FlowFISH with PrimeFlow
Jesse Engreitz, Ronghao Zhou
Abstract
Detect RNA expression in single cell with PrimeFlow
Steps
Before start:
Make sure the heat block temp is set to 40ºC using the digital monitor. The heat block should be ON at least a day in advance to stabilize temperature
Turn on Hyb oven (use to pre-warm target probe diluent)
Make FACS staining buffer: 0.5% BSA in PBS, filter sterilized
- 10% BSA stock: 3g BSA (light sensitive powder in 4ºC MISC) + 30mL PBS
- Store at 4ºC
Set vortex to ~1500RPM
A. Fixation and permeabilization
- leave ~100µL when removing buffer, vortex to resuspend pellet
- invert to mix when adding 1mL and tap the bottom if some cells stuck
- pipette up and down to mix for fixation, permeabilization, and hybridization
- may be in bulk, use volumes that cells don’t exceed 10M cells/mL
- pre-warm PrimeFlow RNA Wash Buffer Wash Buffer to room temperature
Aliquot 5-10M cells in PBS per sample
Add 1mL PBS per sample, pipette to mix, and spin down at 500g at 4ºC for 5min, discard supernatant, resuspend in ~100µL by vortexing gently
Prepare Fixation Buffer 1 by mixing equal parts of PrimeFlow RNA Fixation Buffer 1A and PrimeFlow RNA Fixation Buffer 1B
- need 1mL per sample: 500µL Buffer 1A + 500µL Buffer 1B
- mix gently by inverting, don’t vortex or vigorously shake, prepare fresh
- keep at 4ºC
Add 1mL Fixation Buffer 1 , pipette to mix, incubate in dark (put cardboard box on it) rotating on rotator for 30min at 2–8°C in cold room
- fixation time is critical, do NOT overshoot since the cells will lose integrity
Prepare 1X RNA Permeabilization Buffer with RNase Inhibitors by diluting PrimeFlow RNA Permeabilization Buffer (10X) and RNase Inhibitors (100X) with RNase-free water
- need 2mL per sample: 200µL Perm Buffer + 1.78mL H2O + 20µL RNase Inhibitor 2O + 20µL RNase Inhibitor
- mix gently by inverting, don’t vortex or vigorously shake, prepare fresh
- keep at 4°C
Spin down cells at 800g at 4ºC for 5min, discard supernatant
- speed is critical here: slower spin will lose a lot of cells
Add 1mL RNA Permeabilization Buffer with RNase Inhibitors, pipette to mix, and spin down at 800 g at 4ºC for 5min, discard supernatant
Repeat wash with 1mL RNA Permeabilization Buffer
- set centrifuge to room temp
Prepare 1X RNA Fixation Buffer 2 by diluting PrimeFlow RNA Fixation Buffer 2 (8X) with PrimeFlow RNA Wash Buffer
- need 1mL per sample: 125µL Fix Buffer 2 + 875µL RNA Wash Buffer
- mix gently by inverting, don’t vortex or vigorously shake, prepare fresh
- keep at room temp
Add 1mL RNA Fixation Buffer 2 , pipette to mix, and incubate for 60min in the dark at room temperature while rotating
Spin down cells at 800g at room temp for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
Add 1mL PrimeFlow RNA Wash Buffer , invert to mix, and spin down at 800 g at room temp for 5min, discard supernatant, resuspend (~100µL) by vortexing
