incubation period too long | Follow incubation periods suggested in the protocol | |
| Poor standard curve | Incorrect dilutions | Check your calculations |
| Poor pipetting technique | Review how to pipet serial dilutions | |
| Poor replicate data | Insufficient washing | Follow washing procedure in the protocol |
| Poor assay-to-assay reproducibility | Insufficient washing | Follow washing procedure in the protocol |
| Inconsistent incubation temperature | Review the protocol | |
| Incorrect dilutions | Check your calculations. | |
| Edge effect | Stacked plates | Do not stack plates during incubations |
| Evaporation | Seal the plate completely with sealing tape during incubations |
| Problem | Possible cause | Solution |
|---|---|---|
| Too few cells in the wells | Wash steps were performed too aggressively | Avoid spraying wash solution directly on the wells, and use the magnetic stir bar at lower speed |
| Excess mounting medium or poorly positioned coverslip is causing coverslip slippage, which may disrupt the cellular substrate | Float off the cover slip with a PBS wash and reapply | |
| Hazy or cloudy images | Excessive mounting medium was used | Float off the cover slip with a PBS wash and reapply |
| Long storage of prepared slides in the refrigerator before examination | This may be condensation—allow the slides to warm to room temperature before reading; application of a cover slip adhesive may prevent excess humidity from entering under slide cover, which can also cause problems | |
| Microscope objective or oculars are dirty | Clean with lens paper and an approved solvent | |
| Uneven fluorescence seen in the wells | Specimen was allowed to dry excessively during procedure | Ensure humidity chamber is appropriately moist, and do not permit sample wells to completely dry at any stage |
| Applied samples did not completely cover the slide well | Add appropriate volume of the sample to cover each respective well | |
| Cross-contamination between wells | During wash steps, rinse with a generous volume of wash buffer so that samples are rapidly diluted and flushed to prevent mixing | |
| Salts in the PBS may crystallize on the wells if allowed to dry too much during the procedure | Do not allow slides to dry excessively throughout the procedure | |
| Low or no fluorescence observed | Antibodies in the sample or the conjugate have degraded | Store serum samples in appropriate conditions and use sodium azide or thimerosal for preservation of conjugates; avoid multiple freeze-thaw cycles of samples and conjugate |
| Conjugate was improperly diluted | Ensure appropriate dilution factor is used; do not pre-make this reagent before the necessary protocol step | |
| pH of PBS or other buffer solutions is too acidic | Proper pH is 7.4 ± 0.2 | |
| Increased background fluorescence | Inadequate washing | Use fresh wash buffer; do not crowd slides during washes; increase stir speed slightly |
| Improper mounting medium was used, causing autofluorescence | Phosphate-buffered glycerol used as a mounting medium is appropriate; alternatives need to be confirmed as appropriate before use | |
| Omission of counterstain step, or use of inadequate concentration | Ensure counterstain was added; add more as necessary to mitigate this issue | |
| Increased fluorescence within the cell substrate | Cells were damaged by physical contact with a pipet or through accelerated drying efforts | Avoid touching the wells during the procedure; also, do not wash slides or use a fan; allow them to air dry. |
| Nonspecific binding of the conjugate | Use a higher dilution of the conjugate; re-run the checkerboard system referred to in step 18 if needed. |
Anticipated Results
In addition to the Dunkin Hartley outbred strain, we know of two extant inbred strains: strains 2 and 13. We have tested the flow cytometry panels on the Dunkin Hartley strain and strain 2, and the staining is not noticeably different. If you power your experiments based on preliminary data, we anticipate that strain 2 will provide a given power and significance level with fewer animals due to the lower genetic variability between animals. However, some researchers may prefer using outbred animals for some studies, e.g., vaccine trials, where a relevant immune system combined with genetic diversity will be more informative.
Infecting ticks by microinjection is best done with engorged nymphs that have just dropped from the host within a day. However, we have attempted this with engorged ticks 3-4 days after dropping because of the time taken for shipping. Even if done at the ideal time, there is still some tick mortality, with approximately 25-30% of ticks not undergoing ecdysis. If it is done later, we see mortality of 50%-75%. Plan accordingly and use more ticks than needed for the study.
