Modified protocol for genome-wide mapping of uncapped transcripts (GMUCT) in eukaryotes
Diep R Ganguly, Garima Bhatia, Susheel Sagar Bhat, Brian D Gregory
Abstract
This is a modified protocol to perform genome-wide mapping of uncapped and cleaved transcripts (GMUCT; Willmann et al, 2014 Methods, 10.1016/j.ymeth.2013.07.003).
Before start
Purify DNA-free total RNA samples using method of choice, for example, TRIzol-chloroform extraction (dx.doi.org/10.17504/protocols.io.bt8wnrxe).
Attachments
Steps
Poly-A selection
For each sample, dilute 5 - 30 μg total RNA in 100 μL of nuclease-free water (or 10 mM Tris-Cl pH 7.5).
Perform poly-A RNA selection using Dynabeads mRNA purification kit as per the manufacturer's instructions (see attachments).
Note, the capacity of these beads are 75 μg total RNA. So, if you are starting with < 32.5 μg total RNA, you may want to scale-down all the reactions by one-half in order to conserve reagents.
Determine concentration and purity of poly-A selected RNA using a Nanodrop.
Aliquot a normalized amount of poly-A selected RNA, for 5' adapter, in 9 μL of DEPC-treated water. Ideally, this would be approximately 600 ng poly-A RNA recovered from 30 μg total RNA.
Ligate 5' adapter
Combine the following in 200 μL PCR tubes, for each sample, and mix by pipetting 6 times.
| A | B |
|---|---|
| Reagent | Volume (μL) |
| 25 μM RNA 5' Adapter (RA5) | 1 |
| Poly-A selected RNA (600 ng) | 9 |
Incubate at 70 ºC for 2 min, then 4 ºC for at least 2 min.
Prepare the following ligation mix, multiplying the volumes for the number of samples being processed (+5% for pipetting errors). Mix with gentle pipetting then briefly centrifuge.
| A | B |
|---|---|
| Reagent | Volume (μL) |
| T4 RNA Ligase buffer | 1.5 |
| 10 mM ATP | 1 |
| RNaseOUT | 1 |
| T4 RNA Ligase 1 | 1 |
5' ligation recipe
Add 4.5 μL of the mix to each sample, mix by pipetting 6 times, then briefly spin down.
Incubate at 28 ºC for 1 h.
Proceed directly to the next section or store at -20 ºC overnight.
Another poly-A selection is performed to remove un-ligated adapter using the NEBNext Poly(A) mRNA Magnetic Isolation Module. This kit is much more cost-effective to perform poly-A selection on samples with <5 μg RNA.
Add 35 μL DEPC-treated water to all samples to bring the final volume to 50 μL.
In a second 0.2 mL PCR tube, aliquot 20 μL of well resuspended NEBNext Magnetic Oligo d(T)25 Beads.
Add 100 μL RNA Binding Buffer and pipette mix 6 times.
Place tubes containing beads on a magnetic rack until solution is clear.
Remove supernatant without disturbing the beads.
Remove tubes from the rack and repeat steps 12 - 14.
Resuspend beads in 50 μL RNA Binding Buffer.
Add 50 μL of ligated RNA sample and mix by pipetting 6 times.
Incubate at 65 ºC for 5 minutes then hold at 4 ºC (denature RNA and facilitate binding of the poly-A to oligo dT beads).
Remove tubes from thermal cycler when samples reach 4 ºC.
Resuspend beads by pipetting 6 times, then incubate for 5 minutes at room temperature.
Repeat step 22.
Place tubes on magnetic rack until solution clear (poly-A RNA bound to beads).
Discard all supernatant without disturbing beads.
Remove tubes from magnetic rack.
Wash beads by adding 200 μL Wash Buffer and pipetting 6 times.
Place tubes on the rack and wait for the solution to become clear.
Discard the supernatant then remove tubes from the rack.
Repeat steps 27 - 29.
Add 8 μL of Tris Buffer and mix by pipetting 6 times.
Incubate samples at 80 ºC for 2 minutes, then hold at 25 ºC.
Once samples reach 25 ºC, place immediately on the magnetic rack.
Once solution is completely clear, transfer the supernatant to a clean nuclease-free PCR Tube. Keep tube on ice if continuing with reverse transcription reaction or store at -20 ºC.
Reverse transcription and addition of 3' adapter
Set up the following program in a thermal cycler:
- 65 °C for 5 min
- Hold at 4 °C
- 25 °C for 10 min
- 42 °C for 60 min
- 85 °C for 5 min
- Hold at 4 °C
Run the program to allow the thermal cycler to preheat (pause on step 1).
