New Histone Lysine Acylation Biomarkers and Their Roles in Epigenetic Regulation
Qianyun Fu, Qianyun Fu, Amber Cat, Amber Cat, Y. George Zheng, Y. George Zheng
Abstract
Protein posttranslational modification (PTM) is a biochemical mechanism benefitting cellular adaptation to dynamic intracellular and environmental conditions. Recently, several acylation marks have been identified as new protein PTMs occurring on specific lysine residues in mammalian cells: isobutyrylation, methacrylation, benzoylation, isonicotinylation, and lactylation. These acylation marks were initially discovered to occur on nucleosomal histones, but they potentially occur as prevalent biomarkers on non-histone proteins as well. The existence of these PTMs is a downstream consequence of metabolism and demonstrates the intimate crosstalk between active cellular metabolites and regulation of protein function. Emerging evidence indicates that these acylation marks on histones affect DNA transcription and are functionally distinct from the well-studied lysine acetylation. Herein, we discuss enzymatic regulation and metabolic etiology of these acylations, 'reader' proteins that recognize different acylations, and their possible physiological and pathological functions. Several of these modifications correlate with other well-studied acylations and fine-tune the regulation of gene expression. Overall, findings of these acylation marks reveal new molecular links between metabolism and epigenetics and open up many questions for future investigation. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
INTRODUCTION
Posttranslational modifications (PTMs) of proteins are fundamental mechanisms that regulate cellular processes in all domains of life (L. Meng et al., 2022; Soufi et al., 2012). Fatty acylations on lysine residues have been recognized as crucial PTM marks with various regulatory effects on key biological pathways (Barnes et al., 2019; Sabari et al., 2015; Verdin & Ott, 2015). Lysine acetylation was initially discovered in histones by Phillips (1963), and Allfrey et al. (1964) were the first to show that histone acetylation is linked to regulation of gene expression. Positively charged lysines in the chromatin can be neutralized via acetylation, thus weakening the interaction of negatively charged DNA with histones and thereby increasing levels of gene expression (Allfrey et al., 1964). The roles of lysine acetylation in signal transduction, transcriptional regulation, energy metabolism, and mitochondrial regulation were further demonstrated by identifying a number of non-histone protein substrates, such as tubulin (L'Hernault & Rosenbaum, 1985), the HIV transcriptional regulator Tat (Ott et al., 1999), p53 (Gu et al., 1997), and other diverse substrates across nucleus, cytosol, and mitochondria (S. C. Kim et al., 2006). Furthermore, lysine acetylation has been reported in organisms ranging from prokaryote to protozoa to metazoa (G. W. Kim & Yang, 2011; Soufi et al., 2012), arguing that the biological regulation through lysine acetylation is conserved throughout evolution.
In addition to lysine acetylation, ∼20 different types of lysine acylations have been discovered (Table 1), which include myristylation (C14) (Bursten et al., 1988), formylation (Kfo) (T. Jiang et al., 2007), propionylation (Kpr), butyrylation (Kbu) (Chen et al., 2007), crotonylation (Kcr) (M. Tan et al., 2011), succinylation (Ksucc), malonylation (Kmal) (Z. Y. Xie et al., 2012), methylmalonylation (Kmma) (Head et al., 2022), glutarylation (Kglu) (M. Tan et al., 2014), 2-hydroxyisobutyrylation (Khib) (Dai et al., 2014), β-hydroxybutyrylation (Kbhb) (Z. Xie et al., 2016), 3-hydroxy-3-methylglutarylation (KHMG), 3-methylglutaconylation (KMGc), 3-methylglutarylation (KMG) (Wagner et al., 2017), benzoylation (Kbz) (Huang et al., 2018), lactylation (Kla) (D. Zhang, et al., 2019), isonicotinylation (Kinic) (Y. Jiang et al., 2021), methacrylation (Kmea) (Delaney et al., 2021), and isobutyrylation (Kibu) (Zhu et al., 2021). Abundances of different acylation marks are reliant on the cellular concentrations of corresponding acyl-CoA metabolites, strongly demonstrating the intimate coupling of epigenetic regulation with cellular metabolism (Sabari et al., 2017; Simithy et al., 2017; M. Wang & Lin, 2021). Notably, some lysine acylations can show functions different from lysine acetylation. For instance, p300-catalyzed Kcr stimulates gene transcription to a larger degree than Kac on histones (Sabari et al., 2015). Also, p300 regulates glycolysis via Khib rather than Kac of glycolytic enzymes (Huang et al., 2018).
Molecular regulators of PTM biology can be functionally separated into three kinds of proteins: (1) ‘writers’ such as lysine acetyltransferases (KATs), which attach PTM marks to proteins; (2) ‘erasers’ such as lysine deacetylases (KDACs), which remove PTM marks from proteins; and (3) ‘readers’ such as acetyllysine binders, which selectively interact with PTM marks (He et al., 2018). Various central pathways from metabolism of carbohydrates, proteins, and lipids feed essential precursors for the formation of acyl-coenzyme A (acyl-CoA) for use in lysine acylation reactions by KATs (Baldensperger & Glomb, 2021). Three major families of KATs that are now well-established in eukaryotes: GNAT (the Gcn5 related N -acetyltransferase) members such as PCAF, GCN5, and HAT1; the p300/CBP family members that include p300 and CBP; and MYST (Moz, Ybf2, Sas2, and Tip60) members including MOF, MORF, MOZ, Tip60, and HBO1 (Berndsen & Denu, 2008; Dancy & Cole, 2015). GNAT family members share a similar acetyltransferase domain with ~160 amino acid residues and a conserved BRD domain that can attach to lysine residues (Ud-Din et al., 2016). These enzymes are often involved in transcriptional activation and elongation, and DNA damage repair (Kimura et al., 2005). The MYST family members share the conserved MYST domain of 370 residues which includes a zinc finger domain and an acetyl-CoA binding domain (Avvakumov & Cote, 2007; Sapountzi & Cote, 2011; Voss & Thomas, 2009). The p300/CBP family has two members that contain a 500-amino acid KAT domain with 86% sequence similarity, a bromodomain, and three cysteine-histidine-rich domains (TAZ, PHD, and ZZ) (Dancy & Cole, 2015; X. Liu et al., 2008). P300/CBP mediate acetyl transfer via stable binding of the cofactor acetyl-CoA, followed by transient association of the lysine substrate. This hit-and-run catalytic mechanism allows the broad and promiscuous substrate specificity of p300/CBP (Simon et al., 2016). P300 activity toward catalyzing lysine acylations is negatively correlated with acyl-chain lengthening (Kaczmarska et al., 2017). The majority of the members of the MYST and GNAT families appear to have an even more limited range of acylation activities (Sabari et al., 2017).
18 KDAC members have been identified, which include NAD+-dependent sirtuin deacetylases (SIRT1-7) and Zn2+-dependent histone deacetylases (HDAC1-11). These are the widely reported ‘erasers’ for removing lysine acylation marks in the mammalian proteome (Finkel et al., 2009; Haberland et al., 2009). Some enzymes are more active on removing specific lysine acylation than lysine acetylation. For instance, SIRT5 targets negatively charged lysine acylations especially succinylation and malonylation (Peng et al., 2011; M. Tan et al., 2014), and SIRT6 preferentially removes long-chain fatty acyl marks over acetylation from the lysine side chain (Feldman et al., 2013). The sirtuin orthologs as KDACs are found in both eukaryotes and prokaryotes (the same conservation is seen for the GNAT family members as KATs) (Marsh et al., 2005; Starai & Escalante-Semerena, 2004; Vetting et al., 2005; K. Zhao et al., 2004). In addition to GNATs, YopJ effector proteins also exhibited KAT activities in prokaryotes (Lewis et al., 2011; Ma & Ma, 2016). For KDACs in prokaryotes, only a few proteins showed deacetylase activity and very few protein substrates have been identified (Hildmann et al., 2007). Specifically, CobB, a Sir2 homolog protein deacetylase, was previously reported to exist in Salmonella enterica and Escherichia coli (Starai et al., 2002; K. Zhao et al., 2004).
Histone PTMs are proposed to regulate the chromatin-templated processes through two major mechanisms (Kouzarides, 2007; Ruthenburg et al., 2007): (1) histone PTMs directly regulate chromatin packaging and chromatin structure to modulate the entry of DNA-binding proteins either through changing the net charge of histone molecules or through altering inter-nucleosome interactions; and (2) histone PTM marks serve as docking signals to recruit reader proteins to alter chromatin structure and function (M. Tan et al., 2011). To date, proteins including bromodomains, PHD fingers, YEATS domains (Yaf9, ENL, AF9, Taf14, and Sas5), Tudor domains, and chromodomains have been identified as ‘reader’ proteins (Andrews et al., 2016). Exploring the comprehensive roles of ‘writers’, ‘erasers’, and ‘readers’ in regulating lysine acylations is critical to thoroughly understand mechanisms of lysine acylation in cellular process regulation.
PTMs on cellular proteins can be detected either by specific antibodies or by mass spectrometry (MS) methods. Owing to simplicity, convenience, and affordable cost, antibody-based western blotting (WB) analysis has been the gold standard for measuring relative acylation mark changes. However, this method suffers from several shortcomings: low-throughput detection of acylation marks at a time; low antibody specificity to distinguish acylation marks that have very subtle structure differences; interference by nearby modifications or epitope occlusion; and cross-reactivity to other modification due to homologous protein sequence (Huang et al., 2015; Song et al., 2022). Liquid chromatography (LC)-MS-based approaches are used for detecting acylation marks based on their presumed mass shifts and can address the caveats above by not only detecting multiple acylation marks simultaneously regardless their positions on peptide sequence, but also distinguishing different acylation marks that have similar structures. Unbiased MS screening, without the presumed mass shifts, is an alternative approach to find unknown acylation marks. However, MS methods are difficult to detect modifications that exist at very low stoichiometry and are challenging to distinguish among modifications with the same mass shift, such as structural isomers, which require further data interpretation and validation (Huang et al., 2015). MS/MS and high-performance LC (HPLC) combined, are effective to validate a new acylation mark among multiple structure isomers by pairwise comparison between a synthetic peptide and an in vivo peptide (D. Zhang et al., 2019). Other validation methods include developing specific antibodies against a PTM and identifying a possible precursor for the new PTM mark to confirm it in vivo using immunostaining or WB analysis (Huang et al., 2015). With the use of specific antibodies and unbiased mass spectrometry, several new types of lysine acylation, including isobutyrylation (Kibu) (Zhu et al., 2021), methacrylation (Kmea) (Delaney et al., 2021), benzoylation (Kbz) (Huang et al., 2018), isonicotinylation (Kinic) (Y. Jiang et al., 2021), and lactylation (Kla) (D. Zhang et al., 2019) have been identified in the recent years. These acylation marks are added on different lysine positions in histones (Fig. 1) and exhibit interesting biological functions in epigenetics and metabolism. Signaling pathways involving distinct lysine PTMs are increasingly recognized to be interconnected in eukaryotes (L. Meng et al., 2022). Some of these acylations have been reported and are highly conserved across different species. Below, we highlight recent evidence for these acylations and discuss how they have unique roles for the transcription modulation and chromatin structures. We describe the discovery of these acylations on histones as well as non-histone proteins, and recent studies regarding the ‘writers’, ‘erasers’, and ‘readers’ of these acylation marks. We next discuss the evidence about how these acylations vary according to the relative levels of different acyl-CoAs in the cell. We also discuss how these acylations are functionally distinct from other acylations.
