Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) to Identify MicroRNA Targets
Lauren A. Gay, Lauren A. Gay, Peter C. Turner, Peter C. Turner, Rolf Renne, Rolf Renne
Abstract
This protocol was designed to identify microRNA (miRNA) targetomes from smaller-input samples by performing a simplified workflow of the Cross-Linking and Sequencing of Hybrids (CLASH) technique developed in the Tollervey group. In this ribonomics-based technique, Cross-Linking and Immunoprecipitation (CLIP) of Argonaute (Ago) is combined with an RNA ligase reaction that yields covalently bound “hybrids” between miRNAs and their target RNAs. While this iteration of CLIP identifies “high-confidence” or “unambiguous” miRNA targets, the added ligation step is highly inefficient and therefore requires large numbers of cultured cells. To make this powerful approach applicable to smaller cell numbers, we created qCLASH, incorporating a workflow that performs all enzymatic reactions on bead-bound complexes and omits gel purification of immunoprecipitated Ago complexes associated with major loss of RNA. At a sequencing depth of 100 million reads per library, which is highly feasible with rapidly decreasing sequencing costs, qCLASH, when used with three biological replicates, results in thousands of high-confidence miRNA targets. qCLASH was first developed to identify viral miRNA targetomes of endothelial cells infected with Kaposi's sarcoma−associated herpesvirus. Since then, qCLASH has been applied to Epstein-Barr virus− and MHV68-infected cells, and more recently to metastatic melanoma and breast cancer cells. Currently, protocols are under development to apply qCLASH to human solid tumor specimens. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Quick Cross-Linking and Sequencing of Hybrids (qCLASH)
Support Protocol : Optimization of Ago immunoprecipitation
INTRODUCTION
MicroRNAs (miRNAs) are 19- to 22-nucleotide RNAs involved in the post-transcriptional regulation of gene expression. They bind the protein Argonaute (Ago) to form the RNA-Induced Silencing Complex (RISC), where they recruit and bind mRNAs or other RNA targets, leading to translational repression and eventual degradation (Ambros, 2004). Our laboratory investigates how miRNAs post-transcriptionally regulate whole cellular transcriptomes. An ongoing challenge is to determine miRNA targetomes for all miRNAs that are expressed in a certain cell type or disease, or any physiological condition of interest. This goal can be achieved by Ago-based ribonomics approaches that all require UV crosslinking followed by Ago immunoprecipitation (IP), purification of miRNAs bound to their targets, and analysis by next-generation sequencing. The first such technique, High Throughput Sequencing of RNA isolated by Crosslinking and Immunoprecipitation (HITS-CLIP), was first described by the Darnell laboratory (Chi, Zang, Mele, & Darnell, 2009). A modified version, which labels RNA by incorporating 4-thiouridine, is Photoactivatable Ribonucleoside-enhanced Crosslinking and Immunoprecipitation (PAR-CLIP), developed by the Tuschl group (Hafner et al., 2010). Both these techniques resulted in libraries containing miRNAs and small mRNA fragments that were bound together in RISCs but were separated during cDNA synthesis. To combine miRNAs and their putative targets that were sequenced separately, bioinformatics approaches were used based on current understanding of miRNA targeting, which led to a relatively high false discovery rate. To address this caveat, Helwak and colleagues developed Cross-linking and Sequencing of Hybrids (CLASH; Helwak, Kudla, Dudnakova, & Tollervey, 2013). This protocol adds one extra step in which the mRNA fragment and miRNA present in the RISC, after trimming and IP, are ligated by treatment with RNA ligase. As a result, miRNA and mRNA form “hybrids” (RNAs that contain the miRNA covalently linked to the mRNA fragment that were initially bound through limited Watson-Crick interaction) that can be reverse transcribed and sequenced together. While the ligation step is not very efficient, this step was nevertheless a leap forward in that data analysis did not rely on bioinformatic assumptions and therefore yielded high-confidence targets and a much smaller false-positive discovery rate. A similar protocol was developed by the Rajewski group, which yielded “unambiguous miRNA targets” (Grosswendt et al., 2014).
While all these methods allow the identification of numerous miRNA targets, a major limitation is that they require a large amount of input material. In particular, CLASH was initially performed in double-tagged Ago-engineered 293 cells at approximately 100 million cells per biological replicate. To directly address this limitation, we developed a modified version called quick CLASH that is faster to perform and can be routinely applied to 10 million cells or less from many different sources, since qCLASH is based on endogenous Ago IPs (Gay, Sethuraman, Thomas, Turner, & Renne, 2018). The detailed protocol described below differs from CLASH in two ways. First, traditional CLIP methods gel-purify IPed complexes, which is associated with massive loss of RNA. We omitted this step and dramatically increased the number of sequencing reads to identify sufficient numbers of hybrids. Second, we performed all subsequent enzymatic reactions with bead-bound material, which allowed for stringent washes and reduced nucleic acid precipitations that are associated with loss of small RNAs. So far, qCLASH has been performed to identify targetomes for three DNA tumor viruses that encode viral miRNAs: Kaposi's sarcoma associated herpesvirus (KSHV; Gay et al., 2018), Epstein-Barr virus (EBV; Ungerleider et al., 2020), and murine herpesvirus 68 (MHV68; Bullard et al., 2019). Beyond the field of virology, this protocol has also been applied to breast cancer cells (Stribling et al., 2021) and metastatic melanoma (Kozar et al., 2021). Below we describe two protocols: the Basic Protocol for qCLASH, and a Support Protocol for optimization of Ago immunoprecipitation. The Basic Protocol is the standard qCLASH protocol, while the Support Protocol describes an optional pre-experiment that can be performed prior to qCLASH in order to determine the best conditions for the immunoprecipitation portion of the protocol.
Basic Protocol: QUICK CROSS-LINKING AND SEQUENCING OF HYBRIDS (qCLASH)
The steps for qCLASH are illustrated in Figure 1.Briefly, Ago and its associated RNAs are covalently bound by UV crosslinking of live cells. Ago is immunoprecipitated from the resulting cell lysates and the RNA ends are trimmed with RNase. Intermolecular ligation between the two species of RNA bound to each Ago is carried out through treatment with RNA ligase. Ago is digested so that the RNA can be purified, reverse-transcribed, and sequenced. Since this protocol involves prolonged handling of small quantities of RNA, extra care should be taken to maintain a clean, RNase-free work area and equipment.