Guinea pigs are highly susceptible to SFGR infection and will be expected to demonstrate clinical signs when exposed to virulent bacteria. The percentage of exposed guinea pigs that develop clinical signs will depend on the route of infection or dose, with tick transmission presenting more variability if all the ticks do not attach or not all ticks are known to be infected. On the other hand, the bite of a single American dog tick infected with R. rickettsii is sufficient to cause clinically recognizable illness in most guinea pigs. We have exposed guinea pigs to three adult Gulf Coast ticks infected with R. parkeri to generate clinical signs. Removing the variability relies first on knowing beforehand that ticks are infected; hemolymph testing (Burgdorfer, 1970) is helpful because the same tick can be placed on the guinea pig, as is determining the infectivity rate in a population of the ticks being used for transmission studies. SFGR injection, through needle inoculation, is the most dependable method for obtaining guinea pigs with clinical SFR.
We typically assay for Rickettsia by qPCR at every time point and have only occasionally detected rickettsial DNA in blood samples earlier than the fulminant phase of infection. Conversely, rickettsial DNA is regularly detected in ear-punch biopsies, even in the early stages of infection. The ear is highly vascularized and easy to sample, making it ideal for detecting Rickettsia in active infections (Levin et al., 2016). We optimized the qPCR assays, and you should consistently expect R 2 values of ≥0.985 and efficiencies of 90%-110% (Ross et al., 2022).
When monitoring blood leukocytes by flow cytometry, you should expect the following precision from technical replicates: %T cells, CV% ≤7.6 (x̄ = 5.4); %B cells, CV% ≤12.5 (x̄ = 11.9); %CD4+ cells, CV% ≤3.4 (x̄ = 1.9); %CD8+ cells, CV% ≤8.1 (x̄ = 4.4) (Stokes et al., 2020). We do not have comparable data for the monitoring leukocyte infiltration by flow cytometry because each sample comes from a unique skin biopsy at the site of a particular tick bite and therefore they do not represent true technical replicates (Cross et al., 2022). In this case, confidence in results comes from other controls, e.g., reference controls, negative controls, and the blood samples used as FMO controls.
When we thaw previously harvested and frozen splenocytes, the viability is typically 65%-70%.
When you monitor the antibody titer by ELISA, you should expect the following intra-plate precision from replicates: CV% ≤6.2 (x̄ = 3.3); high % deviation (% deviation of highest single OD from mean OD) ≤11.3 (x̄ = 4.5); low % deviation (% deviation of lowest single OD from mean OD) ≤9.3 (x̄ = 3.9), where OD = optical density. You should expect the following inter-plate precision: CV% ≤12.8 (x̄ = 8.8) (Alugubelly et al., 2021).
Unlike many technical procedures presented in this text, IFA results are more subjective and are semiquantitative. Humans evaluate slides rather than instruments with set parameters and cutoffs, and proficiency in reading slides requires training and experience. Previous studies involving guinea pigs can provide baseline results from which you can form expectations, but many factors will influence the IFA results associated with studies. Some of these factors include the infectious dose, the route of exposure, the age and immune background of the animals in the study, and the time at which the immune response is assessed (Padilla-Carlin et al., 2008). For example, our group assessed male Hartley strain guinea pigs (6-8 months of age) for R. amblyommatis titers at 13 days after inoculation. The inoculation dose was 6.67 × 106 rickettsiae via intraperitoneal (IP) inoculation. Titers were noticeably absent on day 13, except for a 64-endpoint titer for one animal of the six assessed (Snellgrove et al., 2021). We used species-specific antigen to coat the IFA slides, so lowered sensitivity due to a Rickettsia species mismatch in antigen is unlikely. The absence of titers in these animals may be due to our having assessed antibodies too early after infection—a later time point might have been informative in this study. In a similarly designed study, male guinea pigs inoculated with 4 × 106 rickettsiae via the IP route had an endpoint titer of 512 for all animals by 13 days after inoculation. However, the guinea pigs were significantly younger and smaller than the animals used in our previous study, which could account for the variable results (Rivas et al., 2015). Additionally, a different strain of R. amblyommatis was used, which may have resulted in variable titers compared to the previous study. This second study illustrates the point that small changes in study design can provide differential IFA outcomes, despite strong similarities between the two studies.