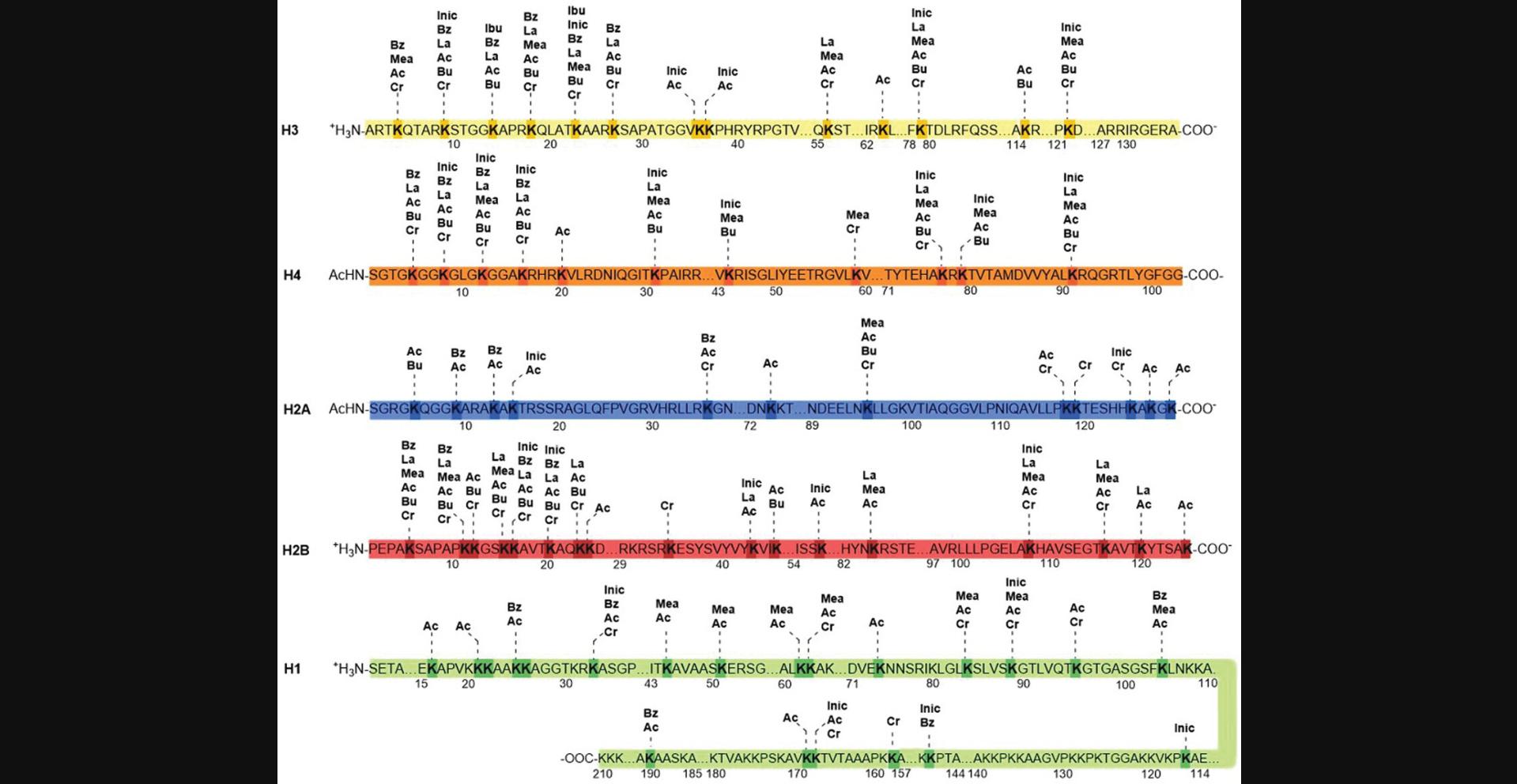
OVERVIEW OF NEW LYSINE ACYLATION BIOMARKERS
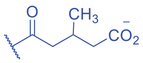
Lysine Isobutyrylation and Butyrylation
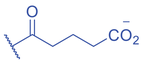
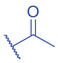
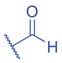
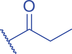
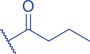
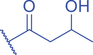
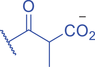
Lysine butyrylation (Kbu) was early reported as a linear acyl mark using n -butyryl-CoA as the acyl donor (Chen et al., 2007). Endogenous n -butyryl-CoA is produced by the β-oxidation of fatty acids in the mitochondria. Mitochondrial carnitine acetyl transferase (CrAT) likely promotes the interconversion of n -butyryl-CoA and butyryl-carnitine to achieve mitochondrial export (Violante et al., 2013). Exogenous uptake of butyrate is another source of intracellular n -butyryl-CoA. Environmental butyrate can be converted to n -butyryl-CoA with the function of acyl-CoA synthetase short chain member 2 (ACSS2) (Abdinejad et al., 1981). Butyryl-CoA can also accumulate under certain condition such as short-chain acyl-CoA dehydrogenase (SCAD) deficiency (Pougovkina et al., 2014). Butyrate metabolism mainly occurs in the liver and intestinal epithelia, and previous studies revealed that butyrate is an abundant short chain fatty acid (SCFA) in the gastrointestinal tract (Cummings et al., 1987). Further studies identified high levels of histone Kbu in intestinal epithelial cells from the cecum and distal mouse intestine (Gates et al., 2022). More in-depth studies report the relative changes of Kbu in different physiological states; starvation enhances histone Kbu in the liver of males, though the basal levels of Kbu is higher in female mice (Goudarzi et al., 2020). Additionally, Kbu levels seem to be sensitive to dietary conditions in liver. One study reported an increase in histone Kbu after the ketogenesis stimulation followed by starvation in liver (Goudarzi et al., 2020). However, H3K18bu levels were downregulated in a high-fat diet-induced obesity model (Nie et al., 2017). Another study revealed that H3K9bu is abundant at all active promoters and negatively regulated by a high-fat diet (Z. Yang et al., 2021).
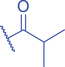
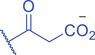
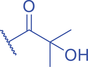
The isomer of n -butyryl-CoA, isobutyryl-CoA, is naturally generated from valine catabolism and branched-chain fatty acid oxidation (Fig. 2) (Robinson et al., 1957). Recently, Zhu et al. (2021) reported that isobutyryl-CoA acts as a donor for a previously unexplored PTM—lysine isobutyrylation (Kibu) in histones of mammalian cells. Exogenous d7-isobutyrate addition to the cells can induce isobutyryl-CoA abundance in a concentration dependent manner, which further contributes to Kibu levels (Zhu et al., 2021). Dairy and ruminant foods rich in branched chain fatty acids are another source of isobutyryl-CoA (Ran-Ressler et al., 2014). Of note, the isobutyryl-CoA abundance was found at a comparative level to n -butyryl-CoA in HEK293T cells (Zhu et al., 2021), suggesting that the cellular butyryl-CoA probably is a mixture of two isomers that give heterogeneous PTMs on cellular proteins. Since both isomers can be derived from food, researching how dietary conditions affect Kibu, and how Kibu differs from Kbu in appropriate response to diet-induced changes in physi(path)ological states would be interesting and warrants further exploration.
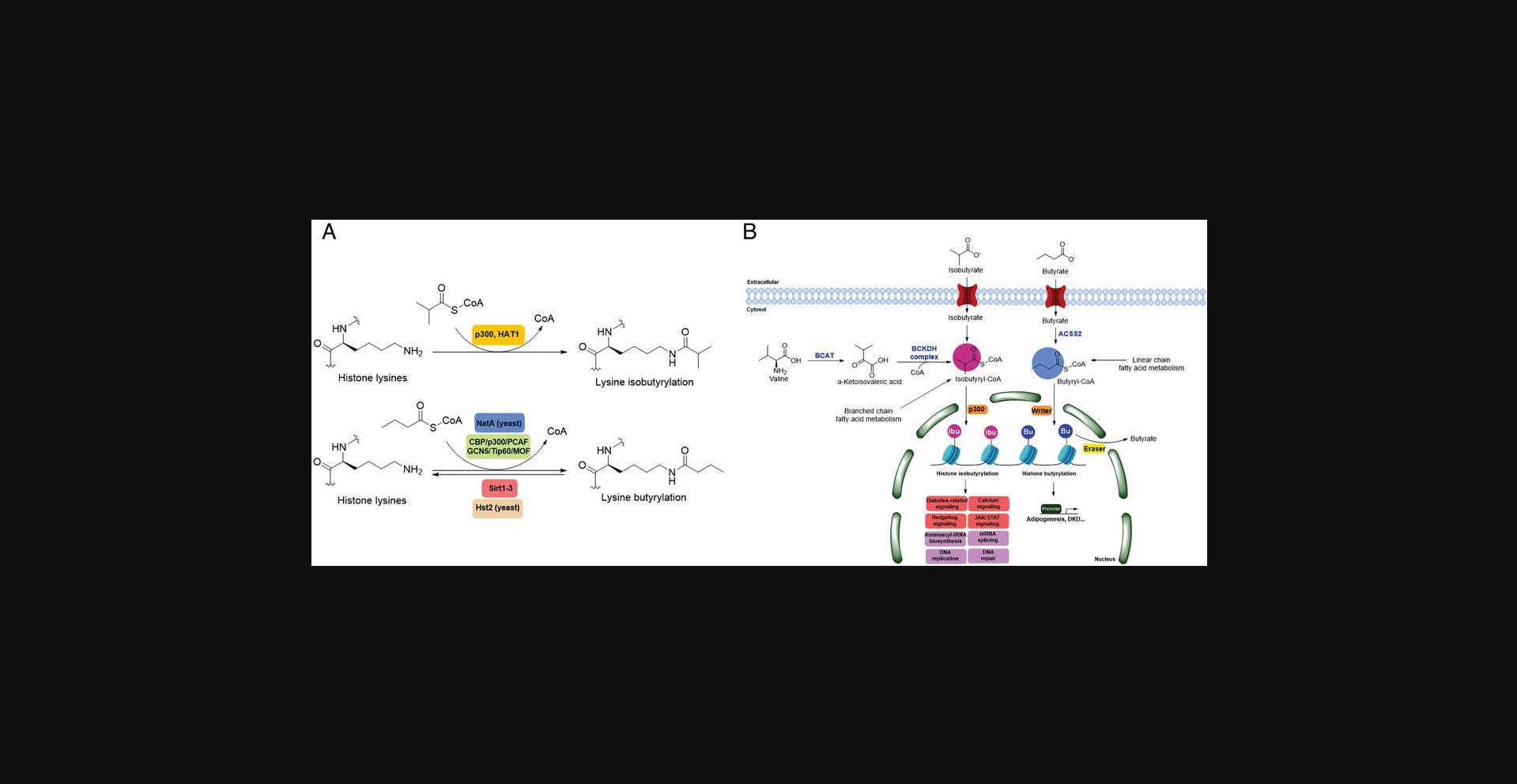
Kbu is evolutionarily conserved among eukaryotes, with multiple Kbu sites identified in histones of yeast and mammalian cells (Chen et al., 2007; K. Zhang et al., 2009). Histone Kbu formation is enzymatically catalyzed by various KATs including GCN5, PCAF, p300, CBP, Tip60, NatA, and MOF (Chen et al., 2007; Simithy et al., 2017). One study compared acylation activities of KATs on peptides showing that GCN5 catalyzes ∼78% of butyrylation on H3K14, whereas acetylation was found at ∼32%. PCAF exhibited ∼88% butyryltransferase activity on H3K14, followed by ∼2% acetylation. Moreover, Tip60 has a preference to utilize butyryl-CoA as a cofactor rather than acetyl-CoA (Simithy et al., 2017). The effects of p300-mediated Kbu formation have been well studied on non-histone proteins as well (Chen et al., 2007). For Kibu, HAT1 exhibited an outstanding Kibu activity (Kcat = 0.4 ± 0.02 min−1, Km = 1.9 ± 0.5 μM, Kcat/Km = 0.21 ± 0.06 min−1μM−1) with ∼25% of its Kac activity (Kcat = 7.4 ± 0.3 min−1, Km = 5.4 ± 0.8 μM, Kcat/Km = 1.4 ± 0.2 min−1μM−1) on peptide; and isobutyrylation activities of p300 (Kcat = 0.32 ± 0.01 min−1, Km = 2.4 ± 0.3 μM, Kcat/Km = 0.13 ± 0.02 min−1μM−1) was ∼13% of its acetylation activity (Kcat = 33.9 ± 2.9 min−1, Km = 17.3 ± 4.8 μM, Kcat/Km = 2.0 ± 0.6 min−1μM−1). However, only p300 was found to regulate histone Kibu in the HEK293T cell model (Fig. 2) (Zhu et al., 2021). P300 shows both butyryltransferase and isobutyryltransferase activity possibly due to its unique binding pocket to accommodate larger acyl-CoA molecules (C. Yang et al., 2013; Y. Yang et al., 2010). A recent study elucidated additional roles of p300-mediated Kbu formation in adipogenesis and the consequent obesity (Bhattacharya et al., 2022). Specifically, histone Kbu increases in the promoters of pro-adipogenic genes resulting in adipogenesis. Garcinol can efficiently inhibit p300-driven acetylation (Kopytko et al., 2021). A specific inhibitor—LTK-14A (13, 14 dimethoxy isogarcinol)—inhibiting the slower p300-mediated butyrylation with minimal effect on faster p300-catalysed acetylation, was developed by screening a series of derivatives of garcinol. The administration with this specific butyrylation inhibitor can abrogate adipogenesis by downregulating pro-adipogenic gene expression and inhibiting H4K5bu (Bhattacharya et al., 2022). P300-mediated Kbu formation is also associated with the regulation of diabetic kidney disease (DKD). As important pathophysiologic processes in DKD, fibrosis and inflammation status can be affected by butyrate (Zhou et al., 2022). However, histone Kbu inhibition induced by p300 inhibitor reverses the anti-fibrosis and anti-inflammatory effects of butyrate, suggesting that butyrate reduces renal inflammatory and fibrosis injury at least in part via histone Kbu (Zhou et al., 2022). Although p300-mediated Kbu has been extensively studied, studies on the role of p300-mediated Kibu have yet to be reported. Therefore, it is necessary to investigate p300-mediated Kibu in a broader scope in the future. Developing specific small molecule inhibitors targeting p300-mediated Kibu and identifying p300-mediated Kibu substrates in vivo may be useful to study the functions of p300-mediated Kibu. In addition to the ‘writers’ that add Kbu and Kibu on proteins, ‘erasers’ that are responsible for removing Kbu on proteins have been studied. Sirt1, Sirt2, and Sirt3 show measurable catalysis rates with Kbu-containing peptides (Smith & Denu, 2007). In addition, Kbu-containing peptides were found to exhibit stronger binding affinity toward Hst2 (KD = 16 ± 3 μM), a HDAC in yeast, compared with the corresponding acetyl-lysine peptide (KD = 21 μM) (Smith & Denu, 2007). Suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor, was shown to induce Kbu on histones and non-histone proteins of human cells (Xu et al., 2014). However, the ‘erasers’ that can remove Kibu marks are poorly understood and require further study.