Materials
- Protein G Dynabeads, 30 mg/ml (Invitrogen, cat. no. 10004D)
- 1× PBS-T, pH 7.2 (see recipe)
- 2.4 µg/ml AffiniPure Rabbit Anti-Mouse IgG, Fc Fragment Specific (bridging antibody; Jackson ImmunoResearch, cat. no. 315-005-008)
- 0.5 µg/ml anti-pan Ago, clone 2A8 (Millipore Sigma, cat. no. MABE56)
- 1× PXL buffer (see recipe)
- Cells of interest
- 1× Dulbecco's phosphate-buffered saline (DPBS; Corning, cat. no. 21-030-CV)
- Lysis buffer (see recipe)
- RQ1 DNase, 1000 U (Promega, cat. no. M610A; reconstitute per manufacturer's instructions)
- RNase T1, 100,000 U (ThermoFisher Scientific, cat. no. EN0541; reconstitute per manufacturer's instructions)
- 5× PXL buffer (see recipe)
- High-stringency buffer (see recipe)
- High-salt buffer (see recipe)
- 1× and 10× PNK buffer (see recipe)
- RNasin Plus, 40 U/µl(Promega, cat. no. N2615)
- ATP, 100 mM (ThermoFisher Scientific, cat. no. R0041)
- T4 polynucleotide kinase (PNK), 10,000 U/ml (NEB, cat. no. M0201S)
- T4 RNA ligase buffer, 10× (packaged with T4 RNA Ligase)
- PEG 8000 (50% w/v; packaged with T4 RNA ligase)
- T4 RNA ligase 1, 10,000 U/ml (NEB, cat. no. M0204L)
- 4 M KCl (Sigma, cat. no. P9541)
- Dephosphorylation buffer, 10× (packaged with alkaline phosphatase)
- Alkaline phosphatase, 1 U/µl (Roche, cat. no. 10713023001)
- 1× PNK-EGTA buffer (see recipe)
- T4 RNA ligase 2, truncated K227Q, 200,000 U/ml (NEB, cat. no. M0351L)
- 3′ adapter, 10 µM (custom order from IDT, see Table 1)
- 500 mM NaHCO3
- SDS (Fisher, cat. no. BP166-100), 10% (w/v)
- 5× PK buffer (see recipe)
- Proteinase K (Roche, cat. no. 03115887001; prepare according to manufacturer's instructions)
- Phenol/chloroform/isoamyl alcohol, 25:24:1, pH 5.2 (Fisher, cat. no. BP1753I-400)
- 3 M sodium acetate (NaOAc), pH 5.2
- GlycoBlue (Invitrogen, cat. no. AM9516)
- Ethanol/isopropanol (1:1)
- 80% and 70% ethanol, ice-cold
- Bovine serum albumin (BSA), 20 mg/ml (NEB, cat. no. B9000S)
- 5′ RNA adapter, 100 pm/µl (custom order from IDT, see Table 1)
- RNA RT primer, 10 µM (custom order from IDT, see Table 1)
- Deoxynucleotide (dNTP) solution mix, 10 mM each (NEB, cat. no. N0447S)
- 5× Super Script RT Buffer (supplied with enzyme)
- 0.1 M DTT (packaged with SuperScript IV)
- SuperScript IV Reverse Transcriptase, 2000 U (ThermoFisher Scientific, cat. no. 18090010)
- Magnesium chloride hexahydrate (Fisher, cat. no. BP214-500)
- 2× Phusion High Fidelity PCR Master Mix with HF Buffer (NEB, cat. no. M0531S)
- RP1 universal primer, 10 µM (custom order from IDT, see Table 1)
- RPI1, RPI2, or RPI3 barcoded primers, 10 µM (custom order from IDT, see Table 1)
- TE buffer: 10 mM Tris⋅HCl/1 mM EDTA
- Agarose PS 1200 (Agarose Unlimited, http://www.agaroseunlimited.com)
- TAE buffer (see recipe)
- 6× DNA loading buffer (see recipe)
- 6× DNA loading buffer, xylene cyanol only (see recipe)
- 50-bp DNA Ladder (NEB, cat. no. N3236)
- QIAquick Gel Extraction Kit (Qiagen, cat. no. 28704)
- 2× SDS Loading Buffer (see recipe)
- Precision Plus Dual Color Color Standard (Bio-Rad, cat. no. 1610374)
- Anti-Ago2 Antibody, clone 11A9 (Millipore-Sigma, cat. no. MABE253)
- Peroxidase-conjugated AffiniPure Goat Anti-Rat IgG, Fcγ Fragment-Specific (Jackson ImmunoResearch, cat. no. 112-035-008)
| Name | Sequence |
|---|---|
| RNA 3′ Adapter (RA3)a | 5′/5rApp/TGGAATTCTCGGGTGCCAAGG/3ddc/3′ |
| RNA 5′ Adapter (RA5)b | 5′/5InvddT/rGrUrUrCrArGrArGrUrUrCrUrArCrArGrUrCrCrGrArCrGrArUrC3′ |
| RNA RT Primer (RTP) | 5′ GCCTTGGCACCCGAGAATTCCA 3′ |
| RNA PCR Primer (RP1) | 5′ AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA3′ |
| RNA PCR Primer, Index 1 (RPI1) | 5′ CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA3′ |
| RNA PCR Primer, Index 2 (RPI2) | 5′ CAAGCAGAAGACGGCATACGAGATACATCGGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA3′ |
| RNA PCR Primer, Index 3 (RPI3) | 5′ CAAGCAGAAGACGGCATACGAGATGCCTAAGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA3′ |
- a
/5rApp/ = 5′ adenylation; /3ddC/ = 3′ dideoxy-cytidine.
- b
/5InvddT/ = 5′ inverted dideoxy-thymidine; lowercase r indicates nucleotide is RNA.
-
Forma Class II A2 Biological Safety Cabinet (Thermo Electron Corporation or equivalent)
-
DNA LoBind Tube, 1.5-ml (Eppendorf, cat. no. 022431021)
-
Sorvall Legend Micro 21R Centrifuge (Thermo Scientific or equivalent)
-
Labquake rotisserie for microcentrifuge tubes (Thermo Scientific)
-
Sorvall Legend RT Centrifuge (or equivalent)
-
Bright-Line Hemacytometer (Hausser Scientific or equivalent)
-
100-mm TC-treated Culture Dishes (Corning, cat. no. 430167)
-
HL-2000 HybriLinker (UVP Laboratory Products or equivalent)
-
50-ml polypropylene centrifuge tubes, conical bottom (Corning, cat. no. 430828)
-
1.5-ml microcentrifuge tubes (USA Scientific, cat. no. 1615-5510)
-
Thermomixer R heat block (Eppendorf or equivalent)
-
PCR thermal cycler
-
Agilent Tape Station
-
Additional reagents and equipment for counting cells (see Current Protocols article: Phelan & May, 2015)
Day 1
Part 1: Bead preparation and antibody conjugation
1.Transfer 600 μl Protein G Dynabeads to a 1.5-ml LoBind tube. Use LoBind tubes throughout the protocol.
2.Wash beads three times, each time with 950 μl cold 1× PBS-T, pH 7.2.Invert tube to resuspend. Microcentrifuge briefly between washes at <775 × g. Carry out all wash steps in this manner throughout the protocol.
3.Add 428 μl 1× PBS-T, pH 7.2, and 72 μl AffiniPure Rabbit Anti-Mouse IgG (bridging antibody) to tube. Resuspend beads.
4.Rotate 50 min at room temperature.
5.Wash three times, each time with 950 μl cold 1× PBS-T, pH 7.2.
6.Add 490 μl PBS-T and 10 μl of anti-pan Ago 2A8 antibody. Resuspend beads and rotate 4 hr at 4°C.
7.Wash 2A8-conjugated beads four times, each time with 950 μl cold 1× PXL buffer.
8.Resuspend in 620 μl 1× PXL and make three aliquots of 200 µl. Store at 4°C until lysates are ready.
Part 2: UV crosslinking of cells
This step can be completed in advance. Crosslinked cells can be stored at −80°C indefinitely.
9.Prepare a single-cell suspension.
10.Centrifuge 5 min at 200 × g , 4°C.
11.Aspirate supernatant and resuspend cells in 10 ml of cold DPBS. If there are multiple cell pellets, they can be combined.
12.Centrifuge 5 min at 200 × g , 4°C, remove supernatant, and resuspend pellet in 10 ml of DPBS. Perform this step twice.
13.Count cells with a hemacytometer or automated cell counting device (see Current Protocols article: Phelan & May, 2015) before transferring the cell suspension to a 10-cm plate on ice.
14.Place the ice tray and plate (with the lid removed) into the HL-2000 HybriLinker. Crosslink once at 400 mJ/cm2, rotate plate 180°, swirl, and crosslink once at 200 mJ/cm2.
15.Swirl plate and transfer cells to a 50-ml conical tube. Rinse the plate with an additional 10 ml of DPBS and add this to the tube. Keep the tube on ice.