- if in bulk, the cells should be transferred to the 1.5mL tubes from kit
Repeat wash with 1mL RNA Wash Buffer
- can store samples overnight at 4ºC, then the last wash with RNase Inhibitors
B. Target Probe hybridization
Thaw Probe Sets (20X), including positive control (RPL13A, Type 4) on ice;
pre-warm PrimeFlow RNA Target Probe Diluent to 40°C
take 2µL cells from “unstained” sample into 18µL PBS -> measure on countess with trypan blue and record cell amount and % live
- critical that the residual volume after all washes be as close to 100μL as possible, use the markings on the 1.5mL tubes to assist
Dilute Probe Sets (20X) 1:20 in PrimeFlow RNA Target Probe Diluent , mix thoroughly by pipetting up and down
- need 100µL per sample: 5µL Target Probe + 5µL RPL13A Probe + 90µL Diluent
- for unstained sample: 100µL Diluent
- keep at 40ºC
Add 100μL diluted Target Probe(s) into the cell suspension (~100µL), pipette to mix, briefly vortex, then incubate for 2h at 40°C
- do not pipette solutions onto the walls of the tubes, and samples should be mixed well before incubating
- vortex samples to mix every 30min
- temp is critical for hybridization
Add 1mL PrimeFlow RNA Wash Buffer , invert to mix, and spin down at 800 g for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
Prepare PrimeFlow RNA Wash Buffer with RNase Inhibitors by diluting RNase Inhibitors (100X) with RNA Wash Buffer
- need 1mL per sample: 10µL RNase Inhibitor + 990µL Wash Buffer
- mix gently by inverting, prepare fresh
- keep at room temp
Add 1mL PrimeFlow RNA Wash Buffer Wash Buffer with RNase Inhibitors, invert to mix, and spin down at 800 g for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
- Store samples overnight in the dark at 4ºC
C. Signal amplification
- Pre-warm samples and PrimeFlow RNA Wash Buffer to room temperature
- Pre-warm PrimeFlow RNA PreAmp Mix , PrimeFlow RNA Amp Mix , and PrimeFlow RNA Label Probe Diluent to 40°C
Add 100μL PrimeFlow RNA PreAmp Mix into the cell suspension (~100µL), pipette to mix, briefly vortex, then incubate for 1.5h at 40°C
- do not pipette solutions onto the walls of the tubes, and samples should be mixed well before incubating
Add 1mL PrimeFlow RNA Wash Buffer , invert to mix, and spin down at 800 g for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
Repeat wash two times with 1mL RNA Wash Buffer , for a total of three washes
Add 100μL PrimeFlow RNA Amp Mix into the cell suspension (~100µL), pipette to mix, briefly vortex, then incubate for 1.5h at 40°C
- do not pipette solutions onto the walls of the tubes, and samples should be mixed well before incubating
Add 1mL PrimeFlow RNA Wash Buffer , invert to mix, and spin down at 800 g for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
Repeat wash with 1mL RNA Wash Buffer