Bromodomain-containing proteins can serve as ‘readers’ for histone Kbu. The second bromodomain of TAF1 and the bromodomains of BRD9 and CECR2 have been identified as Kbu ‘readers’ in vitro and show similar affinities toward Kbu and Kac. Structural analysis revealed that these bromodomains possess a flexible ligand pocket allowing high-affinity binding to Kbu (Flynn et al., 2015). Some ‘readers’ of Kbu can recognize other acylation biomarkers, thus regulating gene transcription. For example, histone H4K5bu at gene promoters competes with H4K5ac to inhibit its binding with BRDT, another bromodomain-containing reader, thereby mediating post-meiotic chromatin reorganization and stage-specific gene expression (Goudarzi et al., 2016). However, it is unclear whether there is any ‘reader’ module that recognizes Kibu or distinguishes it from Kbu and other lysine acylation marks. Given that the isobutyryl mark is structurally specific owing to its branched-chain structure, its binding properties with ‘readers’ may be different from other linear-chain acyl marks. New structural analysis would be of particular value in understanding how ‘reader’ proteins recognize Kibu marks and how the binding correlates with the changes in metabolic pathways.
RNA-Seq profiling initially indicated that isobutyrate modulates the expression of genes involved in many vital biological pathways such as calcium signaling, diabetes-related signaling, hedgehog signaling, mRNA splicing, and DNA repair and replication. A caveat is that the effect of isobutyrate may also come from its inhibitory impact on cellular HDAC activities (Zhu et al., 2021). Therefore, it is necessary to define gene expression changes mediated by Kibu rather than other mediators. The crosstalk between different acylations on lysine of proteins, especially on specific lysine residues, is important to better understand complicated regulation and metabolism in cells. For example, H3K14pr and H3K14bu were previously reported to be preferentially enriched at promoters of active genes and enhance a higher transcriptional output together with acetylation (Kebede et al., 2017). H3K14 was also identified as a site of isobutyrylation in vivo (Zhu et al., 2021). How H3K14ibu interacts with other acylations on the same lysine, such as H3K14pr, H3K14bu, etc., and leads to specific biological impacts needs further study. Isobutyryl-CoA dehydrogenase (IBD) catalyzes the breakdown of isobutyryl-CoA in the valine catabolism (Andresen et al., 2000). IBD deficiency is an inborn error of valine metabolism caused by genetic changes in the ACAD8 gene and causes severe symptoms including developmental delay, dilated cardiomyopathy, anemia, and epilepsy (Lin et al., 2018). Therefore, it will be highly meaningful to elucidate the role of Kibu in pathological states, such as IBD deficiency causing isobutyryl-CoA accumulation (Nguyen et al., 2002).
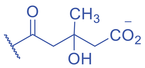
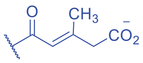
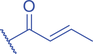
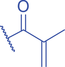
Lysine Methacrylation and Crotonylation
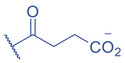
Lysine crotonylation (Kcr) was reported some time ago to influence the epigenetics of gene expression (M. Tan et al., 2011). Histone Kcr occurs broadly in all the core histones, and it is evolutionarily conserved from human to yeast (M. Tan et al., 2011). Recently, Delaney et al. identified a new type of PTM on histones, lysine methacrylation (Kmea), which is a structural isomer of Kcr (Fig. 3) (Delaney et al., 2021). Similar to Kcr, Kmea occurs broadly in all the core histones including H2A, H2B, H3, and H4. Both Kcr and Kmea formation are enzymatic processes and dynamically regulated by intermediate acyl-CoA metabolites in cells. CBP and p300 are likely the major ‘writer’ proteins for Kcr in mammalian cells (Sabari et al., 2015). Furthermore, MOF has been reported as a crotonyltransferase (X. Liu et al., 2017). In yeast, Gcn5- and Esa1-containing ADA and Piccolo NuA4 complexes have been reported as crotonyltransferases (Kollenstart et al., 2019). Distinct from ‘writers’ for Kcr, HAT1 showed a Kcat/Km value for methacrylyl-CoA (Kcat = 0.6 ± 0.2 min−1, Km = 30.3 ± 15.0 μM, Kcat/Km = 0.02 min−1μM−1), which is 10-fold lower than that for acetyl-CoA (Kcat = 1.2 ± 0.1 min−1, Km = 5.9 ± 2.1 μM, Kcat/Km = 0.2 min−1μM−1) on peptide. Further studies identified the role of HAT1 to catalyze lysine methacrylation in mammalian cells (Delaney et al., 2021). HAT1 was reported to regulate cellular metabolism involved in aging phenotypes, influence mitochondrial biogenesis, and act as nutrient sensor (Gruber et al., 2019; Marin et al., 2017; Nagarajan et al., 2019). An interesting question is to what degree HAT1 responds to cellular metabolism and makes its function in part by Kmea. ‘Erasers’ that remove Kcr or Kmea on histones have also been studied; HDAC1-3 and SIRT1-3 can remove crotonyl moieties from lysine residues (Bao et al., 2014; Feldman et al., 2013; M. Tan et al., 2011; Wei et al., 2017). For Kmea, HDAC3 was previously reported to act as an ‘eraser’ for lysine demethacrylation in vitro (Moreno-Yruela et al., 2018). Further study indicated that SIRT1 and SIRT2 exhibited the strongest ‘eraser’ activity among the sirtuin family of enzymes in vitro. The activity of SIRT2 to catalyze the removal of H3K18mea is further confirmed in vivo (Fig. 3) (Delaney et al., 2021). Some evidence suggests that Kcr and Kmea are linked to distinct regulatory circuits. Both histone Kcr and Kmea formation can be metabolically regulated by pathways that affect cellular concentrations of acyl-CoAs, including the thioesters of SCFAs, crotonate, or methacrylate (Delaney et al., 2021; Ntorla & Burgoyne, 2021). One such example is that crotonate promotes H3 and H4 crotonylation in the colon (Fellows et al., 2018). The biosynthesis of crotonyl-CoA or methacrylyl-CoA can occur in the mitochondria or the cytoplasm. Crotonyl-CoA is an intermediate metabolite of fatty acid β-oxidation and the catabolism of tryptophan and lysine in mitochondria (Trefely et al., 2020). Some of the crotonyl-CoA formed by either the fatty acid β-oxidation or amino acid degradation may leak from the mitochondria to generate Kcr formation in the cytosol and the nucleus (Gowans et al., 2019). In the cytoplasm, cellular crotonate is converted to crotonyl-CoA by acyl-CoA synthetase 2 (ACSS2) (Ntorla & Burgoyne, 2021). Different from the biosynthesis sources of crotonyl-CoA, methacrylyl-CoA is an intermediate metabolite of valine catabolism in the mitochondrion (Robinson et al., 1957). In the cytoplasm, cellular uptake of methacrylate can lead to methacrylyl-CoA that in turn drives Kmea formation. However, the mechanisms responsible for producing the nuclear pool of methacrylyl-CoA that fuels histone Kmea production remain unknown.