16.Centrifuge 5 min at 200 × g , 4°C. Resuspend the cells in an appropriate volume of DPBS and make aliquots of 50 million cells in 1.5-ml tubes.
17.Centrifuge 5 min at 200 × g and 4°C. Remove DPBS and freeze the cell pellets at −80°C if not proceeding directly to immunoprecipitation.
Part 3: Lysate preparation
18.Prepare 2 ml complete lysis buffer (containing the protease inhibitor mixture and DTT as described in Reagents and Solutions).
19.Take three aliquots of 5 × 107 cells that were previously UV-crosslinked (see step 17) and thaw them on ice. Resuspend in 500 μl complete lysis buffer per aliquot.
20.Lyse 15 min on ice.
21.Add 10 μl RQ1 DNase to each tube.
22.Incubate 5 min at 37°C while shaking at 1000 rpm.
23.Spin lysates 15 min at 21,000 × g , 4°C.
24.Transfer supernatants to new tubes and discard pellets.
25.Add 0.5 μl RNase T1 to each tube and incubate for 15 min at 22°C.
Part 4: Immunoprecipitation
26.Transfer 5 μl of each lysate to new tubes and store at −20°C.
27.Remove buffer from beads and add lysates. Rotate overnight at 4°C.
Day 2
28.Shortly before ending the 4°C incubation, prepare 14 ml complete lysis buffer (containing the protease inhibitor mixture and DTT as described in Reagents and Solutions).
29.Transfer supernatants to new tubes and store at −20°C.
30.Wash three times (950 μl per wash) with complete lysis buffer. Resuspend beads in 1000 μl complete lysis buffer.
31.Add 0.5 μl RNase T1 to each tube and incubate for 12 min at 22°C, shaking continuously at 700 rpm.
32.Wash three times (950 μl per wash) with each of the following ice-cold buffers: 1× PXL, 5× PXL, high-stringency buffer, high-salt buffer, and 1× PNK buffer. After first wash with high-stringency buffer and high-salt buffer, incubate tubes on ice 1 min.
33.Leave beads in final wash buffer and incubate on ice while preparing the T4 PNK mixture.
Part 5: Phosphorylation
34.Prepare T4 PNK mix, adding the T4 PNK enzyme last (amounts are for 3.5 reactions):
| PNK buffer (10x) | 28 μl |
| RNasin Plus (40 U/μl) | 7 μl |
| ATP (100 mM) | 2.8 μl |
| H2O | 228.2 μl |
| T4 PNK enzyme (10 U/μl) | 14 μl |
| Total | 280 μl |
35.Remove wash buffer and add 80 μl to each tube.
36.Incubate at 10°C for 40 min, shaking every 2 min for 15 s at 1000 rpm.
Part 6: Intermolecular ligation
37.Wash three times, each time with 950 μl of 1× PNK buffer.
38.Leave beads in final wash buffer and incubate on ice while preparing the ligation mix (amounts are for 3.5 reactions):
| T4 RNA ligase buffer (10x) | 175 μl |
| PEG 8000 (50% w/v) | 210 μl |
| KCl (4 M) | 4.38 μl |
| RNAsin Plus (40 U/μl) | 43.75 μl |
| ATP (100 mM) | 17.5 μl |
| H2O | 1124.4 μl |
| Total | 1575 μl |
39.Remove wash buffer and add 450 μl ligation mix followed by 50 μl T4 RNA ligase 1 to each tube.
40.Resuspend beads and rotate overnight at 4°C.
Day 3
Part 7: Dephosphorylation
41.Wash three times, each time with 950 μl of 1× PNK buffer.
42.Leave on ice in buffer from last wash and prepare the dephosphorylation mix (amounts are for 3.5 reactions):
| 10x Dephosphorylation buffer | 28 μl |
| RNasin Plus (40 U/μl) | 7 μl |
| H2O | 234.5 μl |
| Alkaline phosphatase (1 U/μl) | 10.5 μl |
| Total | 280 μl |
43.Remove wash buffer and add 80 μl dephosphorylation mix to tube.
44.Incubate at 10°C for 40 min, shaking every 2 min for 15 s at 1000 rpm.
Part 8: 3′ adapter ligation
45.Wash twice, each time with 950 μl 1× PNK-EGTA buffer. After the first wash, incubate 1 min on ice.
46.Wash three times, each with 950 μl of 1× PNK buffer.
47.Leave samples on ice in the last wash buffer and prepare the ligation mix (amounts are for 3.5 reactions):
| H2O | 147 μl |
| T4 RNA ligase buffer (10x) | 28 μl |
| PEG 8000 (50% w/v) | 56 μl |
| RNAsin Plus (40 U/μl) | 7 μl |
| 3’ adapter (10 μM) | 28 μl |
| T4 RNA ligase 2, truncated, K227Q | 14 μl |
| Total | 280 μl |
48.Remove wash buffer and add 80 μl ligation mix to each tube.
49.Resuspend beads and incubate overnight at 16°C, shaking every 2 min for 15 s at 1000 rpm.
Day 4
Part 9: Complex elution
50.Wash three times, each time with 950 μl 1× PNK buffer.
51.Leave on ice with buffer from last wash while preparing the elution buffer (amounts are for 3.5 reactions):
| NaHCO3 (500 mM) | 130 μl |
| SDS (10%, w/v) | 65 μl |
| H2O | 455 μl |
| Total | 650 μl |
52.Add 100 μl of elution buffer to beads and incubate at room temperature for 15 min, shaking continuously at 1400 rpm. Transfer supernatant to new tube.
53.Repeat previous step, this time adding supernatant to that from previous step.
54.Transfer 2 μl of eluate to new tube and store at −20°C.
Part 10: Proteinase K treatment/RNA extraction
55.During the incubation steps with elution buffer, prepare 1× PK buffer containing 4 mg/ml proteinase K (amounts are for 3.5 reactions):
| PK buffer (5x) | 35 μl |
| Proteinase K | 35 μl |
| H2O | 105 μl |
| Total | 175 μl |
56.Pre-incubate proteinase K solution 20 min at 37°C.
57.Add 50 μl of 1× PK buffer with proteinase K to each tube of eluate and incubate at 37°C for 20 min.
58.Add 250 μl phenol/chloroform/isoamyl alcohol (25:24:1) and incubate at room temperature for 8 min, shaking continuously at 1400 rpm.
59.Centrifuge for 10 min at 18,000 × g , 4°C.
60.Remove approximately 200 μl of the aqueous layer and transfer to a new tube. To this add 20 μl 3 M NaOAc, pH 5.2; 2 μl GlycoBlue; and 500 μl of a 1:1 mixture of ethanol/isopropanol.
61.Incubate overnight at −20°C.
Day 5
Part 11: Phosphorylation
62.Centrifuge for 30 min at 21,000 × g , 4°C.
63.Wash 1× with 900 μl ice cold 80% ethanol and centrifuge for 10 min at 18,000 × g , 4°C.
64.Wash 1× with 200 μl ice cold 80% ethanol and centrifuge for 10 min at 18,000 × g , 4°C.
65.Air dry for approximately 10 min, checking often.
66.Add 10.5 μl ddH2O and incubate on ice for 15 min.
67.Prepare T4 PNK mix, adding the T4 PNK enzyme last (amounts are for 3.5 reactions):
| T4 RNA ligase buffer (10x) | 5.25 μl |
| RNAsin Plus (40 U/μl) | 1.75 μl |
| ATP (10 mM) | 5.25 μl |
| T4 PNK enzyme (10 U/μl) | 3.5 μl |
| Total | 15.75 μl |
68.Add 4.5 μl to each tube.
69.Incubate 40 min at 10°C.