Prepare the following in clean 200 μL PCR tubes for each sample
| A | B |
|---|---|
| Reagent | Volume (uL) |
| 5' adapter-ligated RNA | 8 |
| 20 uM random hexamer primer that includes TruSeq RA3 on the 5' end of the primer (see Materials) | 1 |
| 12.5 mM dNTPs | 1 |
Mix contents of each tube by pipetting 6 times.
Place tubes in pre-heated thermal cycler and run steps 1-2, allows samples to sit at 4 °C for at least 2 minutes.
Prepare the following reverse transcription mix. Multiply volumes per sample and add 5% to account for pipetting errors. Mix gently by pipetting, then briefly centrifuge.
| A | B |
|---|---|
| Reagent | Volume (uL) |
| DEPC-treated water | 2 |
| 5x First Strand Buffer | 4 |
| 100 mM DTT | 2 |
| SuperScript II | 1 |
| RNaseOUT | 1 |
Reverse transcription recipe
Add 10 μL of the mix to each sample (keep on ice) and mix by pipetting up and down 6 times, then centrifuge briefly.
Return samples to the thermal cycler and continue with steps 3-6.
PCR amplification
Set up the following program in a thermal cycler:
- 98 °C for 30 sec
- 98 °C for 10 sec
- 60 °C for 30 sec
- 72 °C for 15 sec
- Back to step 2 (11x)
- 72 °C for 10 min
- Hold at 4 °C
Run the program to allow the thermal cycler to preheat (pause on step 1).
Add 2 μL of a unique RNA PCR Primer Index (RPI#, see materials) to each sample and mix by pipetting 6 times. Use the Illumina Index Adapter Pooling Guide (TruSeq Small RNA Library Prep Kit) to aid with RPI# selection (where # = 1-48).
Prepare the following PCR mix, multiplying the volumes for the number of samples (+5 % extra for pipetting errors). Mix thoroughly by pipetting and briefly centrifuge.
| A | B |
|---|---|
| Reagent | Volume (uL) |
| 2X Phusion Master Mix | 50 |
| RNA PCR Primer 1 (RP1) | 2 |
| 1 M Betaine | 26 |
PCR mix per library