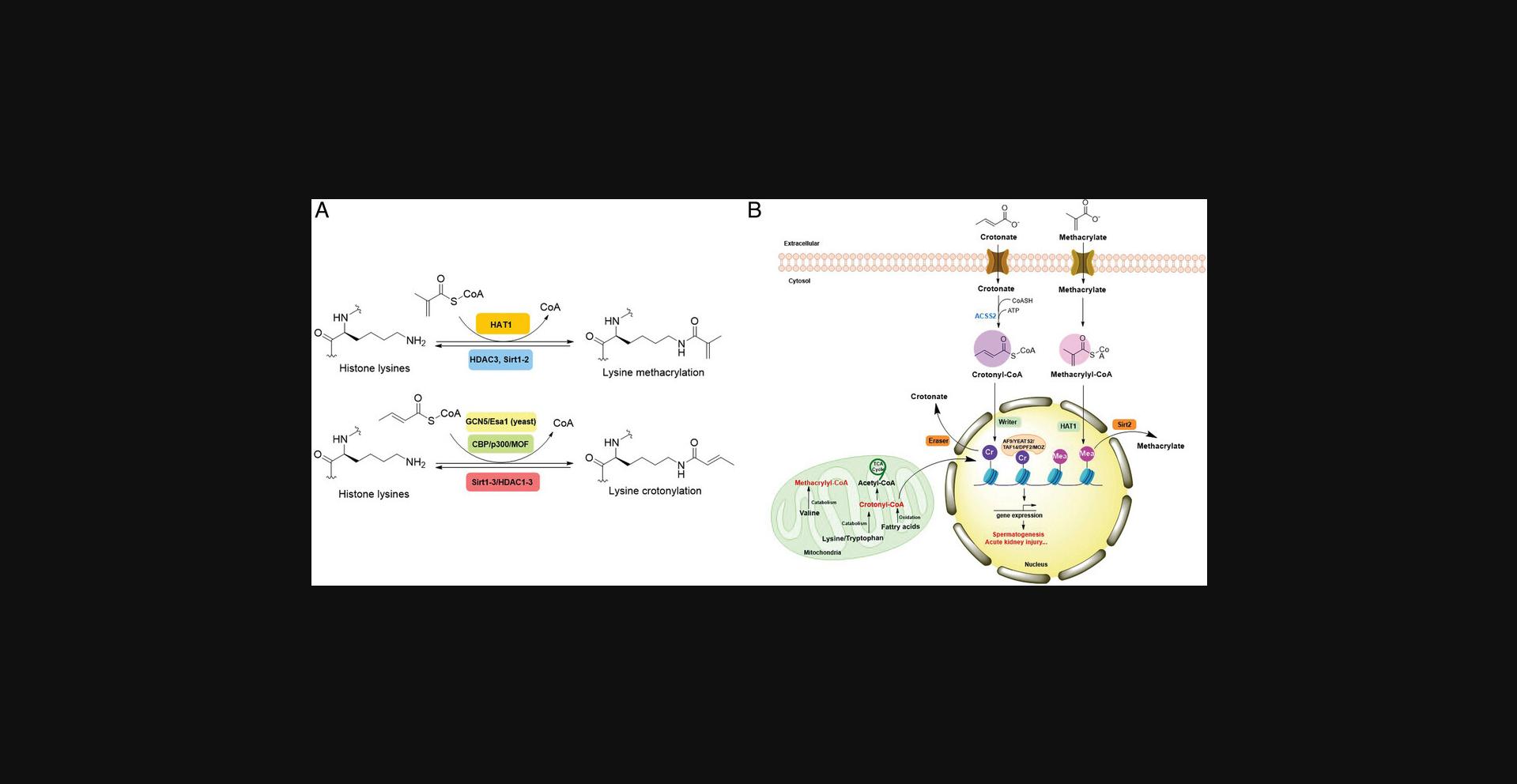
PHD finger- and YEATs domain-containing proteins have been reported as ‘readers’ for Kcr on histones. Notably, the previously identified ‘readers’ for Kac, AF9, Taf14, and double PHD finger (DPF) proteins monocytic leukemic zinc-finger (MOZ) and DPF2, exhibit stronger affinity for Kcr than Kac (Andrews et al., 2016; Gowans et al., 2019; Y. Li et al., 2016; X. Xiong et al., 2016; Q. Zhang et al., 2016; D. Zhao et al., 2016). The YEATS domain seems to preferentially bind to longer acyl chains, with ∼2-7-fold greater affinity for Kcr compared to Kac (Y. Li et al., 2016; D. Zhao et al., 2017). A unique “aromatic-π-aromatic” stacking leads to preferential binding of the YEATS domain to Kcr, with two aromatic residues wrapping the planar crotonamide group and hydrocarbon extension-introduced hydrophobic interactions. This recognition mechanism has been confirmed in the crystal structures of YEATS2, Taf14, and AF9 in complex with Kcr (Andrews et al., 2016; Y. Li et al., 2016; Q. Zhang et al., 2016; D. Zhao et al., 2016). The DPF domains of the MOZ and DPF2 also exhibit highest binding affinity for Kcr, with a 4-8-fold increased affinity compared to Kac (X. Xiong et al., 2016). Distinct from the underlying mechanism of YEATS domain, DPF domains interact with Kcr through an intimate hydrophobic pocket (X. Xiong et al., 2016). Another study found that the DPF domain of the MOZ-related factor (MORF) also has a preferential binding affinity for H3K14cr compared to H3K14ac. However, the binding between the DPF domain and H3K14cr enhances the catalytic activity of MORF to generate H3K23ac (Klein et al., 2019). Recognition of histone Kcr by ‘readers’ links metabolic state to gene expression. P300-mediated histone Kcr formation was found to be more potent than Kac in stimulating gene transcription. In lipopolysaccharide (LPS)-stimulated macrophages, p300 enhances inflammatory gene expression, which is further potentiated by Kcr (Sabari et al., 2015). Further investigation revealed that the YEATS-domain protein AF9 plays an essential role in regulating LPS-stimulated gene transcription mediated by H3K18cr (Y. Li et al., 2016). In addition to enhancing gene expression, histone Kcr is also associated with negative gene regulation. For example, Kcr participates in repressing energy-demanding gene expression after its binding by Taf14 (Gowans et al., 2019). Despite the extensive studies on identifying ‘readers’ of Kcr and its binding consequences, no study yet has described ‘readers’ for Kmea. Due to the structural similarity between Kcr and Kmea, some ‘readers’ that recognize Kcr may also bind to Kmea. However, which ‘reader’ specifically recognizes Kmea and how the binding is distinct from Kcr needs further exploration.
Currently, the role of Kcr has been clarified in acute kidney injury (Ruiz-Andres et al., 2016), stress-induced depression (Y. Liu et al., 2019), spermatogenesis (S. Liu et al., 2017; M. Tan et al., 2011), HIV latency (G. Jiang et al., 2018), DNA damage response (Abu-Zhayia et al., 2019), and telomere maintenance (Fu et al., 2018). Inherited mutations in key enzymes from the valine catabolic pathway such as ECHS1 and HIBCH lead to the development of Leigh syndrome (Brown et al., 1982; Loupatty et al., 2007; Peters et al., 2014). Mechanically, methacrylyl-CoA accumulated in the setting of these mutations may be a causative factor of the disease via its electrophilic capacity to alkylate cysteines (Brown et al., 1982). Elevated levels of acyl-CoA under physiological or pathological changes (e.g., diabetes and hypoxia) correlate with upregulated lysine acylations, which in turn regulate the metabolic state in cells (Z. Xie et al., 2016; D. Zhang et al., 2019). Future studies are needed to elucidate Kmea targets on non-histone proteins as well as their effects on influencing metabolic changes or related pathological states.
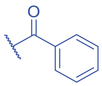
Lysine Benzoylation and Isonicotinylation
Recently, the influence of food and drugs on histone PTMs have become of greater research interest. Sodium benzoate (SB) is a common preservative worldwide and an FDA-approved drug for acute hyperammonemia treatment (Brusilow et al., 1984). Inappropriate uptake of SB can cause human health risks and problems (Park et al., 2011; Pongsavee, 2015; Praphanproj et al., 2000; Yilmaz et al., 2009). Huang et al. (2018) underlined an epigenetic mechanism of SB effects by identifying and characterizing a histone mark, lysine benzoylation (Kbz) in mammalian cells. SB stimulates histone Kbz by generating cellular benzoyl-CoA as an acyl donor (Fig. 4). Addition of isotopic D5-SB to the cells increased D5-benzoyl-CoA in a dose-dependent manner by the MS-based quantification. Further study quantified the changes of Kbz peptide in response to SB treatment by MS after chemically labeling of equal amounts of histones from SB-treated and control cells with 12C- or 13C-propionic anhydride, respectively. The results indicated that SB greatly increases Kbz abundance at low concentration, e.g., 5 mM. Several sites such as H3K23bz, H4K8bz, H4K5bz, H2AK9bz increased from 26.25-fold to >50-fold. In mammalian cells, SIRT2 was reported to exhibit deacetylase activity of Kbz on peptide with Kcat/Km (Kcat = 0.17 ± 0.01 min−1, Km = 0.92 ± 0.16 μM, Kcat/Km = 3162 s−1M−1) approximately 1/6 of that for deacetylation (Kcat = 9.71 ± 0.29 min−1, Km = 8.12 ± 0.90 μM, Kcat/Km = 19,926 s−1M−1). The debenzoylase activity of SIRT2 has further confirmed in vivo (Huang et al., 2018). The DPF domain and YEATS domain, rather than bromodomains, are ‘readers’ for histone Kbz (Fig. 4). The DPF and YEATS family members display unique features in flexible recognition of different acylation marks. For example, YEATS2 shows highest affinity to Kbz over Kcr or Kac (Ren et al., 2021). Distinct from Kac and Kcr, the Kbz mark is aromatic, hydrophobic, and larger in size. These structural characteristics may explain the unique regulatory role of Kbz, which mainly occurs at the N-termini of histones (Huang et al., 2018; Ren et al., 2021). RNA-seq and CHIP-seq analysis did show the association of the Kbz epigenetic mark with gene expression, which is distinct from histone Kac (Huang et al., 2018). In addition, increased Kbz level is positively related to ovarian steroidogenesis, glycerophospholipid metabolism, and phospholipase D signaling pathways. ChIP-qPCR results further confirmed that the Kbz levels at the promoters of genes from signaling above increased after SB treatment, however, their corresponding Kac levels were reduced, indicating Kbz mediated specific gene upregulation (Huang et al., 2018). A recent study has revealed that Kbz is stimulated by SB on histone proteins in Saccharomyces cerevisiae , suggesting that Kbz is conserved among species (D. Wang et al., 2022). In S. cerevisiae , histone Kbz can be enzymatically added by the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex and removed by NAD+-dependent histone deacetylase Hst2. Unlike ‘readers’ identified in mammalian cells, bromodomains and YEATS domains seem to serve as Kbz ‘readers’ in yeast (Fig. 4) (D. Wang et al., 2022).