Part 12: 5′ adapter ligation
70.Prepare a ligation solution, adding the T4 RNA ligase last (amounts are for 3.5 reactions):
| T4 RNA Ligase Buffer (10x) | 1.75 μl |
| BSA (1 mg/ml) | 7 μl |
| ATP (10 mM) | 1.75 μl |
| 5’ RNA adapter (100 pmol/μl) | 3.5 μl |
| T4 RNA ligase 1 (10 U/μl) | 3.5 μl |
| Total | 17.5 μl |
71.Add 5 μl ligation solution to each tube. Mix by pipetting.
72.Incubate overnight at 16°C.
Day 6
73.Add 200 μl ddH2O and 250 μl phenol/chloroform/isoamyl alcohol to each tube.
74.Vortex 8 min at 1400 rpm at room temperature.
75.Centrifuge 10 min at 18,000 × g , 4°C.
76.Remove approximately 200 μl of the aqueous layer and transfer to a new tube. To this add 20 μl 3 M NaOAc, pH 5.2; 1 μl Glycoblue; and 500 μl of a 1:1 mixture of ethanol/isopropanol.
77.Incubate overnight at −20°C.
Day 7
Part 13: Reverse transcription
78.Centrifuge 30 min at 21,000 × g , 4°C.
79.Wash twice, each time with 200 μl ice-cold 80% ethanol. Centrifuge 10 min at 18,000 × g , 4°C, after each wash.
80.Air dry and resuspend in 11 μl ddH2O.
81.Add to each tube:
| RT primer (RTP) (10 μM) | 1 μl |
| dNTPs (10 mM) | 1 μl |
82.Incubate at 65°C for 5 min, then chill on ice and centrifuge briefly.
83.Prepare an RT mix (amounts are for 3.5 reactions):
| 5× Super Script RT Buffer | 14 μl |
| DTT (0.1 M) | 3.5 μl |
| RNasin Plus (40 U/μl) | 3.5 μl |
| Super Script Enzyme (200 U/μl) | 3.5 μl |
| Total | 24.5 μl |
84.Add 7 μl of the RT mix to each tube of RNA.
85.Incubate at 50°C for 45 min, 55°C for 15 min, and 95°C for 5 min.
86.Chill on ice while preparing PCR mix.
Part 14: PCR
87.Prepare a PCR mix (amounts are for 7.5 reactions):
| 2× Phusion High Fidelity Master Mix | 75 μl |
| RPI1, 2, or 3 (10 μM) | 7.5 μl |
| RP1 (10 μM) | 7.5 μl |
| Water | 45 μl |
| Total | 135 μl |
88.Add 18 μl of the PCR mix to 2 μl of each RT product. Set up two reactions for each biological replicate and include a water control. Store the remaining RT product at −80°C.
89.Preheat PCR thermal cycler block to 98°C. Place tubes in block and run the following cycle:
- 98°C for 30 s
- 98°C for 10 s
- 52°C for 30 s
- 72°C for 30 s
- 72°C for 5 min
- 4°C hold.
Part 15: Size selection (option 1)
This optional size-selection step is intended to eliminate excess adapters. Protocol users may choose to perform this step, or its alternative described in Part 16.
90.Prepare a mixture of 30% PEG 8000/29.25 mM MgCl2:
| PEG 8000 (50%, w/v) | 240 μl |
| MgCl2 (1 M) | 11.8 μl |
| H2O | 148.2 μl |
| Total | 500 μl |
91.Add 60 μl of TE buffer to each PCR product, followed by 40 μl of the PEG 8000/MgCl2 mixture.
92.Vortex and incubate 10 min at room temperature.
93.Centrifuge 15 min at 10,000 × g , room temperature.
94.Transfer supernatants to new tubes and add 12 μl 3M NaOAc, 300 μl 100% ethanol, and 1 μl Glycoblue.
95.Incubate overnight at −20°C.
96.Centrifuge 30 min at 21,000 × g , 4°C.
97.Wash twice with 900 μl and once with 200 μl of 70% ice-cold ethanol. Centrifuge 10 min at 18,000 × g , 4°C after each wash.
98.Air dry and resuspend in 15 μl ddH2O.
Part 16: Agarose gel purification (option 2)
This optional gel purification step is intended to eliminate excess adapters. Protocol users may choose to perform this step, or its alternative described in Part 15.
99.Prepare a 2% agarose gel by dissolving 200 × g of agarose in 100 ml of 1× TAE buffer.
100.Add 2 μl of 6× DNA loading buffer with xylene cyanol to each PCR product.
101.Run products on gel at 100 V for approximately 1 hr alongside 50-bp ladder, prepared by combining 1 µl of 6× DNA loading buffer (containing both xylene cyanol and bromophenol blue), 5 µl of NEB 50-bp DNA Ladder, and 4 µl of water. Stop gel when bromophenol blue marker is approximately 2 cm from the bottom.
102.Excise regions from gel corresponding to approximately 175-300 bp and transfer gel slices to low-bind tubes.
103.Perform gel extraction with Qiaquick Gel Extraction Kit according to the provided protocol. Do not skip optional steps.
Part 17: Western blot (optional)
104.Prepare 300 μl of 2× SDS Loading Buffer with DTT (60 μl 1 M DTT, 240 μl 2× SDS Loading Buffer).
105.Prepare samples by combining volumes specified in Table 2. Each lane will contain the equivalent of 500,000 cells. M = Precision Plus marker, I = input, FT = flow-through, E = eluate.
| M | I | FT | E | |
|---|---|---|---|---|
| Sample (µl) | 5 | 5 | 5 | 2 |
| H2O (µl) | 10 | 10 | 10 | 13 |
| 2× Buffer (µl) | 15 | 15 | 15 | 15 |
| Total (µl) | 30 | 30 | 30 | 30 |
106.Incubate at 95°C for 5 min, vortex briefly, and centrifuge briefly at room temperature.