Add 78 μL of the PCR mix to each sample and mix by pipetting up and down 6 times. Centrifuge briefly and keep samples on ice.
Distribute 100 μL across 4 PCR tubes (i.e. 4 x 25 μL reactions per sample). This is done to increase the PCR efficiency.
Place all samples into the pre-heated thermal cycler and run the program.
Ethanol precipitation
Combine 25 μL aliquots into a single 1.5 mL microtube.
Precipitate DNA by adding 500 μL ethanol, 3.5 μL GlycoBlue, and 10 μL 3 M NaOAc (pH 5.5).
Leave samples at -80 °C for at least 2 hours.
Centrifuge at max speed for 45 min at 4 °C.
Remove supernatant and rinse with 750 μL 80% ethanol.
Centrifuge at max speed for 5 min at 4 °C.
Remove supernatant without disturbing pellet and allow to air dry for 1-2 min.
Resuspend in 10 μL of nuclease-free water and allow samples to resuspend for 20 minutes on ice.
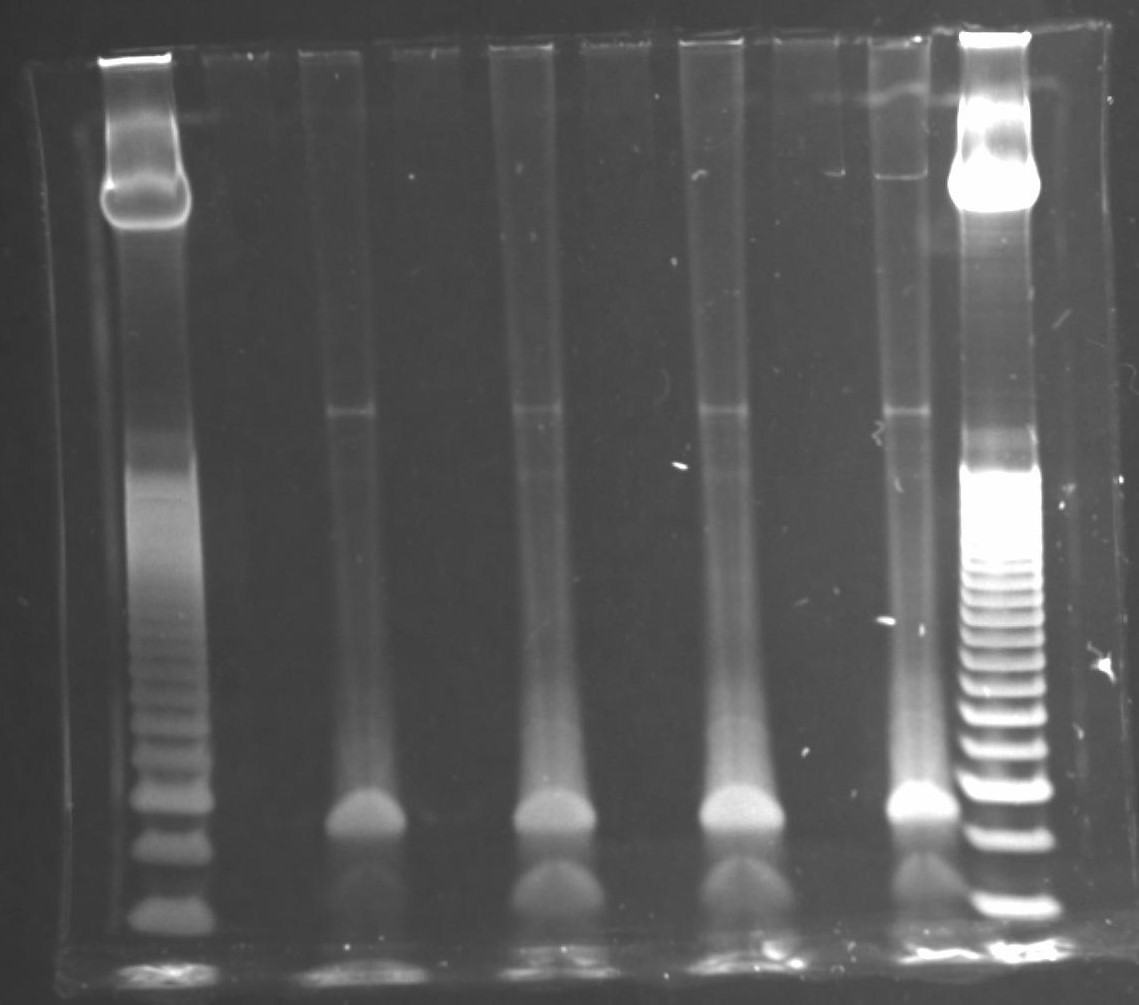
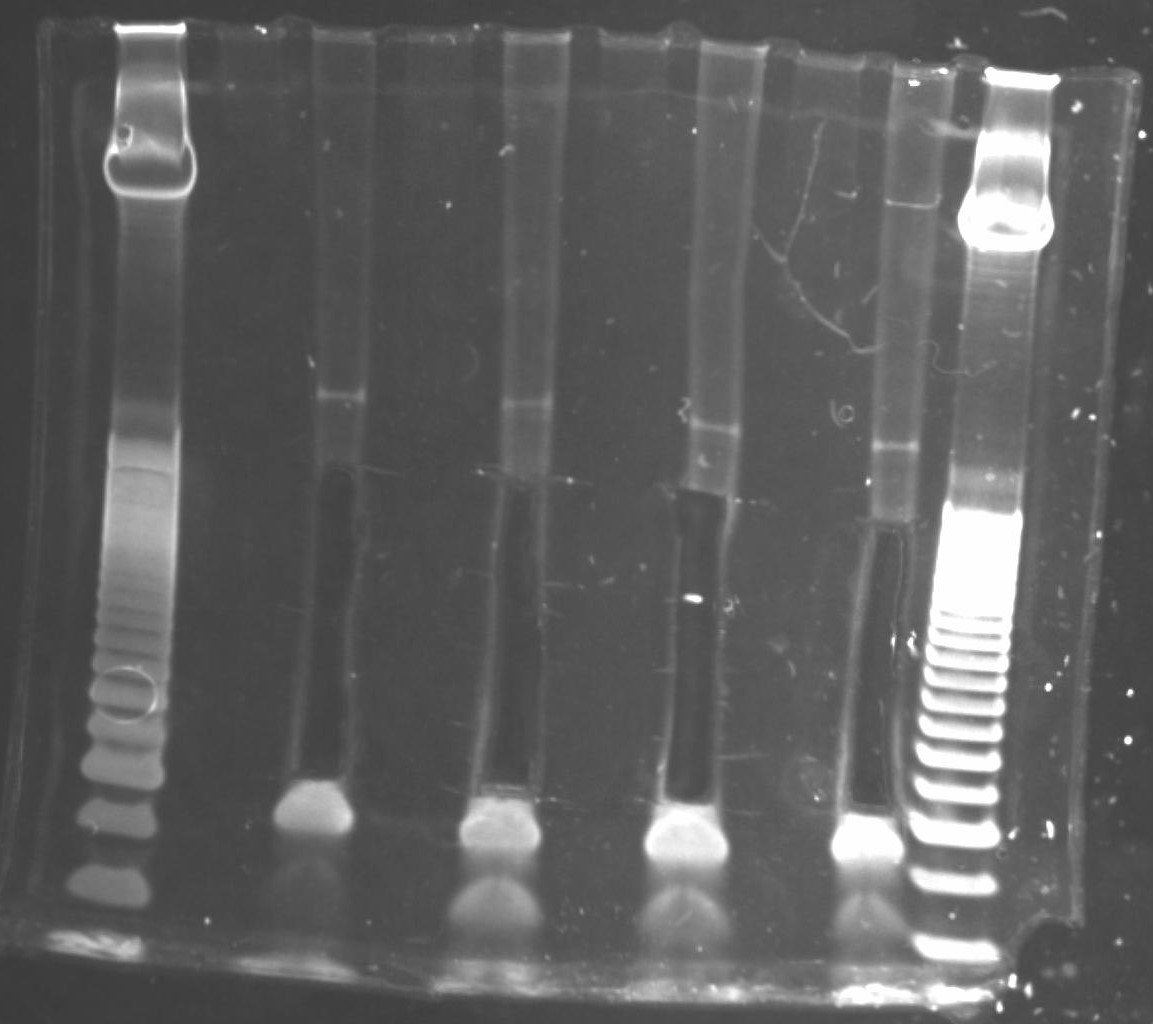
Size selection
Setup a 6% TBE polyacrylamide gel (Invitrogen) in a gel running tank with 1x TBE buffer (89 mM Tris base, 89 mM boric acid, 2 mM EDTA pH 8).
Prepare ladder and samples:
-
Mix 1.5 μL 25 bp ladder (Invitrogen) with 8.5 μL DEPC water and 10 μL gel loading buffer II (Invitrogen or NEB).
-
Mix 10 μL sample with 10 μL gel loading buffer II.
Load ladder and and samples into TBE gel, keeping one lane free between each sample to minimize likelihood of cross-contamination and make gel excision easier.
Run gel at 155 V for approximately 30 minutes (dye front should be just touching the bottom of the gel).
Note, you may prefer to run the gel at 80-100 V for 1-1.5 hours to achieve better separation of adapter-adapter clones (see below). You could also use a higher percentage of polyacrylamide.
While the gel is running, prepare gel breaker tubes for each sample to be size selected (or nuclease-free 0.5 mL tubes).