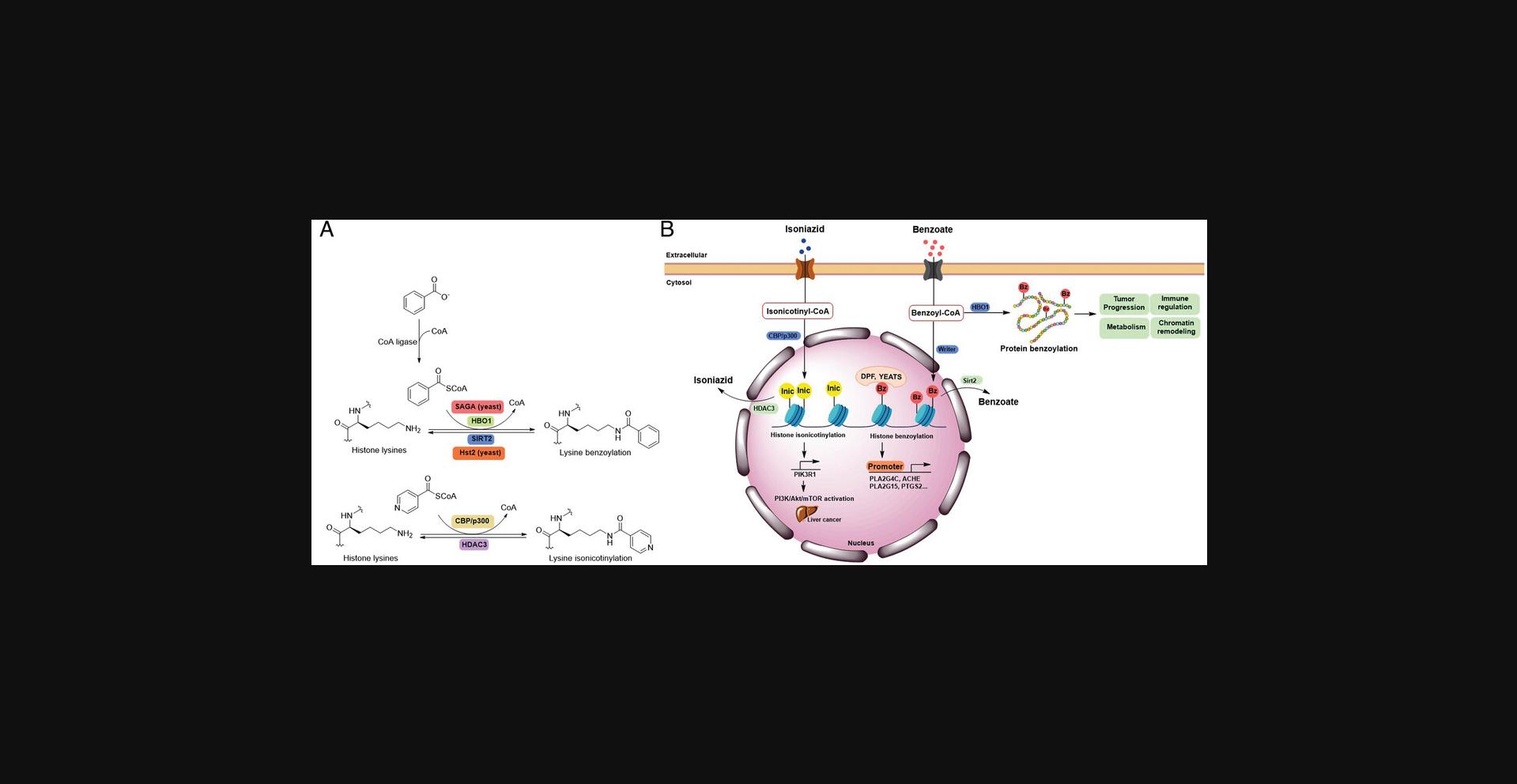
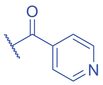
Isoniazid (INH) has been a first-line anti-tuberculosis drug since 1952 (Bernstein et al., 1952). INH is also widely known for its side effects such as hepatotoxicity and liver failure risk (Centers for Disease & Prevention, 2010; F. Li et al., 2016). However, the molecular mechanisms of the side effects caused by INH remains largely unknown. One study recently reported that similar to SB, INH can also induce a novel PTM on histones, lysine isonicotinylation (Kinic), thereby changing chromatin structure and promoting gene transcription (Y. Jiang et al., 2021). Kinic and Kbz share some structural similarity, the former with an N atom in the pyridine ring instead of a -CH in the benzene ring. However, Kinic and Kbz as histone marks are distinct from each other. Kbz marks are predominantly present on N-terminal tails, whereas Kinic marks are widely distributed from the N- to C-termini of histones. Diversity in the distribution of modified lysine sites may reflect their distinct functions in epigenetic chromatin regulation and gene expression. Histone Kinic is an enzymatic process and widely exists in eukaryotic cells. However, different from SB-induced Kbz, INH induces Kinic by generating intracellular isonicotinyl-CoA as the acyl donor, which is utilized by CBP/p300 for transferring isonicotinyl groups onto lysine residues of histones. The removal of isonicotinyl marks relies on the deacetylase HDAC3 as histone deisonicotinylase (Fig. 4). Histone Kinic was reported to relax chromatin structure and enhance gene transcription. INH was reported to induce liver cancer in rats (Szende et al., 1967). Intriguingly, INH-mediated histone Kinic is involved in the upregulation of PIK3R1 gene expression and activation of the PI3K/Akt/mTOR signaling pathway in liver cancer cells, explaining the effects of INH on tumorigenicity in the liver (Y. Jiang et al., 2021). Additionally, the deficiency of NAT2, the primary enzyme that metabolizes INH, was previously shown to increase the risk of INH-induced liver injury (Yuliwulandari et al., 2016). Further study revealed that histone Kinic levels were elevated when NAT2 activity was inhibited (Y. Jiang et al., 2021). This evidence indicates that histone Kinic formation may partially account for the induction of hepatotoxicity and tumorigenicity by INH.
In addition to modification on the core histones, Kbz and Kinic also modify non-histone proteins. Over 1,700 Kbz sites on cellular proteins in mammalian cells were discovered (D. Tan et al., 2022). Kbz marks formed by HBO1, a reported ‘writer’ of Kbz in mammalian cells, have been reported to be involved in multiple critical processes such as transcription regulation, chromatin remodeling, tumor growth, and immune regulation (D. Tan et al., 2022). In yeast, Kbz-modified non-histone proteins are enriched in glycolysis/gluconeogenesis, ribosome biogenesis, and rRNA processing pathways (D. Wang et al., 2022). Excessive intake of SB can increase the risk of diseases, including ADHD symptoms and motor coordination impairment (D. Tan et al., 2022). In terms of the role of Kinic on non-histone proteins, another study demonstrated the high association of INH-induced Kinic with certain types of cancer (Y. Jiang et al., 2021). However, how Kinic on these proteins may be oncogenic and the protein identities remain unknown. The identification of Kbz and Kinic sheds light on how the intake of an exogenous preservative or pyridine-containing drug can affect the gene expression profile and may cause physiological consequences by altering the human epigenome. Further characterization of ‘writer’, ‘eraser’, and ‘reader’ proteins of Kbz or Kinic will be important. Due to their structural characteristics, Kbz and Kinic represent a type of PTM containing pyridine/benzene rings and may endow unique biological functions, which need to be studied further.
The genetic code expansion technique has been applied to study the defined physiological roles of PTMs at desired lysine residues (C. H. Kim et al., 2012; Neumann et al., 2008; Xiao et al., 2015). A recent study genetically encoded benzoyllysine and its analogue fluorinated benzoyllysines into full-length histone H3 in a site-specific manner in live cells to investigate molecular mechanisms of interactions between site-specific Kbz and interactomes. The developed strategy allowed the elucidation of distinct dynamics of debenzoylation in the presence of SIRT2 (Tian et al., 2021). However, how the site-specific Kbz/Kinic modification on key proteins involved in physiological settings or even pathological diseases has not been well-studied and should be a goal of future work. Furthermore, identifying possible endogenous sources of Kbz/Kinic in cells would be interesting.
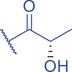
Lysine Lactylation
Glucose is metabolized into pyruvate, which is oxidized to acetyl-CoA for further metabolism to generate energy. However, incomplete oxidation of glucose produces the metabolite lactate through a phenomenon known as the Warburg effect (Liberti & Locasale, 2020). The Warburg effect is associated with various cellular processes such as polarization of macrophages, hypoxia, cancer, angiogenesis, and activation of T cells (Palsson-McDermott & O'Neill, 2013; Pavlova & Thompson, 2016). The functions of the Warburg effect have been extensively studied, involving cell signaling, ATP and macromolecule synthesis, as well as tumor microenvironment restructuring (Liberti & Locasale, 2016).
Lactate is widely known for its role as an energy source and metabolic by-product. Recently, a non-metabolic function of lactate in biology was reported in human cells: either exogenous lactate or endogenous lactate from glycolysis modifies histones by lysine lactylation (Kla) (D. Zhang et al., 2019). Hypoxia, interferon (IFN)-γ, LPS, and bacterial challenges also stimulate histone lactylation by inducing the production of lactate as a precursor (Fig. 5) (D. Zhang et al., 2019). Twenty-eight Kla sites on core histones were identified in human HeLa cells and mouse bone marrow-derived macrophages (BMDMs) (D. Zhang et al., 2019). Lactyl-CoA, the donor for Kla, was detected by MS in mammalian cells and tissues (Varner et al., 2020). Histone Kla can be installed and removed by regulatory enzymes: p300 functions as a potential histone Kla ‘writer’ (D. Zhang et al., 2019), and HDAC1-3 serve as delactylases (Fig. 5) (Moreno-Yruela et al., 2022). In particular, HDAC3 exhibited the most efficient delactylase activity at the H3K9 (Kcat = 0.24 ± 0.02 s−1, Km = 90 ± 20 μM, Kcat/Km = (2.6 ± 0.6) × 103 s−1M−1) and H4K12 sites (Kcat = 0.14 ± 0.01 s−1, Km = 120 ± 10 μM, Kcat/Km = (1.1 ± 0.1) × 103 s−1M−1) when compared to its deacetylase activity at the H3K9 (Kcat = 0.32 ± 0.03 s−1, Km = 23 ± 8 μM, Kcat/Km = (14 ± 5) × 103 s−1M−1) and H4K12 (Kcat = 0.29 ± 0.03 s−1, Km = 100 ± 20 μM, Kcat/Km = (2.9 ± 0.8) × 103 s−1M−1) (Moreno-Yruela et al., 2022). Another study revealed that SIRT2 removes the lactyl group from pyruvate kinase M2 related peptides (Jennings et al., 2021). In a recent study, histone H2B was produced by semisynthesis with an engineered sortase transpeptidase and incorporated into nucleosomes to investigate HDAC1 complexes and sirtuins selectivity toward site-specifically acylated H2B nucleosomes (Z. A. Wang et al., 2022). Among these deacylases, MiDAC (V/[E] = 0.0034 min–1) and RERE (V/[E] = 0.0018 min–1) measurably removed Kla marks from semisynthetic H2BK11la nucleosomes. But the deacylation rates of H2BK11la were comparable or even slower than the corresponding rates of H2BK11ac removal by RERE (V/[E] = 0.0023 min–1) or MiDAC (V/[E] = 0.0023 min–1), suggesting that the lactyl-Lys group is a nonprimary target of the sirtuins or HDAC1 complexes (Z. A. Wang et al., 2022). In addition to enzymatic reactions involving Kla, non-enzymatic reactions of Kla have been demonstrated as well (Gaffney et al., 2020; Varner et al., 2020). Lactoylglutathione (LGSH) is produced through the reaction between the glycolytic byproduct methylglyoxal and glutathione by the thioesterase glyoxalase 1 (GLO1). LGSH then is hydrolyzed to D-lactate and glutathione by GLO2. A “LactoylLys” modification on proteins was then generated non-enzymatically by transferring a lactyl group from LGSH to protein lysine residues (Gaffney et al., 2020).