Time Considerations
Basic Protocol 1: ∼15-25 min (steps 3-7); ∼5-15 min (steps 13-19 and 21-22) per guinea pig
Support Protocol 1: ∼3-4 hr
Alternate Protocol 1: ∼30-60 min (steps 4a-11a or 4b-9b and 12-14)
Basic Protocol 2: ∼5-15 min per guinea pig
Basic Protocol 3: ∼0.5-1 hr per guinea pig
Support Protocol 2: ∼ 15-20 min (steps 2-8)
Basic Protocol 4: ∼ 1.5 hr (steps 3-12)
Basic Protocol 5: ∼ 6.5 hr (steps 8-36)
Support Protocol 3: ∼2 hr (steps 3-11)
Basic Protocol 6: ∼ 9 hr (steps 12-77)
Basic Protocol 7: ∼6.5 hr (steps 4-26)
Support Protocol 4: ∼6 hr (day 1); ∼1 hr (day 2); ∼0.5 hr (day 3)
Alternate Protocol 2: 3.5-5 hr (steps 20-57); time for reading results is dependent on the number of slides and the experience level of the reader.
Acknowledgments
The authors acknowledge the assistance and support of researchers who recognize the advantages of the guinea pig model and who have promoted the continued development of their potential as a biomedical model. The authors thank Jamie Walker in the Laboratory Animal Resources and Care at Mississippi State University College of Veterinary Medicine for her exceptional skills with all guinea pig techniques and her general assistance. The authors also thank Dr. David Walker at the University of Texas Medical Branch for his continued mentorship in research and professional development and Dr. William Nicholson for his invaluable technical expertise and vast knowledge of IFA that he has imparted both within and outside of the Centers for Disease Control and Prevention (CDC) over the years. The work throughout this manuscript was supported by funding sources, including the US National Institutes of Health (P20 GM103646/GM/NIGMS) and CDC.
Author Contributions
John V. Stokes : Conceptualization, methodology, writing—original draft, and editing; Michael L. Levin : Methodology, writing—original draft, and editing; Claire E. Cross : Methodology, writing—original draft, and editing; Anne-Marie L. Ross : Methodology, writing—original draft, and editing; Alyssa N. Snellgrove : Methodology, writing—original draft, and editing; Bridget V. Willeford : Writing—original draft and editing; Navatha Alugubelly : Methodology, writing—original draft, and editing; Andrea S. Varela-Stokes : Conceptualization, methodology, writing—original draft, and editing
Conflict of Interest
The authors declare no conflict of interest. The views expressed in this article are those of the authors and do not reflect the official policy or position of the Centers for Disease Control and Prevention or the U.S. Government.
Open Research
Data Availability Statement
Data available upon reasonable request from the authors.
Supporting Information
| Filename | Description |
|---|---|
| cpz1584-sup-0001-SuppMat.pdf108.7 KB | Suppinfo |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Alugubelly, N., Stokes, J. V., Cross, C. E., Ross, A. L., Crawford, A. E., Fiihr, G. F., & Varela-Stokes, A. S. (2021). Beyond the IFA: Revisiting the ELISA as a more sensitive, objective, and quantitative evaluation of spotted fever group rickettsia exposure. Pathogens , 10(2), 88. doi: 10.3390/pathogens10020088
- American Committee of Medical Entomology and American Society of Tropical Medicine & Hygiene. (2019). Arthropod containment guidelines, Version 3.2. Vector-Borne and Zoonotic Diseases , 19(3), 152–173. doi: 10.1089/vbz.2018.2431
- Ammerman, N. C., Beier-Sexton, M., & Azad, A. F. (2008). Laboratory maintenance of Rickettsia rickettsii. Current Protocols in Microbiology , 11, 3A.5.1–3A.5.21. doi: 10.1002/9780471729259.mc03a05s11
- Araya-Anchetta, A., Busch, J. D., Scoles, G. A., & Wagner, D. M. (2015). Thirty years of tick population genetics: A comprehensive review. Infection, Genetics and Evolution , 29, 164–179. doi: 10.1016/j.meegid.2014.11.008
- Baldridge, G. D., Kurtti, T. J., Burkhardt, N., Baldridge, A. S., Nelson, C. M., Oliva, A. S., & Munderloh, U. G. (2007). Infection of Ixodes scapularis ticks with Rickettsia monacensis expressing green fluorescent protein: A model system. Journal of Invertebrate Pathology , 94(3), 163–174. doi: 10.1016/j.jip.2006.10.003
- Birck, M. M., Tveden-Nyborg, P., Lindblad, M. M., & Lykkesfeldt, J. (2014). Non-terminal blood sampling techniques in guinea pigs. Journal of Visualized Experiments , 0(92), e51982. doi: 10.3791/51982
- Blanton, L. S., Mendell, N. L., Walker, D. H., & Bouyer, D. H. (2014). "Rickettsia amblyommii " induces cross protection against lethal Rocky Mountain spotted fever in a guinea pig model. Vector Borne Zoonotic Diseases , 14(8), 557–562. doi: 10.1089/vbz.2014.1575
- Broad_Institute. (2022). Guinea Pig Genome Project. Retrieved from https://www.broadinstitute.org/guinea-pig/guinea-pig-genome-project
- Burgdorfer, W. (1970). Hemolymph test. A technique for detection of rickettsiae in ticks. Am J Trop Med Hyg , 19(6), 1010–1014. doi: 10.4269/ajtmh.1970.19.1010
- CDC. (2021). Spotted Fever Rickettsiosis (including Rocky Mountain Spotted Fever) (SFR, including RMSF) 2020 Case Definition. Retrieved from https://ndc.services.cdc.gov/case-definitions/spotted-fever-rickettsiosis-2020/
- Cross, C. E., Stokes, J. V., Alugubelly, N., Ross, A. L., Willeford, B. V., Walker, J. D., & Varela-Stokes, A. S. (2022). Skin in the game: An assay to monitor leukocyte infiltration in dermal lesions of a guinea pig model for tick-borne Rickettsiosis. Pathogens , 11(2), 119. doi: 10.3390/pathogens11020119
- Eisenberg, G. H., Jr., & Osterman, J. V. (1979). Gamma-irradiated scrub typhus immunogens: Broad-spectrum immunity with combinations of rickettsial strains. Infection and Immunity , 26(1), 131–136. doi: 10.1128/iai.26.1.131-136.1979
- Embers, M. E., Grasperge, B. J., Jacobs, M. B., & Philipp, M. T. (2013). Feeding of ticks on animals for transmission and xenodiagnosis in Lyme disease research. Journal of Visualized Experiments , 0(78), 50617. doi: 10.3791/50617
- Frickmann, H., & Dobler, G. (2013). Inactivation of rickettsiae. European Journal of Microbiology and Immunology (BP) , 3(3), 188–193. doi: 10.1556/EuJMI.3.2013.3.6
- Goddard, J. (2003). Experimental infection of lone star ticks, Amblyomma americanum (L.), with Rickettsia parkeri and exposure of guinea pigs to the agent. Journal of Medical Entomology , 40(5), 686–689. doi: 10.1603/0022-2585-40.5.686
- Lee, J. K., Moraru, G. M., Stokes, J. V., Wills, R. W., Mitchell, E., Unz, E., … Varela-Stokes, A. S. (2017). Rickettsia parkeri and "Candidatus Rickettsia andeanae" in questing Amblyomma maculatum (Acari: Ixodidae) from Mississippi. Journal of Medical Entomology , 54(2), 476–480. doi: 10.1093/jme/tjw175
- Levin, M. L., Ford, S. L., Hartzer, K., Krapiunaya, L., Stanley, H., & Snellgrove, A. N. (2020). Minimal duration of tick attachment sufficient for transmission of infectious Rickettsia rickettsii (Rickettsiales: Rickettsiaceae) by its primary vector Dermacentor variabilis (Acari: Ixodidae): Duration of Rickettsial reactivation in the vector revisited. Journal of Medical Entomology , 57(2), 585–594. doi: 10.1093/jme/tjz191
- Levin, M. L., Killmaster, L., Zemtsova, G., Grant, D., Mumcuoglu, K. Y., Eremeeva, M. E., & Dasch, G. A. (2009). Incongruent effects of two isolates of Rickettsia conorii on the survival of Rhipicephalus sanguineus ticks. Experimental and Applied Acarology , 49(4), 347–359. doi: 10.1007/s10493-009-9268-9
- Levin, M. L., & Schumacher, L. B. M. (2016). Manual for maintenance of multi-host ixodid ticks in the laboratory. Experimental and Applied Acarology , 70(3), 343–367. doi: 10.1007/s10493-016-0084-8
- Levin, M. L., Snellgrove, A. N., & Zemtsova, G. E. (2016). Comparative value of blood and skin samples for diagnosis of spotted fever group rickettsial infection in model animals. Ticks and Tick-borne Diseases , 7(5), 1029–1034. doi: 10.1016/j.ttbdis.2016.05.011
- Levin, M. L., Zemtsova, G. E., Killmaster, L. F., Snellgrove, A., & Schumacher, L. B. M. (2017). Vector competence of Amblyomma americanum (Acari: Ixodidae) for Rickettsia rickettsii. Ticks and Tick-borne Diseases , 8(4), 615–622. doi: 10.1016/j.ttbdis.2017.04.006
- Londono, A. F., Mendell, N. L., Walker, D. H., & Bouyer, D. H. (2019). A biosafety level-2 dose-dependent lethal mouse model of spotted fever rickettsiosis: Rickettsia parkeri Atlantic Rainforest strain. PLOS Neglected Tropical Diseases , 13(6), e0007054. doi: 10.1371/journal.pntd.0007054
- Matsumoto, K., Brouqui, P., Raoult, D., & Parola, P. (2005). Experimental infection models of ticks of the Rhipicephalus sanguineus group with Rickettsia conorii. Vector-Borne and Zoonotic Diseases , 5(4), 363–372. doi: 10.1089/vbz.2005.5.363
- McDade, J. E., Black, C. M., Roumillat, L. F., Redus, M. A., & Spruill, C. L. (1988). Addition of monoclonal antibodies specific for Rickettsia akari to the rickettsial diagnostic panel. Journal of Clinical Microbiology , 26(10), 2221–2223. doi: 10.1128/jcm.26.10.2221-2223.1988
- Meechan, P. J., Hatcher, B., & Potts, J. (2020). Biosafety in Microbiological and Biomedical Laboratories ( 6 edn). Atlanta, Georgia: U.S. Centers for Disease Control.
- Mestas, J., & Hughes, C. C. (2004). Of mice and not men: Differences between mouse and human immunology. The Journal of Immunology , 172(5), 2731–2738. doi: 10.4049/jimmunol.172.5.2731
- Monzon, J. D., Atkinson, E. G., Henn, B. M., & Benach, J. L. (2016). Population and evolutionary genomics of Amblyomma americanum , an expanding arthropod disease vector. Genome Biology and Evolution , 8(5), 1351–1360. doi: 10.1093/gbe/evw080
- Newman, B. C., Sutton, W. B., Wang, Y., Schweitzer, C. J., Moncayo, A. C., & Miller, B. T. (2019). A standardized method for the construction of a tick drag/flag sampling approach and evaluation of sampling efficacy. Experimental and Applied Acarology , 79(3-4), 433–446. doi: 10.1007/s10493-019-00429-6
- Padilla-Carlin, D. J., McMurray, D. N., & Hickey, A. J. (2008). The guinea pig as a model of infectious diseases. Comparative Medicine , 58(4), 324–340. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/18724774
- Pearson, T., Greiner, D. L., & Shultz, L. D. (2008). Creation of "humanized" mice to study human immunity. Current Protocols in Immunology , 81, 15.21.1–15.21.21. doi: 10.1002/0471142735.im1521s81
- Philip, R. N., Lane, R. S., & Casper, E. A. (1981). Serotypes of tick-borne spotted fever group Rickettsiae from western California. American Journal of Tropical Medicine and Hygiene , 30(3), 722–727. doi: 10.4269/ajtmh.1981.30.722
- Ricketts, H. T. (1906). The study of "Rocky Mountain spotted fever" (tick fever?) by means of animal inoculations: A preliminary communication. JAMA , 47, 33–36. doi: 10.1001/jama.1906.25210010033001j
- Rivas, J. J., Moreira-Soto, A., Alvarado, G., Taylor, L., Calderon-Arguedas, O., Hun, L., … Troyo, A. (2015). Pathogenic potential of a Costa Rican strain of 'Candidatus Rickettsia amblyommii ' in guinea pigs (Cavia porcellus) and protective immunity against Rickettsia rickettsii. Ticks and Tick-borne Diseases , 6(6), 805–811. doi: 10.1016/j.ttbdis.2015.07.008
- Romanenko, S. A., Perelman, P. L., Trifonov, V. A., Serdyukova, N. A., Li, T., Fu, B., … Yang, F. (2015). A first generation comparative chromosome map between guinea pig (Cavia porcellus) and humans. PLoS One , 10(5), e0127937. doi: 10.1371/journal.pone.0127937
- Ross, A. L., Stokes, J. V., Cross, C. E., Alugubelly, N., & Varela-Stokes, A. S. (2022). Multiplex TaqManR Quantitative PCR assays for host-tick-pathogen studies using the guinea pig-tick-rickettsia system. Pathogens , 11(5), 594. doi: 10.3390/pathogens11050594
- Salomon, J., Hamer, S. A., & Swei, A. (2020). A beginner's guide to collecting questing hard ticks (Acari: Ixodidae): A standardized tick dragging protocol. Journal of Insect Science , 20(6). doi: 10.1093/jisesa/ieaa073
- Schumacher, L., Snellgrove, A., & Levin, M. L. (2016). Effect of Rickettsia rickettsii (Rickettsiales: Rickettsiaceae) infection on the biological parameters and survival of its tick vector— Dermacentor variabilis (Acari: Ixodidae). Journal of Medical Entomology , 53(1), 172–176. doi: 10.1093/jme/tjv166
- Shomer, N. H., Holcombe, H., & Harkness, J. E. (2015). Biology and diseases of guinea pigs. Laboratory Animal Medicine , 247–283. doi: 10.1016/B978-0-12-409527-4.00006-7
- Snellgrove, A. N., Krapiunaya, I., Scott, P., & Levin, M. L. (2021). Assessment of the Pathogenicity of Rickettsia amblyommatis , Rickettsia bellii , and Rickettsia montanensis in a guinea pig model. Vector-Borne and Zoonotic Diseases , 21(4), 232–241. doi: 10.1089/vbz.2020.2695
- Stanley, H. M., Ford, S. L., Snellgrove, A. N., Hartzer, K., Smith, E. B., Krapiunaya, I., & Levin, M. L. (2020). The ability of the invasive Asian longhorned tick Haemaphysalis longicornis (Acari: Ixodidae) to acquire and transmit Rickettsia rickettsii (Rickettsiales: Rickettsiaceae), the agent of rocky mountain spotted fever, under laboratory conditions. Journal of Medical Entomology , 57(5), 1635–1639. doi: 10.1093/jme/tjaa076
- Stokes, J. V., Crawford, A. E., Cross, C. E., Ross, A. L., Walker, J. D., Willeford, B. V., & Varela-Stokes, A. S. (2020). An optimized five-color/seven-parameter flow cytometry panel for immunophenotyping guinea pig peripheral blood lymphocytes. Journal of Immunological Methods , 476, 112682. doi: 10.1016/j.jim.2019.112682
- Stokes, J. V., Walker, D. H., & Varela-Stokes, A. S. (2020). The guinea pig model for tick-borne spotted fever rickettsioses: A second look. Ticks and Tick-borne Diseases , 11(6), 101538. doi: 10.1016/j.ttbdis.2020.101538
- Troughton, D. R., & Levin, M. L. (2007). Life cycles of seven ixodid tick species (Acari: Ixodidae) under standardized laboratory conditions. Journal of Medical Entomology , 44(5), 732–740. doi: 10.1603/0022-2585(2007)44%5b732:LCOSIT%5d2.0.CO;2
- Tsuji, M., & Akkina, R. (2019). Development of humanized mouse models for infectious diseases and cancer. Frontiers in Immunology , 10, 3051. doi: 10.3389/fimmu.2019.03051
- Walker, D. H., Harrison, A., Henderson, F., & Murphy, F. A. (1977). Identification of Rickettsia rickettsii in a guinea pig model by immunofluorescent and electron microscopic techniques. The American Journal of Pathology , 86(2), 343–358. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/402079
- Walker, D. H., & Henderson, F. W. (1978). Effect of immunosuppression on Rickettsia rickettsii infection in guinea pigs. Infection and Immunity , 20(1), 221–227. doi: 10.1128/iai.20.1.221-227.1978
- Williams, W. R., & Kendall, L. V. (2015). Blood collection in the guinea pig (Cavia porcellus). Lab Anim (NY) , 44(6), 207–208. doi: 10.1038/laban.787
- Yang, T. S., Reichard, M. V., Marr, H. S., Cohn, L. A., Nafe, L., Whitehurst, N., & Birkenheuer, A. J. (2022). Direct injection of Amblyomma americanum ticks with Cytauxzoon felis. Ticks and Tick-Borne Diseases , 13(1), 101847. doi: 10.1016/j.ttbdis.2021.101847
- Ye, X., Sun, Y., Ju, W., Wang, X., Cao, W., & Wu, M. (2014). Vector competence of the tick Ixodes sinensis (Acari: Ixodidae) for Rickettsia monacensis. Parasit Vectors , 7, 512. doi: 10.1186/s13071-014-0512-8
- Zemtsova, G., Killmaster, L. F., Mumcuoglu, K. Y., & Levin, M. L. (2010). Co-feeding as a route for transmission of Rickettsia conorii israelensis between Rhipicephalus sanguineus ticks. Experimental and Applied Acarology , 52(4), 383–392. doi: 10.1007/s10493-010-9375-7