Use a 21G needle to make holes in the bottom of the tubes (or, in the case of gel breaker tubes, to increase their size) and place within a nuclease free 2 mL microtube.
Once gel run has completed, incubate gel for 10 minutes with 1X GelStar Nucleic Acid Stain in 100-200 mL 1X TBE in a nuclease-free tray.
Rinse gel with distilled water 3 times then image on a UV transilluminator.
For each sample, excise the region corresponding to 135 - 500 bp (can include larger fragments if there is visible product) and place inside gel breaker tubes (nested in a 2 mL microtube). Keep the excised region consistent among samples within an experiment.
Note: a minimum size of 135 bp is recommended since this corresponds to a 17 bp insert and anything smaller becomes much more difficult to map uniquely to the genome. This also avoids adapter-adapter clones (produced from random hexamer binding to 5' adapter), the longest of which would be 118 bp.
Centrifuge at max speed for 2 minutes and ensure that the gel completely passes through the holes (repeat spin and add more holes if necessary).
Add 300 μL NEB Buffer 2 (50 mM NaCl, 10 mM Tris-Cl, 10 mM MgCl2. 1 mM DTT, pH 7.9) and rotate for at least 2 hours (or overnight at 4 ºC).
Precipitate RNA by adding 5 μL GlycoBlue, 30 μL 3 M NaOAc pH 5.5, and 900 μL ethanol.
Incubate at -80 ºC for at least 2 hours (can leave overnight to maximize recovery).
Centrifuge at max speed for 1 hour at 4 ºC.
Remove supernatant and wash with 750 μL 70% ethanol.
Centrifuge at max speed for 5 min at 4 ºC.
Remove as much supernatant as possible, and allow tubes to air dry for 1 -2 minutes.
Quantify library concentration using a Qubit fluorometer with Qubit dsDNA HS Assay Kit (Invitrogen). Alternatively, run libraries on a LabChip GXII or Bioanalyzer to give information on concentration and fragment sizes.