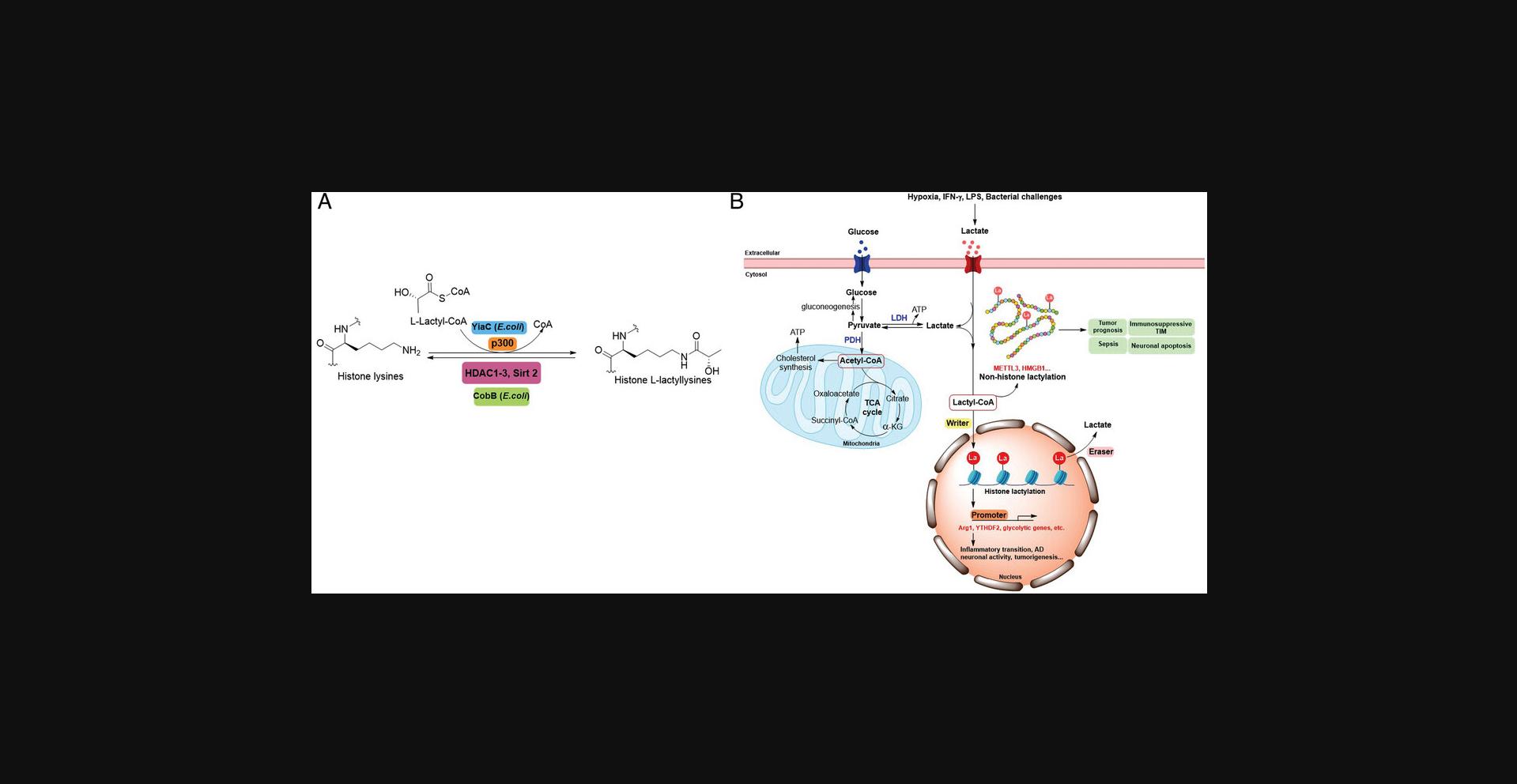
Histone Kla levels are sensitive to metabolic stress and Kla can regulate gene expression during altered macrophage states. Histone Kla is different from Kac in terms of temporal dynamics. Histone Kla increased over 24 hr, whereas histone Kac reached a peak at 3 hr and is steady at 6 hr. Moreover, upregulated histone Kla induces homeostatic genes that are involved in wound healing, such as Arg1 in the late phase of M1 macrophage polarization when exposed to bacteria. This suggests that histone Kla may act as an endogenous ‘lactate clock’ to promote the transition of inflammatory phenotype to homeostatic phenotype in macrophages (X. L. Li et al., 2022; D. Zhang et al., 2019). Macrophages can initiate inflammation to eliminate invading microbes, then protect host tissues from damage by downregulating inflammatory genes and expressing repair genes. Kla was reported to be involved in reparative macrophage transition after clearance of inflammatory stimuli states (Irizarry-Caro et al., 2020). Specifically, B-cell adapter for PI3K (BCAP), which is a signal adapter for Toll-like receptors (TLRs), is vital to facilitating the transition from inflammatory to reparative macrophages. BCAP deficiency reduced lactate production as well as histone Kla and led to defective aerobic glycolysis. However, exogenous lactate addition to BCAP-deficient BMDMs increased histone Kla and reversed the effects mediated by BCAP deficiency (Irizarry-Caro et al., 2020). In another study, lactate produced by probiotic S. cerevisiae was reported to inhibit BMDM activation and reduce ulcerative colitis in BMDMs by H3K9ac and H3K18la (S.Y. Sun et al., 2021). Additionally, lung myofibroblasts promote profibrotic mediator expression in macrophages through lactate-induced histone Kla (Cui et al., 2021). Kla is also associated with dysfunction in Alzheimer's disease (Pan et al., 2022); specifically, H4K12la level is upregulated in Aβ plaque-adjacent microglia and activates gene transcription to increase glycolytic activity. Eventually, the glycolysis/H4K12la/PKM2 positive feedback loop accelerates microglial dysfunction in AD (Pan et al., 2022). Furthermore, Kla on histones is involved in mediating neuronal activity (Hagihara et al., 2021). Other studies revealed that histone Kla can affect cellular metabolic reprogramming by mediating gene expression in pluripotent stem cells or in non-small cell lung cancer (J. Jiang et al., 2021; L. Li et al., 2020). Functions of histone Kla in promoting tumor development were examined recently. YTHDF2 plays a critical role in promoting tumorigenesis in ocular melanoma. Kla on histones exacerbates tumorigenesis of ocular melanoma and is related to the poor prognosis by activating YTHDF2 expression (Yu et al., 2021). In non-small cell lung cancer, histone Kla alters gene expression of critical metabolic enzymes or other factors such as glycolysis-related enzymes hexokinase (HK)-1 and TCA cycle-related enzymes like isocitrate dehydrogenase (IDH) to attenuate glucose uptake and glycolysis (J. Jiang et al., 2021). To date, histone Kla has been reported in other tumor cell lines, including MCF-7 cells, HeLa cells and HepG2 cells (X. L Li et al., 2022).
In addition to being an epigenetic mark on histones, Kla is identified on non-histone proteins in mammalian cells. High mobility group box-1 (HMGB1) is a nuclear protein released by activated macrophages to orchestrate inflammatory responses (Deng et al., 2018; H. Yang et al., 2020). High circulating levels of HMGB1 and lactate are positively related to the severity and mortality of sepsis (Broder & Weil, 1964; Sunden-Cullberg et al., 2005). Macrophages can promote p300/CBP-dependent lactylation on HMGB1 by taking up environmental lactate. The lactylated HMGB1 is released from macrophages through exosome secretion which increases endothelium permeability, contributing to dysfunction of the endothelial barrier and promoting the development of sepsis (K. Yang et al., 2022). Furthermore, Kla on non-histone proteins appears to play a critical role in cancer biology. One study identified 2,375 Kla sites in 1,014 proteins from gastric cancer AGS cells (D. W. Yang et al., 2022). Further studies found that Kla levels were greatly higher in tumor tissues than their corresponding adjacent tissues. Additionally, higher levels of Kla were detected in poorly differentiated tumors and tumors with lymph node metastasis, suggesting that Kla was positively related to a poor prognosis (D. W. Yang et al., 2022). Another study revealed that lactylation increased methyltransferase-like 3 (METTL3) expression in tumor-infiltrating myeloid cells (TIMs), leading to N 6-methyladenosine (m6A) modification on Jak1 mRNA and further METTL3/m6A/JAK1/STAT3 axis activation to promote the immunosuppressive capacity of TIMs (J. Xiong et al., 2022).
The role of Kla in cerebral ischemia–reperfusion injury (CIRI) has been elucidated recently. Extensive Kla modifications were identified on proteins from cortical proteins of a CIRI rat, and most of the Kla modified proteins mediate neuronal apoptosis by their involvement in Ca2+ signaling pathways (Yao et al., 2022). In addition to antibody-based immunoprecipitation coupled with MS on proteomic studies, the bioorthogonal chemical reporter strategy has emerged as a powerful method for studying protein PTMs (Grammel & Hang, 2013; Parker & Pratt, 2020; Prescher & Bertozzi, 2005). Specifically, chemical reporters, such as the azido or alkynyl derivatives of PTM donor precursors can be metabolically incorporated into proteins for labeling with bioorthogonal groups, allowing for subsequently conjugation with affinity tags through bioorthogonal reactions for proteomic identification (Sletten & Bertozzi, 2009; Song & Zheng, 2020). One recent study developed YnLac, an alkynyl-functionalized L-lactate analogue bioorthogonal chemical reporter, for metabolic labeling and proteomic profiling of Kla protein substrates in mammalian cells (Y. A. Sun et al., 2022). With this approach, 114 lactylation sites across 62 proteins including several previously unidentified lactylation sites were identified. Specially, lactylation of PARP1, a newly identified substrate protein, was found to regulate its ADP-ribosylation activity (Y. A. Sun et al., 2022). In another study, non-enzymatic Kla was reported by using an alkyne-tagged methylglyoxal analog (Gaffney et al., 2020). 350 Kla modified proteins were identified, in which most are enriched on glycolytic enzymes and play important roles in regulating glycolysis (Gaffney et al., 2020).
To date, Kla has been extensively identified in eukaryotes, including humans, mice, Botrytis cinerea and rice (Gao et al., 2020; X. X. Meng et al., 2021; D. Zhang et al., 2019). A recent study reported Kla in prokaryotes, indicating that the regulatory functions of Kla are well-conserved (Dong et al., 2022). YiaC was identified to act as a lysine lactylase (Kcat = 0.97 ± 0.03 s−1, Km = 26.6 ± 1.1 μM, Kcat/Km = 3.6 × 104 s−1M−1), which was more effective than its acetylase activity (Kcat = 0.23 ± 0.04 s−1, Km = 28.7 ± 1.2 μM, Kcat/Km = 8.0 × 103 s−1M−1). CobB functions as a lysine delactylase (Kcat = 0.99 ± 0.04 s−1, Km = 25.4 ± 1.5 μM, Kcat/Km = 3.9 × 104 s−1M−1), which is similar to that of deacetylation (Kcat = 0.97 ± 0.02 s−1, Km = 18.7 ± 1.0 μM, Kcat/Km = 5.2 × 104 s−1M−1) in E. coli MG1655. In addition, YdiF catalyzes the formation of lactyl-CoA, which is the acyl donor for Kla. By performing stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics approach, 79 YiaC-mediated Kla sites and 446 CobB-targeted Kla candidates were identified in vivo after normalizing the detected abundances to their corresponding proteins. Most lactylated proteins were associated with metabolism. Specifically, Kla marks on K382 of PykF can be removed by CobB to promote glycolysis and the growth of E. coli (Dong et al., 2022).
CONCLUDING REMARKS
Precision, high throughput MS-based proteomics has driven the recent discovery of novel acylation marks on histone and non-histone proteins and, equally important, reveals how lysine acylations respond to environmental conditions and metabolic changes. Since the discovery of these novel histone lysine acylations, a major question has been how prevalent they are and what are their functional consequences. From a structural or metabolic perspective, these acylations are believed to be functionally different from previously reported acylations and can exhibit unique impacts in terms of the regulation of chromatin structure and gene transcription. However, regulation of these acylations in cells is complicated and involves intersection with other acylations, both at the metabolite precursor level and on target proteins. For example, butyryl-CoA can be reduced to crotonyl-CoA by acyl-CoA dehydrogenase short chain (ACADS) in the last step of the β-oxidation spiral (Houten et al., 2016). Kbu and Kcr exhibit similar genome-wide occupancy patterns and have been connected to the stimulation of gene transcription in mammals and yeast (Sabari et al., 2017). Their respective isomers, isobutyryl-CoA and methacrylyl-CoA are metabolites generated in the valine pathway. Owing to the same metabolic sources, they appear to share overlapping characteristics in their participation in enzymatic pathways and biological functions. Nevertheless, how the dynamic regulation in cells affects the abundance of these metabolites to alter the corresponding acylation marks and how one connects to another in specific physiological and pathological conditions are largely unknow.
In addition, molecular characterization of pertinent regulatory enzymes such as acyl-CoA synthetases, acyltransferases, deacylases, and ‘readers’ are only in the very early stage. Studies on their regulatory enzymes and corresponding substrates are necessary to provide an in-depth understanding on the biological functions of these novel lysine acylation marks. Studies on lysine acylation are frequently and easily performed on synthetic peptide fragments containing acyllysine residues as substrates to identify ‘writers’, ‘erasers’, ‘readers’, determine kinetics, and investigate site-specific molecular recognition as well as binding profiling of acylated peptides with interactomes. However, studies using fragment peptides cannot reveal interactions or functions of lysine acylation under biological environments. Therefore, there is always a need to develop and incorporate novel approaches applied to acylated proteins to gain insight into their interactions with the interactome and clarify molecular mechanisms in physiological contexts. Furthermore, several acylations rely on the same enzymatic mechanism to modify cellular proteins and can be recognized by the same ‘readers’ for downstream functionality. For example, SIRT2 was reported as an ‘eraser’ to remove acetyl, crotonyl, methacrylyl, and benzoyl marks on histones as well as non-histone proteins (Bao et al., 2014; Bhaskar et al., 2020; Delaney et al., 2021; Huang et al., 2018; Ntorla & Burgoyne, 2021; Serrano et al., 2013; Yi et al., 2021). Some studies have found that metabolic enzymes are involved in the pathogenesis of diseases by targeting more than one modification (Shang et al., 2022). Therefore, profiling dynamic changes of different modifications and elucidating how they compete with regulatory enzymes for downstream functionality is important to understand the underlying mechanisms of diseases.