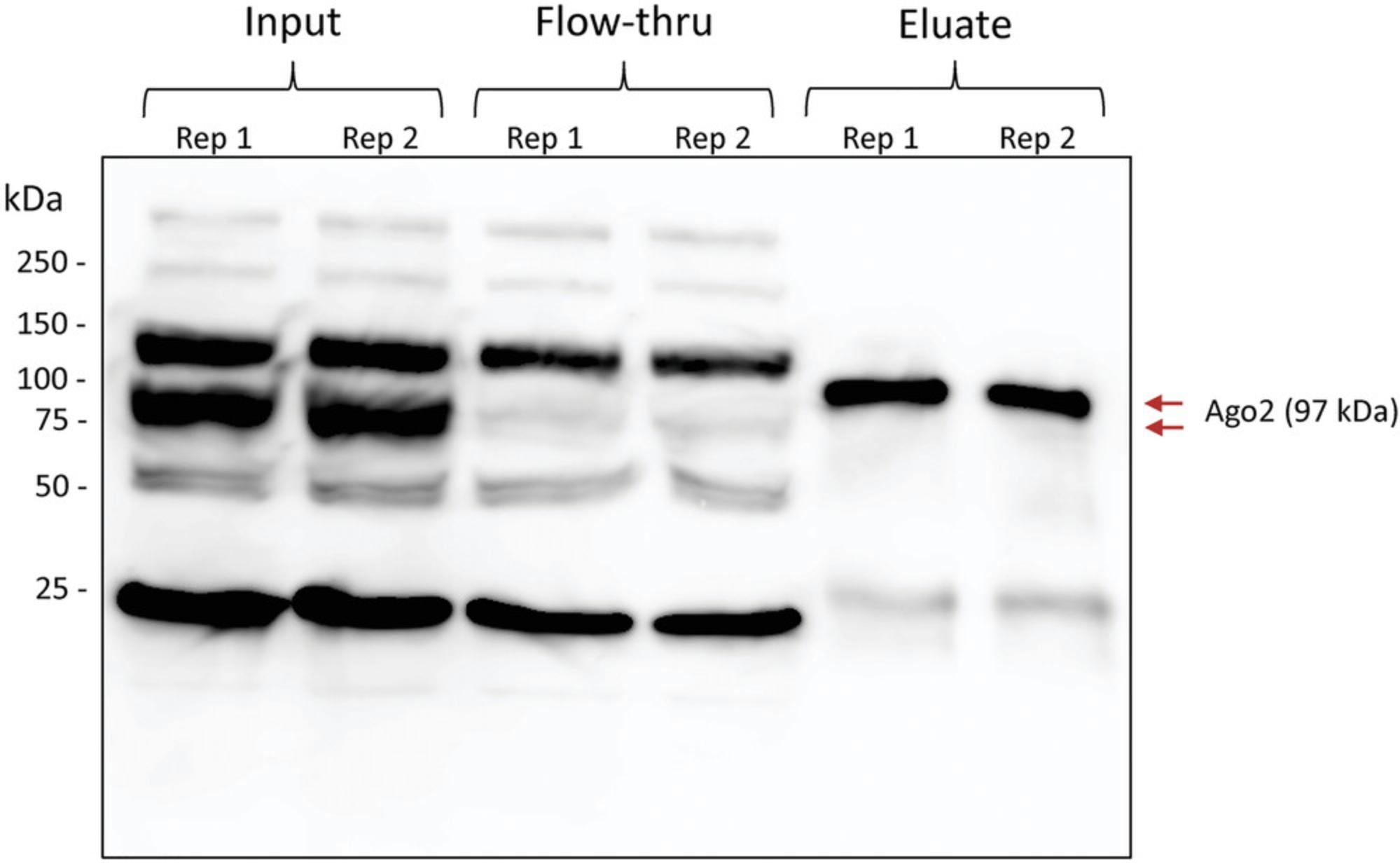
107.Proceed with a standard Western blot. Use Anti-Ago2 11A9 diluted 1:1000 as the primary antibody and peroxidase-conjugated goat anti-Rat IgG diluted 1:5000 as the secondary antibody. See Figure 2 for a representative Western blot.
Part 17: Quality control and sequencing
108.Accurately quantify the newly prepared qCLASH libraries.
109.Assess the size distribution of the qCLASH libraries on an Agilent TapeStation.

110.Perform next-generation sequencing. At a minimum, 100 cycles should be used. Sequencing can be single or paired-end, depending on the bioinformatics analysis to be performed.
Support Protocol: OPTIMIZATION OF AGO IMMUNOPRECIPITATION
This Support Protocol is a scaled-down version of the immunoprecipitation portion of the qCLASH protocol, using only 500,000 cells per replicate. It is intended to be completed prior to starting the main protocol in order to optimize the conditions of the immunoprecipitation. We have found that different cell lines may have different pulldown efficiencies, and therefore it is important to determine the most appropriate conditions for each cell line. The main factor that can be adjusted here is the concentration of the anti-Ago antibody. As indicated in the annotations of the Basic Protocol, the amount of anti-Ago 2A8 used is not fully saturating, so it should be possible to increase its concentration without adjusting the concentration of beads or bridging antibody used.
For materials, see Basic Protocol.
Day 1
Bead preparation and antibody conjugation
1.Transfer 120 μl Protein G Dynabeads to a Lo-Bind 1.5-ml tube.
2.Wash beads three times, each time with 450 μl cold 1× PBS-T, pH 7.2.Invert tube to resuspend. Microcentrifuge briefly between washes at <775 × g. Carry out all wash steps in this manner throughout the protocol.
3.Add 485 μl 1× PBS-T, pH 7.2, and 15 μl rabbit anti-mouse bridging antibody to tube. Resuspend beads.
4.Rotate 1 hr at room temperature.
5.Wash three times, each time with 450 μl cold 1× PBS-T, pH 7.2.
6.Resuspend in 620 µl PBS-T and divide into six aliquots of 100 µl.
7.Prepare the six tubes as indicated in Table 3.
| Tube 1 | Tube 2 | Tube 3 | Tube 4 | Tube 5 | Tube 6 | |
|---|---|---|---|---|---|---|
| 2A8 (µl) | 2 | 2 | 3 | 3 | 4 | 4 |
| PBS-T (µl) | 398 | 398 | 397 | 397 | 396 | 396 |
8.Rotate 4 hr at 4°C.
9.Wash 2A8-conjugated beads four times, each time with 450 μl cold 1× PXL buffer.
10.Resuspend each aliquot in 200 μl of 1× PXL. Store at 4°C until lysates are ready.
Part 2: Lysate preparation
11.Prepare 500 µl complete lysis buffer containing the protease inhibitor mixture and DTT as described in Reagents and Solutions.
12.Take one aliquot of 3 × 107 cells and thaw on ice. Resuspend in 300 μl complete lysis buffer.
13.Lyse 15 min on ice.
14.Spin lysate 15 min at 21,000 × g , 4°C.
15.Transfer supernatant to new tube and discard pellet.
Part 3: Immunoprecipitation
16.Transfer 10 μl of lysate to new tube and store at −20°C.
17.Remove buffer from beads and transfer 48 µl lysate to each of the six tubes. Add 252 µl complete lysis buffer to each tube. Rotate overnight at 4°C.
Day 2
18.Transfer supernatants to new tubes and store at −20°C.
19.Wash three times (450 μl per wash) with each of the following ice-cold buffers: 1× PXL, 5× PXL, high-stringency buffer, high-salt buffer, and 1× PNK buffer. After first wash with high-stringency buffer and high-salt buffer, incubate tubes onice 1 min.
20.Leave beads in final wash buffer and incubate on ice while preparing 400 μl of 1× SDS loading buffer with DTT (40 μl 1 M DTT, 160 μl 2× SDS loading buffer, 200 μl H2O).
21.Remove wash buffer and add 50 μl of 1× SDS loading buffer to each tube.
22.Store samples at −20°C if not proceeding directly to Western blot.
Part 4: Western blot
23.Prepare 300 μl of 2× SDS loading buffer with DTT (60 μl 1M DTT, 240 μl 2× SDS loading buffer).
24.Prepare samples by combining volumes specified in Table 4.Each lane will contain the equivalent of 250,000 cells. M = Precision Plus marker, I = input, FT = flow-through, E = eluate.
| M | I | FT | E | |
|---|---|---|---|---|
| Sample (µl) | 5 | 2.5 | 15 | 2.5 |
| H2O (µl) | 10 | 12.5 | 0 | 12.5 |
| 2× Buffer (µl) | 15 | 15 | 15 | 15 |
| Total (µl) | 30 | 30 | 30 | 30 |
25.Incubate at 95°C for 5 min, vortex briefly, and centrifuge briefly at room temperature.
26.Proceed with a standard Western blot. Use anti-Ago2 11A9 diluted 1:1000 as the primary antibody and peroxidase-conjugated goat anti-rat IgG diluted 1:5000 as the secondary antibody. See Figure 2 for a representative Western blot.
REAGENTS AND SOLUTIONS
Unless otherwise noted, stock solutions should be passed through a 0.45-µm filter prior to buffer preparation. Use only distilled, deionized, DEPC-treated water (abbreviated DEPC ddH2O; Current Protocols , 2001 ) unless otherwise indicated.
DNA loading buffer, 6×
- 3.3 ml glycerol (Fisher, cat. no. BP229-1)
- 200 µl 1% (w/v) bromophenol blue (Sigma, cat. no. B6131)
- 200 µl 1% (w/v) xylene cyanol (Fisher, cat. no. BP565-10)
- 6.3 ml ddH2O
- Water does not need to be DEPC-treated
- Store at room temperature indefinitely
DNA loading buffer, 6× (xylene cyanol only)
- 3.3 ml glycerol (Fisher, cat. no. BP229-1)
- 200 µl 1% xylene cyanol (Fisher, cat. no. BP229-1)
- 6.5 ml ddH2O
- Water does not need to be DEPC-treated
- Store at room temperature indefinitely
High-salt buffer
- 0.75 ml 1 M Tris·HCl, pH 7.5 (Current Protocols, 2001)
- 0.5 ml 0.5 M EDTA (Sigma, cat. no. E9884)
- 0.25 ml 0.5 M EGTA (Sigma, cat. no, E3889) ml
- 10% (v/v) Triton X-100 (Fisher, cat. no. BP151-100)
- 5 ml 10% (v/v) sodium deoxycholate (Sigma, cat. no. D6750)
- 0.5 ml 10% (w/v) SDS (Fisher, cat. no. BP166-100)
- 10 ml 5 M NaCl (Fisher, cat. no. S271-500)
- 28 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4°C for up to 1 year
Also see http://cshprotocols.cshlp.org/content/2018/12/pdb.prot097956.full#sec-1.
High-stringency buffer
- 0.75 ml 1 M Tris·HCl, pH 7.5 (Current Protocols, 2001)
- 0.5 ml 0.5 M EDTA (Sigma, cat. no. E9884)
- 0.25 ml 0.5 M EGTA (Sigma, cat. no, E3889)
- 5 ml 10% (v/v) Triton X-100 (Fisher, cat. no. BP151-100)
- 5 ml 10% (v/v) sodium deoxycholate (Sigma, cat. no. D6750)
- 0.5 ml 10% (w/v) SDS (Fisher, cat. no. BP166-100)
- 1.2 ml 5 M NaCl (Fisher, cat. no. S271-500)
- 0.3125 ml 4 M KCl (Sigma, cat. no. P9541)
- 36.488 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4°C for up to 1 year
Also see http://cshprotocols.cshlp.org/content/2018/12/pdb.prot097956.full#sec-1.
Lysis buffer
- 2.5 ml 1 M HEPES-KOH, pH 7.5 (HEPES, Fisher, cat. no. S271-500; KOH, Fisher, cat. no. BP310-100)
- 1.875 ml 4 M KCl (Sigma, cat. no. P9541)
- 200 μl 0.5 M EDTA (Sigma, cat. no. E9884)
- 100 μl 0.5 M NaF (Fisher, cat. no. S299-500)
- 2.5 ml 10% (v/v) NP-40 (IGEPAL CA-630; Sigma, cat. no. I3021)
- 42.825 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4°C for up to 1 year
- Immediately before use, add 5 µl/1 ml 0.1 M DTT (packaged with SuperScript IV )and 40 μl/1 ml 25× Complete Protease Inhibitor Mix (Roche, cat. no. 11836170001
PBS, pH 7.4 (w/o Ca+/ Mg+), 10×
- 274 ml 5 M NaCl (Fisher, cat. no. S271-500)
- 6.75 ml 4 M KCl (Sigma, cat. no. P9541)
- 11.36 g Na2HPO4 (Fisher, cat. no. S375-12)
- 2.72 g KH2PO4 (Fisher, cat. no. P285-500)
- DEPC ddH2O (Current Protocols, 2001) up to 1 L
- Store at room temperature for up to 1 year
PBS-T, 1×
- 5 ml 10× PBS, pH 7.4 w/o Ca++/Mg++ (see recipe)
- 100 μl Tween-20 (Fisher, cat. no. BP337-100)
- 44.9 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4°C for up to 1 year
PK buffer, 5×
- 25 ml 1 M Tris·HCl, pH 7.5 (Current Protocols, 2001)
- 2.5 ml 5 M NaCl (Fisher, cat. no. S271-500)
- 5 ml 0.5 M EDTA (Sigma, cat. no. E9884)
- 17.5 ml DEPC ddH2O (Current Protocols, 2001)
- Store at room temperature for up to 1 year
PNK, 1× and 10×
- 2.5 ml 1 M Tris·HCl, pH 7.5 (Current Protocols, 2001)
- 0.5 ml 1 M MgCl2 (Fisher, cat. no. BP214-500)
- 2.5 ml 10% (v/v) NP-40 (Sigma, cat. no. I3021)
- 44.5 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4° C for up to 1 year
Also see http://cshprotocols.cshlp.org/content/2018/12/pdb.prot097956.full#sec-1.
PNK-EGTA, 1×
- 2.5 ml 1 M Tris·HCl, pH 7.5 (Current Protocols, 2001)
- 2 ml 0.5 M EGTA (Sigma, cat. no, E3889)
- 2.5 ml 10% (v/v) NP-40 (Sigma, cat. no. I3021)
- 43 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4° C for up to 1 year
PXL, 1×
- 5 ml 10× PBS, pH 7.4 w/o Ca++/Mg++ (see recipe)
- 0.5 ml 10% (w/v) SDS (Fisher, cat. no. BP166-100)
- 2.5 ml 10% (v/v) sodium deoxycholate (Sigma, cat. no. D6750)
- 2.5 ml 10% (v/v) NP-40 (Sigma, cat. no. I3021)
- 35 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4° C for up to 1 year
Also see http://cshprotocols.cshlp.org/content/2018/12/pdb.prot097956.full#sec-1.
PXL, 5×
- 25 ml 10× PBS, pH 7.4 w/o Ca++/Mg++ (see recipe)
- 0.5 ml 10% (w/v) SDS (Fisher, cat. no. BP166-100)
- 2.5 ml 10% (v/v) sodium deoxycholate (Sigma, cat. no. D6750)
- 2.5 ml 10% (v/v) NP-40 (Sigma, cat. no. I3021)
- 19.5 ml DEPC ddH2O (Current Protocols, 2001)
- Store at 4° C for up to 1 year
Also see http://cshprotocols.cshlp.org/content/2018/12/pdb.prot097956.full#sec-1.
SDS loading buffer, 2×
- 500 μl 0.5 1 M Tris·HCl, pH 8.0 (Current Protocols, 2001)
- 200 mg SDS (Fisher, cat. no. BP166-100)
- 2 ml 50% (v/v) glycerol (Fisher, cat. no. BP229-1)
- 1.25 ml 1% (w/v) bromophenol blue (Sigma, cat. no. B6131)
- 50 µl ddH2O
- Stock solutions do not need to be filtered
- Water does not need to be DEPC-treated
- Store at room temperature indefinitely
- Immediately before use, add DTT to a final concentration of 200 mM. For example, to prepare 100 µl, combine 20 µl 1 M DTT with 80 µl of the above mixture.
TAE, 50×
- 242 g Tris base (Fisher, cat. no. BP152-5)
- 57.1 ml acetic acid (Fisher, cat. no. A38S-500)
- 7.43 g EDTA (Sigma, cat. no. E9884)
- ddH2O up to 1 L
- Store at room temperature indefinitely
Also see http://cshprotocols.cshlp.org/content/2013/5/pdb.rec074294.full.
COMMENTARY
Background Information
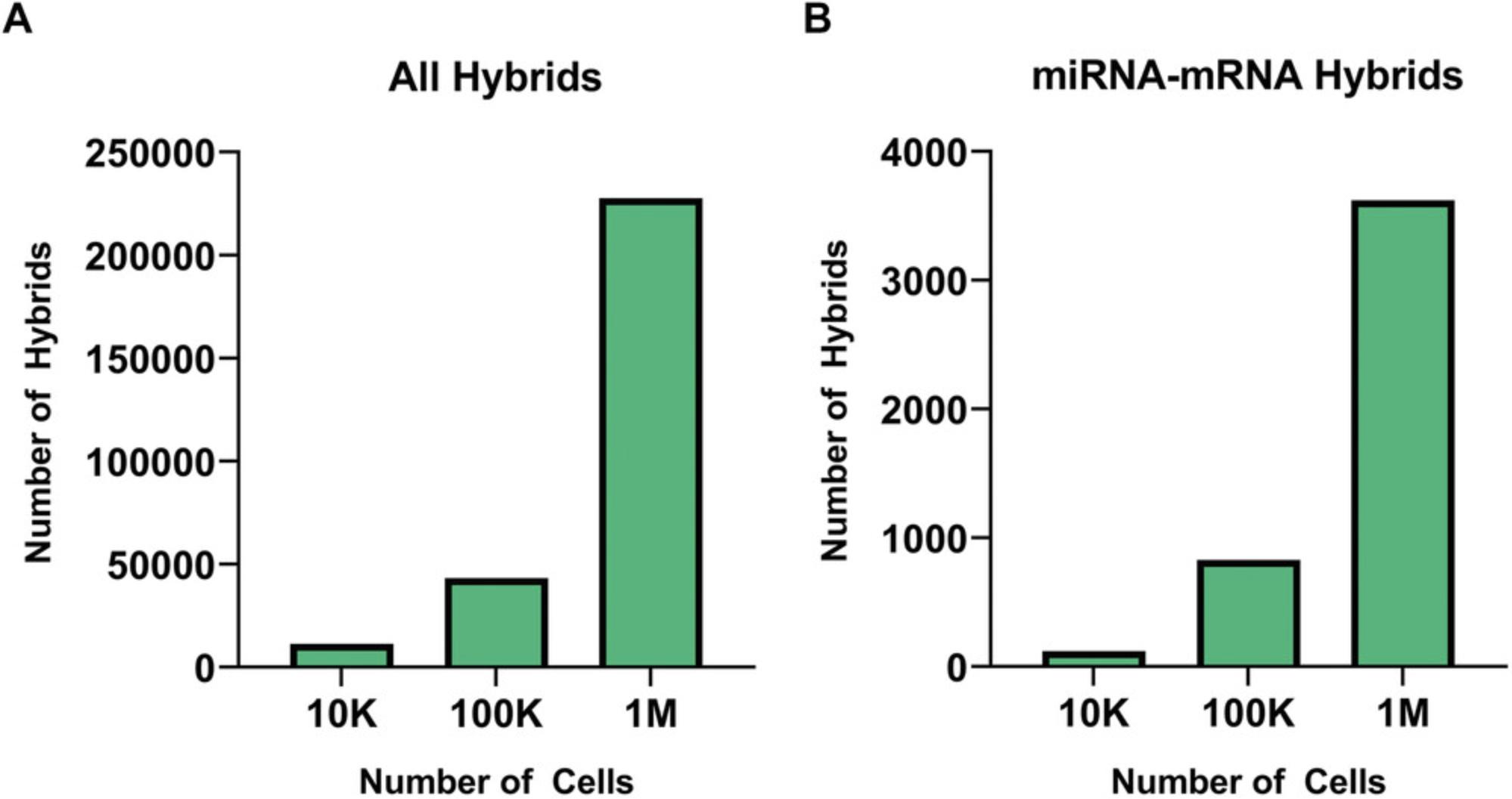
The original idea of CLASH was to covalently link miRNAs and RNA fragments by RNA ligation and thereby identify high-confidence miRNA targets (Helwak & Tollervey, 2014). In essence, our modified qCLASH protocol is streamlined and allows us to apply the method to a smaller number of cells. The primary differences are omitting the gel purification step of immunoprecipitated Ago/RNA complexes, and performing several steps with bead-bound biochemistry. qCLASH generates libraries that contain many RNAs, including miRNA/RNA hybrids that can be identified by next-generation sequencing at higher read depths, which is now fiscally feasible. We initially applied this to endothelial cell cultures, which are expensive to grow to very high cell numbers, and now have performed qCLASH using less than 1 million cells (Fig. 4). The long-term goal is to apply this technique to human tumor samples to assess miRNA-dependent post-transcriptional regulation in vivo. While this protocol was developed to study KSHV, EBV, and MHV-68 miRNA targetomes, it has already been applied to non-viral cancers such as metastatic melanoma (Bullard et al., 2019; Gay et al., 2018; Kozar et al., 2021; Ungerleider et al., 2020). Moreover, in combination with innovative bioinformatic approaches, qCLASH has identified a novel human miRNA called miR-SNAR that is up-regulated in breast cancer cells and regulates migration (Stribling et al., 2021). In summary, this protocol simplifies the application of state-of-the-art ribonomics methods to identify high-confidence miRNA targetomes in disease-relevant models and tissues.
Critical Parameters
Although shorter than its predecessor, qCLASH is a lengthy protocol with few opportunities to monitor progress during the procedure. This makes it difficult to pinpoint the source of a problem when the protocol does not yield the desired results. As a consequence, there are quite a few parameters to consider when attempting to perform troubleshooting or optimization. The most significant considerations when planning or performing a qCLASH experiment are discussed below.
The Ago IP is essentially the foundation of the qCLASH protocol, and is therefore absolutely critical to its success. Since qCLASH pulls down native Ago, the IP is completely unlike that of the original CLASH. Rather, it is more similar to the IP from Ago HITS-CLIP. With this method, we are routinely able to achieve Ago pulldown efficiencies of 90% or greater (Fig. 3). However, we have noticed that there is some variability in the efficiency between different cell lines. For this reason, we recommend performing an IP optimization prior to attempting the full-length qCLASH protocol. The test IP can be scaled down from the full experiment to save on materials, but the total volume of the IP sample during overnight incubation should be large enough to ensure that the beads remain in suspension (approximately 300 µl; see Support Protocol). Since the amount of anti-Ago 2A8 used in qCLASH is not saturating, increasing the concentration of this antibody is the most straightforward adjustment which can be made to the IP. Several different concentrations can be tested side-by-side during optimization in order to ensure the best result.
Another important consideration before beginning a qCLASH experiment is the amount of starting material that will be used. One of the main benefits of qCLASH is the lower amount of input required. We originally used 50 million cells per replicate, although we have tested inputs as low as 10,000 cells. Hybrids can still be found even when very few cells are used initially, but their number is quite limited (Fig. 4). Based on our observations, 10 million cells per replicate should provide a sufficient number of hybrids for most purposes.
Although it is always a good idea to be careful when working with delicate RNA, it is absolutely essential when performing qCLASH. All work surfaces and equipment should be thoroughly cleaned with freshly-prepared 10% bleach or other RNase-destroying product. All water used during the protocol should be treated with DEPC in order to remove RNases. The RNA-containing samples should always be kept on ice when they are not being handled. Lastly, common-sense precautions, such as wearing a lab coat and gloves, should not be neglected.
Troubleshooting
See Table 5 for troubleshooting recommendations.
| Problem | Possible cause | Solution |
|---|---|---|
| Inadequate pulldown of Argonaute | Concentration of anti-Ago 2A8 too low | Perform the Support Protocol in order to optimize the amount of anti-Ago 2A8 used |
| Low concentration of final PCR product | Insufficient amplification | Increase the number of PCR cycles used in Part 14 |
| Insufficient starting material | Increase the number of cells used as input in Part 2 | |
| Strong peak around 120-130 bp on TapeStation electropherogram | Formation of primer-dimers | Perform the optional size selection described in Part 15 |
| Alternatively, perform the optional gel purification described in step 16 | ||
| Decrease the concentration of the 3′ and 5′ adapters added in Parts 8 and 12, respectively |
Understanding Results
While the analysis of sequencing data falls outside the scope of this protocol, we will discuss the general characteristics of the data we obtained so that protocol users may determine if their own data are consistent with expectations. Table 6 shows statistics for nine qCLASH libraries (see Data Availability Statement). The raw sequencing data were analyzed with Hyb, software specifically developed for CLASH analysis by Travis, Moody, Helwak, Tollervey, & Kudla (2014). In all, hybrids make up approximately 2% of reads. However, with an average sequencing depth of approximately 125 million reads, hundreds of thousands of hybrids could be identified in each library. In addition to miRNA-mRNA hybrids, numerous hybrids consisting of miRNA and long noncoding RNA (lncRNA) have been found (Sethuraman, Thomas, Gay, & Renne, 2018).
| Filtered reads | Hybrids | miRNA-mRNA hybrids | |
|---|---|---|---|
| Wild-Type BR1a | 35,787,224 | 366,007 | 113,926 |
| Wild-Type BR2 | 47,918,368 | 403,582 | 144,792 |
| Wild-Type BR3 | 141,007,968 | 804,314 | 215,276 |
| ΔmiR-K12-11 BR1 | 129,696,332 | 754,887 | 178,805 |
| ΔmiR-K12-11 BR2 | 158,797,064 | 911,100 | 178,413 |
| ΔmiR-K12-11 BR3 | 67,441,408 | 354,417 | 70,108 |
| Uninfected BR1 | 120,656,016 | 530,682 | 97,309 |
| Uninfected BR2 | 220,792,548 | 983,058 | 171,062 |
| Uninfected BR3 | 218,518,836 | 695,475 | 96,751 |
- a BR1, biological replicate 1.
Time Considerations
Seven days are required to complete all the wet lab steps of qCLASH (Basic Protocol). The time required for each of the major components of the protocol is indicated in the Basic Protocol. Additional time is needed for sequencing and bioinformatic analysis.
Acknowledgments
The work described here was supported by the National Cancer Institute, NIH (5F31 CA180522 to L.A.G.; and 5R01 CA119917 and 5P01 CA214091 to R.R.).
Author Contributions
Lauren Gay : conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, validation, visualization, writing original draft, writing review and editing; Peter Turner : writing review and editing; Rolf Renne : conceptualization, funding acquisition, methodology, resources, writing original draft
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The raw data that support the protocol are openly available in the NCBI Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/, accession number GSE101978. Additional data (Excel tables) resulting from this protocol are available in the supplementary material of Gay et al. (2018).
Literature Cited
- Ambros, V. (2004). The functions of animal microRNAs. Nature , 431, 7006, 350–355.
- Current Protocols. (2001). Common stock solutions, buffers, and media. Current Protocols in Cell Biology , 00, A.2A.1−A.2A.10. doi: 10.1002/0471143030.cba02as00.
- Bullard, W. L., Kara, M., Gay, L. A., Sethuraman, S., Wang, Y., Nirmalan, S. … Tibbetts, S. A. (2019). Identification of murine gammaherpesvirus 68 miRNA-mRNA hybrids reveals miRNA target conservation among gammaherpesviruses including host translation and protein modification machinery. PLoS Pathogens , 15(8), e1007843. doi: 10.1371/journal.ppat.1007843.
- Chi, S. W., Zang, J. B., Mele, A., & Darnell, R. B. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature , 460(7254), 479–486. doi: 10.1038/nature08170.
- Gay, L. A., Sethuraman, S., Thomas, M., Turner, P. C., & Renne, R. (2018). Modified cross-linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi's sarcoma-associated herpesvirus microRNA targets in endothelial cells. Journal of Virology , 92(8), e02138–02117. doi: 10.1128/JVI.02138-17.
- Grosswendt, S., Filipchyk, A., Manzano, M., Klironomos, F., Schilling, M., Herzog, M. … Rajewsky, N. (2014). Unambiguous identification of miRNA: Target site interactions by different types of ligation reactions. Molecular Cell , 54(6), 1042–1054. doi: 10.1016/j.molcel.2014.03.049.
- Hafner, M., Landthaler, M., Burger, L., Khorshid, M., Hausser, J., Berninger, P. … Tuschl, T. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell , 141(1), 129–141. doi: 10.1016/j.cell.2010.03.009.
- Helwak, A., Kudla, G., Dudnakova, T., & Tollervey, D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell , 153(3), 654–665. doi: 10.1016/j.cell.2013.03.043.
- Helwak, A., & Tollervey, D. (2014). Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH). Nature Protocols , 9(3), 711–728. doi: 10.1038/nprot.2014.043.
- Kozar, I., Philippidou, D., Margue, C., Gay, L. A., Renne, R., & Kreis, S. (2021). Cross-Linking Ligation and Sequencing of Hybrids (qCLASH) reveals an unpredicted miRNA targetome in melanoma cells. Cancers , 13(5), 1096. doi: 10.3390/cancers13051096.
- Moore, M. J., Scheel, T. K., Luna, J. M., Park, C. Y., Fak, J. J., Nishiuchi, E. … Darnell, R. B. (2015). miRNA-target chimeras reveal miRNA 3'-end pairing as a major determinant of Argonaute target specificity. Nature Communications , 6, 8864. doi: 10.1038/ncomms9864.
- Sethuraman, S., Thomas, M., Gay, L. A., & Renne, R. (2018). Computational analysis of ribonomics datasets identifies long non-coding RNA targets of gamma-herpesviral miRNAs. Nucleic Acids Research , doi: 10.1093/nar/gky459.
- Stribling, D., Lei, Y., Guardia, C. M., Li, L., Fields, C. J., Nowialis, P. … Xie, M. (2021). A non-canonical microRNA derived from the snaR-A non-coding RNA targets a metastasis inhibitor. RNA , 27(6): 694–709. doi: 10.1261/rna.078694.121.
- Travis, A. J., Moody, J., Helwak, A., Tollervey, D., & Kudla, G. (2014). Hyb: A bioinformatics pipeline for the analysis of CLASH (crosslinking, ligation and sequencing of hybrids) data. Methods , 65(3), 263–273. doi: 10.1016/j.ymeth.2013.10.015.
- Ungerleider, N., Bullard, W., Kara, M., Wang, X., Roberts, C., Renne, R. … Flemington, E. (2020). EBV miRNAs are potent effectors of tumor cell transcriptome remodeling in promoting immune escape. bioRxiv , 2020.2012.2021.423766. doi: 10.1101/2020.12.21.423766 bioRxiv.
Internet Resources
ThermoFisher Dynabeads Protein G for Immunoprecipitation product page.
Cold Spring Harbor Protocols immunoprecipitation and SDS-PAGE for cross-linking immunoprecipitation (CLIP).
ThermoFisher RNase T1 product information page.
NEB T4 RNA Ligase 1 product information page.
Roche APMB-RO Alkaline Phosphatase product information page, from Sigma Aldrich.
NEB T4 RNA Ligase 2, truncated K227Q product information page.
Recipe for 50× TAE.
Citing Literature
Number of times cited according to CrossRef: 3
- Zhaokang Shen, Muhammad Naveed, Jianqiang Bao, Untacking small RNA profiling and RNA fragment footprinting: Approaches and challenges in library construction, WIREs RNA, 10.1002/wrna.1852, 15 , 3, (2024).
- Kylie I Krohmaly, Robert J Freishtat, Andrea L Hahn, Bioinformatic and experimental methods to identify and validate bacterial RNA-human RNA interactions, Journal of Investigative Medicine, 10.1136/jim-2022-002509, 71 , 1, (23-31), (2023).
- Siyu Chen, Yue Deng, Dongli Pan, MicroRNA Regulation of Human Herpesvirus Latency, Viruses, 10.3390/v14061215, 14 , 6, (1215), (2022).