Dilute PrimeFlow RNA Label Probes (100X) 1:100 in PrimeFlow RNA Label Probe Diluent
- need 100µL per sample: 1µL Label Probes + 99µL Diluent
- keep at 40ºC
Add 100μL diluted Label Probes into the cell suspension (~100µL), pipette to mix, briefly vortex, then incubate for 1h at 40°C
- do not pipette solutions onto the walls of the tubes, and samples should be mixed well before incubating
Add 1mL PrimeFlow RNA Wash Buffer at room temp, invert to mix, and spin down at 800 g for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
Repeat washes with warm (35ºC) 1mL RNA Wash Buffer 5 times
Add 1mL FACS staining buffer (or PrimeFlow RNA storage buffer ), invert to mix, and spin down at 800 g for 5min, discard supernatant, resuspend (~100µL) by vortexing gently
- Samples can be stored in the dark at 4ºC for up to three days before analysis
- QC: take stained vs unstained samples and check fluorescence with microscope
D. Flow cytometric analysis
Add 100µL FACS buffer to cells, transfer all to filter, quick spin
Add 100µL FACS buffer to cap to wash remaining cells from filter, quick spin
FACS:
- 400-500µL of an optimally concentrated sample (20M cells/mL) take ~30min
- when low volume left, add ~200µL staining buffer can recover more cells
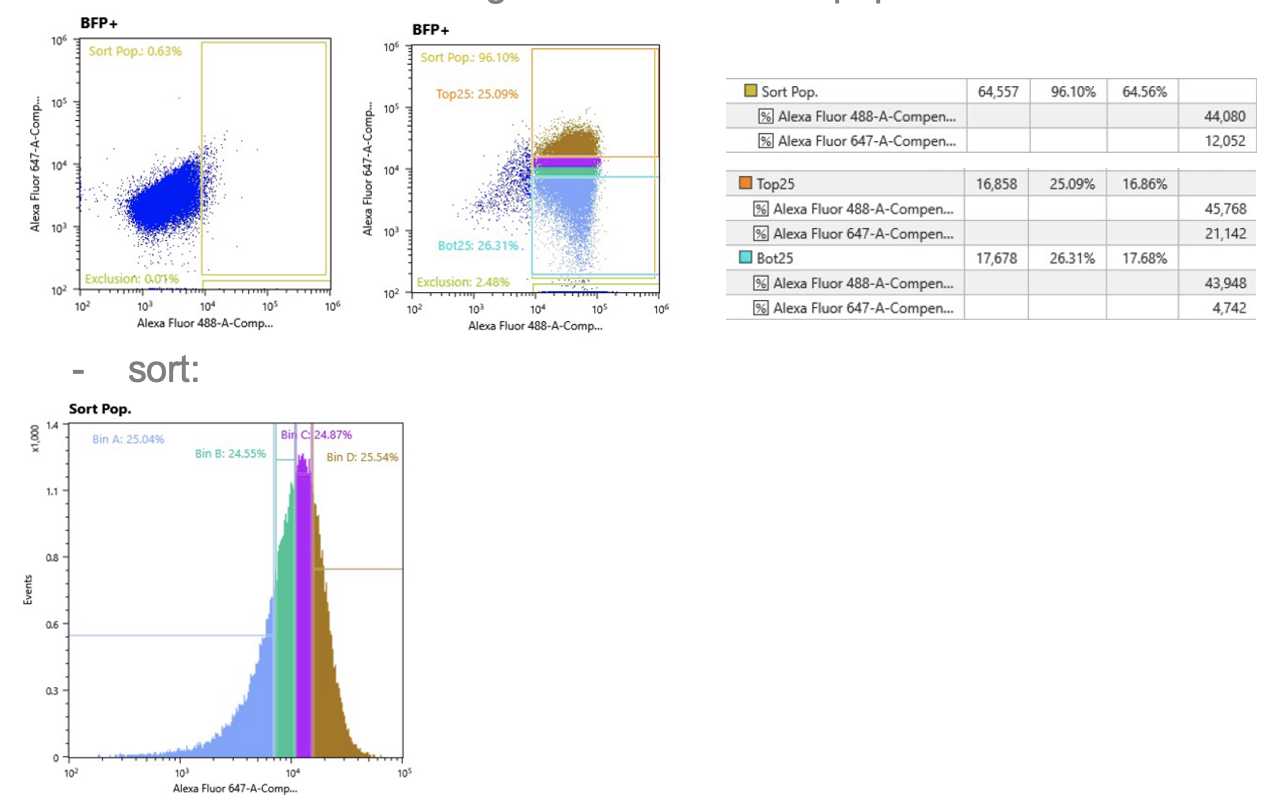
- setup:
o sort population should be <10% of unstained sample
o exclusion should be <5% of stained sample
§ if more than 5%, increase voltage or decrease compensation
o compensate for each gene: AF647 over AF488
§ RPL13A (AF488) expression = total RNA, compensate gene of interest expression (AF647) relative to total RNA
§ average AF488 for Top & Bottom 25% AF647 should be within 10% of average AF488 for all sort population
E. gDNA Extraction after FlowFISH
- Homemade ChIP Lysis Buffer :
o 1% SDS, 10mM EDTA, 50mM Tris-HCl, pH7.5
o store at 4ºC
o for 50mL
- Make Lysis Buffer fresh or warm at room temp 30-60min to solubilize precipitate
- Always include a no cell control until PCR to check contamination
- Set thermocycler:
o 65ºC Hold -> 65ºC 10min -> 37ºC Hold -> 37ºC 30min -> 65ºC Hold -> 65ºC 2h -> 95ºC 20min -> 4ºC Hold
- Protocol for 1M cells
Spin cells down for 10min at 800 g at 4ºC, remove supernatant
Resuspend cells in 70µL ChIP Lysis Buffer , transfer to 96-well plate
Incubate at 65ºC for 10min
When samples cool to 37ºC, add 2µL RNase Cocktail (Invitrogen AM2286) mix well by pipetting
Incubate at 37ºC for 30min
Add 10µL Proteinase K (Thermo 25530049), mix well by pipetting
Incubate at 65ºC for 2h, then 95ºC for 20min
Store at 4ºC (can store overnight)
Beads clean with 0.7X AMPure beads: 70µL AMPure beads for ~100µL samples
a. warm AMPure beads to room temp
b. add beads, pipette to mix, and bind DNA for 2min
c. wash 3 times with 150µL 70% EtOH
d. let beads dry for 5-10min after 3rd wash
e. elute with ~40-50µL H2O, elute for 2min, transfer to a new plate/tube