Last but not the least, a dynamic abundance of SCFAs greatly determines acyl-CoA levels for acylations. However, some SCFAs such as butyrate and isobutyrate, also showed inhibitory effects on HDAC enzymes (Chriett et al., 2019; Zhu et al., 2021). One relevant example is that gut microbiota-derived butyrate was reported to affect histone decrotonylation (Fellows et al., 2018). Therefore, it would be interesting to investigate how SCFAs are balanced as inducers for corresponding acylations on proteins versus as inhibitory mediators for other acylations.
AUTHOR CONTRIBUTIONS
Qianyun Fu : Writing - original draft, writing - review and editing, Amber Cat : Writing - original draft, writing - review and editing, Y. George Zheng : Funding acquisition, writing - original draft, writing - review and editing.
CONFLICT OF INTEREST
The authors have no conflict of interest to claim.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated.
LITERATURE CITED
- Abdinejad, A., Fisher, A. M., & Kumar, S. (1981). Production and utilization of Butyryl-Coa by fatty-acid synthetase from mammalian-tissues. Archives of Biochemistry and Biophysics , 208(1), 135–145. https://doi.org/10.1016/0003-9861(81)90132-6
- Abu-Zhayia, E. R., Machour, F. E., & Ayoub, N. (2019). HDAC-dependent decrease in histone crotonylation during DNA damage. Journal of Molecular Cell Biology , 11(9), 804–806. https://doi.org/10.1093/jmcb/mjz019
- Allfrey, V. G., Faulkner, R., & Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of Rna synthesis. Proceedings of the National Academy of Sciences of the United States of America , 51, 786–794.
- Andresen, B. S., Christensen, E., Corydon, T. J., Bross, P., Pilgaard, B., Wanders, R. J., Ruiter, J. P., Simonsen, H., Winter, V., Knudsen, I., Schroeder, L. D., Gregersen, N., & Skovby, F. (2000). Isolated 2-methylbutyrylglycinuria caused by short/branched-chain acyl-CoA dehydrogenase deficiency: Identification of a new enzyme defect, resolution of its molecular basis, and evidence for distinct acyl-CoA dehydrogenases in isoleucine and valine metabolism. American Journal of Human Genetics , 67(5), 1095–1103. https://doi.org/10.1086/303105
- Andrews, F. H., Shinsky, S. A., Shanle, E. K., Bridgers, J. B., Gest, A., Tsun, I. K., Krajewski, K., Shi, X., Strahl, B. D., & Kutateladze, T. G. (2016). The Taf14 YEATS domain is a reader of histone crotonylation. Nature Chemical Biology , 12(6), 396–U333. https://doi.org/10.1038/nchembio.2065
- Andrews, F. H., Strahl, B. D., & Kutateladze, T. G. (2016). Insights into newly discovered marks and readers of epigenetic information. Nature Chemical Biology , 12(9), 662–668. https://doi.org/10.1038/nchembio.2149
- Avvakumov, N., & Cote, J. (2007). The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene , 26(37), 5395–5407.
- Baldensperger, T., & Glomb, M. A. (2021). Pathways of non-enzymatic lysine acylation. Frontiers in Cell and Developmental Biology , 9, 664553. https://doi.org/10.3389/fcell.2021.664553
- Bao, X. C., Wang, Y., Li, X., Li, X. M., Liu, Z., Yang, T. P., Wong, C. F., Zhang, J., Hao, Q., & Li, X. D. (2014). Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. Elife , 3, e02999.https://doi.org/10.7554/eLife.02999
- Barnes, C. E., English, D. M., & Cowley, S. M. (2019). Acetylation & Co: An expanding repertoire of histone acylations regulates chromatin and transcription. Essays in Biochemistry , 63(1), 97–107. https://doi.org/10.1042/EBC20180061
- Berndsen, C. E., & Denu, J. M. (2008). Catalysis and substrate selection by histone/protein lysine acetyltransferases. Current Opinion in Structural Biology , 18(6), 682–689.
- Bernstein, J., Lott, W. A., Steinberg, B. A., & Yale, H. L. (1952). Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. American Review of Tuberculosis , 65(4), 357–364. https://doi.org/10.1164/art.1952.65.4.357
- Bhaskar, A., Kumar, S., Khan, M. Z., Singh, A., Dwivedi, V. P., & Nandicoori, V. K. (2020). Host sirtuin 2 as an immunotherapeutic target against tuberculosis. Elife , 9, e55415. https://doi.org/10.7554/eLife.55415
- Bhattacharya, A., Chatterjee, S., Bhaduri, U., Singh, A. K., Vasudevan, M., Sashidhara, K. V., Guha, R., Natesh, N., & Kundu, T. K. (2022). EP300 (p300) mediated histone butyrylation is critical for adipogenesis. BioRxiv , 2021.2008.2001.454641. https://doi.org/10.1101/2021.08.01.454641
- Broder, G., & Weil, M. H. (1964). Excess lactate: An index of reversibility of shock in human patients. Science , 143(3613), 1457–1459. https://doi.org/10.1126/science.143.3613.1457
- Brown, G. K., Hunt, S. M., Scholem, R., Fowler, K., Grimes, A., Mercer, J. F., Truscott, R. M., Cotton, R. G., Rogers, J. G., & Danks, D. M. (1982). Beta-hydroxyisobutyryl coenzyme-a deacylase deficiency - a defect in valine metabolism associated with physical malformations. Pediatrics , 70(4), 532–538.
- Brusilow, S. W., Danney, M., Waber, L. J., Batshaw, M., Burton, B., Levitsky, L., Roth, K., Mckeethren, C., & Ward, J. (1984). Treatment of episodic hyperammonemia in children with inborn errors of urea synthesis. New England Journal of Medicine , 310(25), 1630–1634. https://doi.org/10.1056/NEJM198406213102503
- Bursten, S. L., Locksley, R. M., Ryan, J. L., & Lovett, D. H. (1988). Acylation of monocyte and glomerular mesangial cell-proteins - Myristyl acylation of the interleukin-1 precursors. Journal of Clinical Investigation , 82(5), 1479–1488. https://doi.org/10.1172/Jci113755
- Centers for Disease Control and Prevention (CDC). (2010). Severe isoniazid-associated liver injuries among persons being treated for latent tuberculosis infection - United States, 2004-2008. Mmwr Morbidity and Mortality Weekly Report , 59(8), 224–229.
- Chen, Y., Sprung, R., Tang, Y., Ball, H., Sangras, B., Kim, S. C., Falck, J. R., Peng, J., Gu, W., & Zhao, Y. M. (2007). Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & Cellular Proteomics, 6(5), 812–819. https://doi.org/10.1074/mcp.M700021-MCP200
- Chriett, S., Dabek, A., Wojtala, M., Vidal, H., Balcerczyk, A., & Pirola, L. (2019). Prominent action of butyrate over beta-hydroxybutyrate as histone deacetylase inhibitor, transcriptional modulator and anti-inflammatory molecule. Scientific Reports , 9(1), 742. https://doi.org/10.1038/s41598-018-36941-9
- Cui, H. C., Xie, N., Banerjee, S., Ge, J., Jiang, D. Y., Dey, T., Matthews, Q. L., Liu, R. - M., & Liu, G. (2021). Lung myofibroblasts promote macrophage profibrotic activity through lactate-induced histone lactylation. American Journal of Respiratory Cell and Molecular Biology , 64(1), 115–125. https://doi.org/10.1165/rcmb.2020-0360OC
- Cummings, J. H., Pomare, E. W., Branch, W. J., Naylor, C. P. E., & Macfarlane, G. T. (1987). Short chain fatty-acids in human large-intestine, portal, hepatic and venous-blood. Gut , 28(10), 1221–1227. https://doi.org/10.1136/gut.28.10.1221
- Dai, L. Z., Peng, C., Montellier, E., Lu, Z. K., Chen, Y., Ishii, H., Debernardi, A., Buchou, T., Rousseaux, S., Jin, F., Sabari, B. R., Deng, Z., Allis, C. D., Ren, B., Khochbin, S., & Zhao, Y. M. (2014). Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nature Chemical Biology , 10(5), 365–U373. https://doi.org/10.1038/Nchembio.1497
- Dancy, B. M., & Cole, P. A. (2015). Protein lysine acetylation by p300/CBP. Chemical Reviews , 115(6), 2419–2452. https://doi.org/10.1021/cr500452k
- Delaney, K., Tan, M. J., Zhu, Z. S., Gao, J. J., Dai, L. Z., Kim, S., Ding, J., He, M., Halabelian, L., Yang, L., Nagarajan, P., Parthun, M. R., Lee, S., Khochbin, S., Zheng, Y. G., & Zhao, Y. M. (2021). Histone lysine methacrylation is a dynamic post-translational modification regulated by HAT1 and SIRT2. Cell Discovery , 7(1), https://doi.org/10.1038/s41421-021-00344-4
- Deng, M., Tang, Y., Li, W., Wang, X., Zhang, R., Zhang, X., Zhao, X., Liu, J., Tang, C., Liu, Z., Huang, Y., Peng, H., Xiao, L., Tang, D., Scott, M. J., Wang, Q., Liu, J., Xiao, X., Watkins, S., … Lu, B. (2018). The endotoxin delivery protein HMGB1 mediates caspase-11-dependent lethality in sepsis. Immunity , 49(4), 740–753.e747. https://doi.org/10.1016/j.immuni.2018.08.016
- Dong, H., Zhang, J., Zhang, H., Han, Y., Lu, C., Chen, C., Tan, X., Wang, S., Bai, X., Zhai, G., Tian, S., Zhang, T., Cheng, Z., Li, E., Xu, L., & Zhang, K. (2022). YiaC and CobB regulate lysine lactylation in Escherichia coli. Nature Communications , 13(1), 6628. https://doi.org/10.1038/s41467-022-34399-y
- Feldman, J. L., Baeza, J., & Denu, J. M. (2013). Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. Journal of Biological Chemistry , 288(43), 31350–31356. https://doi.org/10.1074/jbc.C113.511261
- Fellows, R., Denizot, J., Stellato, C., Cuomo, A., Jain, P., Stoyanova, E., Balázsi, S., Hajnády, Z., Liebert, A., Kazakevych, J., Blackburn, H., Corrêa, R. O., Fachi, J. L., Sato, F. T., Ribeiro, W. R., Ferreira, C. M., Perée, H., Spagnuolo, M., Mattiuz, R., … Varga-Weisz, P. (2018). Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nature Communications , 9, 105. https://doi.org/10.1038/s41467-017-02651-5
- Finkel, T., Deng, C. X., & Mostoslavsky, R. (2009). Recent progress in the biology and physiology of sirtuins. Nature , 460(7255), 587–591. https://doi.org/10.1038/nature08197
- Flynn, E. M., Huang, O. W., Poy, F., Oppikofer, M., Bellon, S. F., Tang, Y., & Cochran, A. G. (2015). A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure (London, England) , 23(10), 1801–1814. https://doi.org/10.1016/j.str.2015.08.004
- Fu, H. F., Tian, C. L., Ye, X. Y., Sheng, X. Y., Wang, H., Liu, Y. F., & Liu, L. (2018). Dynamics of telomere rejuvenation during chemical induction to pluripotent stem cells. Stem Cell Reports , 11(1), 70–87. https://doi.org/10.1016/j.stemcr.2018.05.003
- Gaffney, D. O., Jennings, E. Q., Anderson, C. C., Marentette, J. O., Shi, T., Oxvig, A. M. S., Streeter, M. D., Johannsen, M., Spiegel, D. A., Chapman, E., Roede, J. R., & Galligan, J. J. (2020). Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chemical Biology , 27(2), 206–+. https://doi.org/10.1016/j.chembiol.2019.11.005
- Gao, M. M., Zhang, N., & Liang, W. X. (2020). Systematic analysis of lysine lactylation in the plant fungal pathogen botrytis cinerea. Frontiers in Microbiology , 11, 594743. https://doi.org/10.3389/fmicb.2020.594743
- Gates, L. A., Reis, B. S., Lund, P. J., Paul, M. R., Leboeuf, M., Nadeem, Z., Carroll, T. S., Garcia, B. A., Mucida, D., & Allis, C. D. (2022). Microbiota-dependent histone butyrylation in the mammalian intestine. BioRxiv , 2022.2009.2029.510184. https://doi.org/10.1101/2022.09.29.510184
- Goudarzi, A., Hosseinmardi, N., Salami, S., Mehdikhani, F., Derakhshan, S., & Aminishakib, P. (2020). Starvation promotes histone lysine butyrylation in the liver of male but not female mice. Gene , 745, 144647. https://doi.org/10.1016/j.gene.2020.144647
- Goudarzi, A., Zhang, D., Huang, H., Barral, S., Kwon, O. K., Qi, S. K., Tang, Z., Buchou, T., Vitte, A. - L., He, T., Cheng, Z., Montellier, E., Gaucher, J., Curtet, S., Debernardi, A., Charbonnier, G., Puthier, D., Petosa, C., Panne, D., … Khochbin, S. (2016). Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Molecular Cell , 62(2), 169–180. https://doi.org/10.1016/j.molcel.2016.03.014
- Gowans, G. J., Bridgers, J. B., Zhang, J., Dronamraju, R., Burnetti, A., King, D. A., Thiengmany, A. V., Shinsky, S. A., Bhanu, N. V., Garcia, B. A., Buchler, N. E., Strahl, B. D., & Morrison, A. J. (2019). Recognition of histone crotonylation by Taf14 links metabolic state to gene expression. Molecular Cell , 76(6), 909–+. https://doi.org/10.1016/j.molcel.2019.09.029
- Grammel, M., & Hang, H. C. (2013). Chemical reporters for biological discovery. Nature Chemical Biology , 9(8), 475–484. https://doi.org/10.1038/nchembio.1296
- Gruber, J. J., Geller, B., Lipchik, A. M., Chen, J., Salahudeen, A. A., Ram, A. N., Ford, J. M., Kuo, C. J., & Snyder, M. P. (2019). HAT1 coordinates histone production and acetylation via H4 promoter binding. Molecular Cell , 75(4), 711–+. https://doi.org/10.1016/j.molcel.2019.05.034
- Gu, W., Shi, X. L., & Roeder, R. G. (1997). Synergistic activation of transcription by CBP and p53. Nature , 387(6635), 819–823.
- Haberland, M., Montgomery, R. L., & Olson, E. N. (2009). The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nature Reviews Genetics , 10(1), 32–42. https://doi.org/10.1038/nrg2485
- Hagihara, H., Shoji, H., Otabi, H., Toyoda, A., Katoh, K., Namihira, M., & Miyakawa, T. (2021). Protein lactylation induced by neural excitation. Cell Reports , 37(2), 109820. https://doi.org/10.1016/j.celrep.2021.109820
- He, M., Han, Z., Liu, L., & Zheng, Y. G. (2018). Chemical biology approaches for investigating the functions of lysine acetyltransferases. Angewandte Chemie , 57(5), 1162–1184. https://doi.org/10.1002/anie.201704745
- Head, P. E., Myung, S., Chen, Y., Schneller, J. L., Wang, C., Duncan, N., Hoffman, P., Chang, D., Gebremariam, A., Gucek, M., Manoli, I., & Venditti, C. P. (2022). Aberrant methylmalonylation underlies methylmalonic acidemia and is attenuated by an engineered sirtuin. Science Translational Medicine , 14(646), eabn4772. https://doi.org/10.1126/scitranslmed.abn4772
- Hildmann, C., Riester, D., & Schwienhorst, A. (2007). Histone deacetylases–an important class of cellular regulators with a variety of functions. Applied Microbiology and Biotechnology , 75(3), 487–497.
- Houten, S. M., Violante, S., Ventura, F. V., & Wanders, R. J. (2016). The biochemistry and physiology of mitochondrial fatty acid beta-oxidation and its genetic disorders. Annual Review of Physiology , 78, 23–44. https://doi.org/10.1146/annurev-physiol-021115-105045
- Huang, H., Lin, S., Garcia, B. A., & Zhao, Y. (2015). Quantitative proteomic analysis of histone modifications. Chemical Reviews , 115(6), 2376–2418. https://doi.org/10.1021/cr500491u
- Huang, H., Tang, S., Ji, M., Tang, Z. Y., Shimada, M., Liu, X. J., Qi, S., Locasale, J. W., Roeder, R. G., Zhao, Y., & Li, X. L. (2018). p300-Mediated lysine 2-hydroxyisobutyrylation regulates glycolysis (vol 70, pg 663, 2018). Molecular Cell , 70(5), 984–984. https://doi.org/10.1016/j.molcel.2018.05.035
- Huang, H., Zhang, D., Wang, Y., Perez-Neut, M., Han, Z., Zheng, Y. G., Hao, Q., & Zhao, Y. (2018). Lysine benzoylation is a histone mark regulated by SIRT2. Nature Communications , 9, 3374. https://doi.org/10.1038/s41467-018-05567-w
- Irizarry-Caro, R. A., McDaniel, M. M., Overcast, G. R., Jain, V. G., Troutman, T. D., & Pasare, C. (2020). TLR signaling adapter BCAP regulates inflammatory to reparatory macrophage transition by promoting histone lactylation. Proceedings of the National Academy of Sciences of the United States of America , 117(48), 30628–30638. https://doi.org/10.1073/pnas.2009778117
- Jennings, E. Q., Ray, J. D., Zerio, C. J., Trujillo, M. N., Mcdonald, D. M., Chapman, E., Spiegel, D. A., & Galligan, J. J. (2021). Sirtuin 2 regulates protein LactoylLys modifications. Chembiochem , 22(12), 2102–2106. https://doi.org/10.1002/cbic.202000883
- Jiang, G., Nguyen, D., Archin, N. M., Yukl, S. A., Méndez-Lagares, G., Tang, Y., Elsheikh, M., Thompson, G. R., Hartigan-O'connor, D. J., Margolis, D. M., Wong, J. K., … Dandekar, S. (2018). HIV latency is reversed by ACSS2-driven histone crotonylation. Journal of Clinical Investigation , 128(3), 1190–1198. https://doi.org/10.1172/Jci98071
- Jiang, J., Huang, D., Jiang, Y., Hou, J., Tian, M., Li, J., Sun, L., Zhang, Y., Zhang, T., Li, Z., Li, Z., Tong, S., & Ma, Y. (2021). Lactate modulates cellular metabolism through histone lactylation-mediated gene expression in non-small cell lung cancer. Frontiers in Oncology , 11, 647559. https://doi.org/10.3389/fonc.2021.647559
- Jiang, T., Zhou, X., Taghizadeh, K., Dong, M., & Dedon, P. C. (2007). N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proceedings of the National Academy of Sciences of the United States of America , 104(1), 60–65.
- Jiang, Y., Li, Y., Liu, C., Zhang, L., Lv, D., Weng, Y., Cheng, Z., Chen, X., Zhan, J., & Zhang, H. (2021). Isonicotinylation is a histone mark induced by the anti-tuberculosis first-line drug isoniazid. Nature Communications , 12(1), 5548. https://doi.org/10.1038/s41467-021-25867-y
- Kaczmarska, Z., Ortega, E., Goudarzi, A., Huang, H., Kim, S., Márquez, J. A., Zhao, Y., Khochbin, S., & Panne, D. (2017). Structure of p300 in complex with acyl-CoA variants. Nature Chemical Biology , 13(1), 21–29. https://doi.org/10.1038/Nchembio.2217
- Kebede, A. F., Nieborak, A., Shahidian, L. Z., Le Gras, S., Richter, F., Gomez, D. A., Baltissen, M. P., Meszaros, G., de Fatima Magliarelli, H., Taudt, A., Margueron, R., Colomé-Tatché, M., Ricci, R., Daujat, S., Vermeulen, M., Mittler, G., & Schneider, R. (2017). Histone propionylation is a mark of active chromatin. Nature Structural & Molecular Biology, 24(12), 1048–+. https://doi.org/10.1038/nsmb.3490
- Kim, C. H., Kang, M., Kim, H. J., Chatterjee, A., & Schultz, P. G. (2012). Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angewandte Chemie (International ed in English) , 51(29), 7246–7249. https://doi.org/10.1002/anie.201203349
- Kim, G. W., & Yang, X. J. (2011). Comprehensive lysine acetylomes emerging from bacteria to humans. Trends in Biochemical Sciences , 36(4), 211–220. https://doi.org/10.1016/j.tibs.2010.10.001
- Kim, S. C., Sprung, R., Chen, Y., Xu, Y., Ball, H., Pei, J., Cheng, T., Kho, Y., Xiao, H., Xiao, L., Grishin, N. V., White, M., Yang, X. - J., & Zhao, Y. (2006). Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular Cell , 23(4), 607–618. https://doi.org/10.1016/j.molcel.2006.06.026
- Kimura, A., Matsubara, K., & Horikoshi, M. (2005). A decade of histone acetylation: Marking eukaryotic chromosomes with specific codes. Journal of Biochemistry , 138(6), 647–662. https://doi.org/10.1093/jb/mvi184
- Klein, B. J., Jang, S. M., Lachance, C., Mi, W. Y., Lyu, J., Sakuraba, S., Krajewski, K., Wang, W. W., Sidoli, S., Liu, J., Zhang, Y., Wang, X., Warfield, B. M., Kueh, A. J., Voss, A. K., Thomas, T., Garcia, B. A., Liu, W. R., Strahl, B. D., … Kutateladze, T. G. (2019). Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nature Communications , 10, 4724. https://doi.org/10.1038/s41467-019-12551-5
- Kollenstart, L., De Groot, A. J. L., Janssen, G. M. C., Cheng, X., Vreeken, K., Martino, F., Côté, J., Van Veelen, P. A., … Van Attikum, H. (2019). Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. Journal of Biological Chemistry , 294(52), 20122–20134. https://doi.org/10.1074/jbc.RA119.010302
- Kopytko, P., Piotrowska, K., Janisiak, J., & Tarnowski, M. (2021). Garcinol-A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug. International Journal of Molecular Sciences , 22(6), 2828. https://doi.org/10.3390/ijms22062828
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell , 128(4), 693–705. https://doi.org/10.1016/j.cell.2007.02.005
- L'Hernault, S. W., & Rosenbaum, J. L. (1985). Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry , 24(2), 473–478. https://doi.org/10.1021/bi00323a034
- Lewis, J. D., Lee, A., Ma, W., Zhou, H., Guttman, D. S., & Desveaux, D. (2011). The YopJ superfamily in plant-associated bacteria. Molecular Plant Pathology , 12(9), 928–937. https://doi.org/10.1111/j.1364-3703.2011.00719.x
- Li, F., Wang, P., Liu, K., Tarrago, M. G., Lu, J., Chini, E. N., & Ma, X. (2016). A high dose of isoniazid disturbs endobiotic homeostasis in mouse liver. Drug Metabolism and Disposition , 44(11), 1742–1751. https://doi.org/10.1124/dmd.116.070920
- Li, L., Chen, K., Wang, T., Wu, