Phage Display Library Prep Method
Sabrina A Mann, Sara Vazquez, Caleigh Mandel-Brehm, Lillian Khan, Joseph Derisi
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
This protocol was designed to be used after the Scaled High Throughput Vacuum PhIP Protocol or the Scaled Moderate Throughput Multichannel PhIP Protocol. The immunoprecipitated phage targets are prepared for sequencing through a two PCR amplification rounds which first amplify the peptide target and second, add on UMIs.
Before start
Steps
Lysis
Dilute phage lysis product in water at a 1:4 ratio. Phage lysis should be used either immediately after IP or within 1 week of being stored at 4°C.
- NOTE: we have done several tests and noticed that diluting the phage lysate 1:4 in water results in the most product at the end. Other dilutions ranging from 1:2 to 1:16 also worked. Using undiluted lysate also works, however there is the possibility that too much input can inhibit the PCR.
Heat lysate in preparation for PCR at 70°C for 0h 15m 0s (can be done on the PCR machine if aliquoted into PCR tubes or plates). Keep on ice afterwards.
PCR 1- Amplifying Insert
- Add the following reagents. Reaction volumes of
50µL,25µLor12.5µLall work. - Multiply the number of samples by either the
50µL,25µLor12.5µLreaction coefficients to get the total volumes of each reagent to add to the master mix. - Make master mix and aliquot into a new PCR plate.
- Add pre-heated phage lysis to PCR I master mix.
| A | B | C | D |
|---|---|---|---|
| COMPONENT | 50uL REACTION | 25uL REACTION | 12.5uL REACTION |
| Nuclease-free water | 34.5 | 16.25 | 8.125 |
| 5X Phusion HF or GC Buffer | 10 | 5 | 2.5 |
| 10 mM dNTPs | 1 | 0.5 | 0.25 |
| 10 uM Forward Primer | 1 | 0.5 | 0.25 |
| 10 uM Reverse Primer | 1 | 0.5 | 0.25 |
| Polymerase | 0.5 | 0.25 | 0.125 |
| Template DNA (lysis) | 2 | 2 | 1 |
| Total amount of Master Mix | 48 | 23 | 11.5 |
Program PCR Machine for the following steps:
| A | B | C |
|---|---|---|
| STEP | TEMP | TIME |
| Initial Denaturation | 98°C | 30 seconds |
| 13-16 Cycles | 98°C | 5 seconds |
| 70°C | 20 seconds | |
| 72°C | 15 seconds | |
| Final Extension | 72°C | 2 minutes |
| Hold | 10°C |
PCR 2- Adding on UMI
- Multiply number of samples by either the
50µL,25µLor12.5µLreaction number coefficients to get total volume of each reagent to add to master mix. - Combine the following reagents (except barcode primers and PCR I product) to make PCR II master mix, then aliquot master mix volume into new PCR plate
- Next, add barcode primers and PCR I product, making sure to keep track of which barcode IDs corresponds to which sample well.
| A | B | C | D |
|---|---|---|---|
| COMPONENT | 50uL REACTION | 25uL REACTION | 12.5uL REACTION |
| Nuclease-free water | 33.5 | 16.25 | 8.125 |
| 5X Phusion HF or GC Buffer | 10 | 5 | 2.5 |
| 10 mM dNTPs | 1 | 0.5 | 0.25 |
| Polymerase | 0.5 | 0.25 | 0.125 |
| 5 uM Forward and Reverse Primer Barcodes | 4 | 2 | 1 |
| Template DNA (product of PCR I) | 1 | 1 | 0.5 |
| Total amount of Master Mix | 46 | 22 | 11 |
Program the PCR machine for the following steps:
| A | B | C |
|---|---|---|
| STEP | TEMP | TIME |
| Initial Denaturation | 98°C | 30 seconds |
| 5 Cycles | 98°C | 5 seconds |
| 70°C | 20 seconds | |
| 72°C | 15 seconds | |
| Final Extension | 72°C | 2 minutes |
| Hold | 10°C |
Pool and Bead Clean
Amplified samples are now uniquely labeled and are very similar in concentration and library size, so equal volume of each sample can be pooled together. If there is concern that samples are dissimilar, skip this step, bead clean, BioAnalyze and Qubit each sample individually. However, in our experience it is easier to pool first and then bead clean everything together prior to the QC check via BioAnalyzer and Qubit.
If pooling, it is recommend to pool anywhere between 5µL-10µL of each sample together by using a multichannel and combining all of the rows into a reservoir.
- NOTE: It is also recommended to pool each plate separately from other plates. This allows us to protect individual plates from phage contamination.
*Allow beads to sit in 70Room temperature for 0h 30m 0s prior
*Ratio of beads changes based on application and library size.
- Determine the total volume of the pool. (i.e. 100uL)
- Use Agencourt AmPure XP Beads 0.9x ratio of beads-to-total volume of sample. Prepare 80% EtOH.
- Add 0.9x (i.e. 90uL) beads of room temperature AmPure Beads to pool. Mix well by pipetting up and down gently.
- Pulse spin the tube but do not spin down beads. Incubate for
0h 5m 0sat70Room temperature. - Place samples on magnetic rack and incubate for
0h 5m 0son the rack. - Remove supernatant without disturbing the beads.
- Add 2x original volume of 80% EtOH or enough to submerge bead pellet while on the magnetic rack. Incubate at
70Room temperaturefor0h 0m 30sthen remove the supernatant. - Repeat EtOH wash (step 7) for a total of 2 times.
- Air dry the beads for
0h 5m 0swhile on the magnetic rack. - Remove tube from magnetic rack. Elute DNA from beads in desired volume of 0.1x TE Buffer, 10mM Tris-HCl, or Nuclease free water plus
3µLof dead volume. - Vortex to mix. Pulse spin tubes and incubate for
0h 2m 0sat70Room temperatureoff the magnetic rack. - Place on magnetic rack until solution is clear and bead pellet has formed ~
0h 5m 0s. - Remove desired volume of eluant and transfer to a clean nuclease-free PCR tube. Do not disturbe the bead pellet.
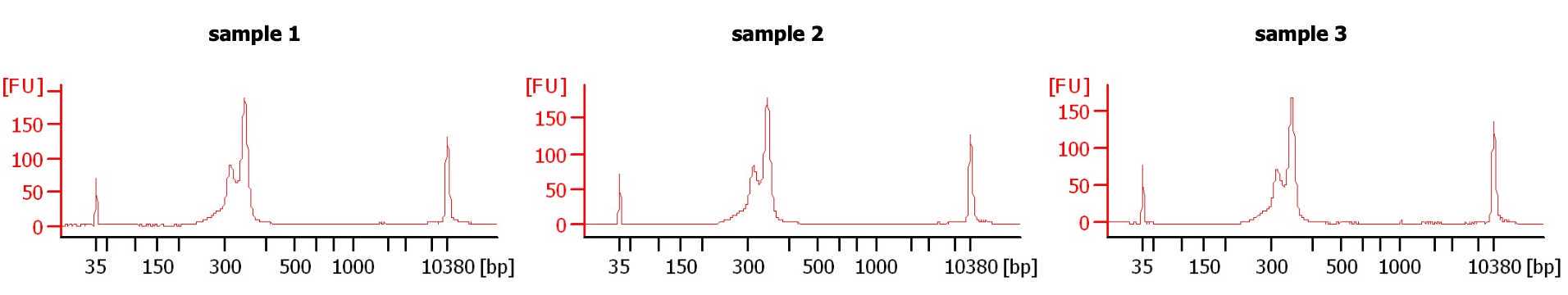
Quantify and Qualify Library
- Qubit the samples for accurate concentration. BioAnalyzer also gives the concentration of sample but in practice this number not as accurate as the Qubit result.The Qubit is our gold standard for confirming DNA concentration for dilution prior to using the BioAnalyzer as to not overload it.
- BioAnalyze or TapeStation library pools.
- If the library size looks as expected, pool separate plate pools together at equal concentration or at desired ratio for read depth. Final libraries can be stored at
4°Cfor a few weeks. For longterm storage, samples should be kept at-20°C.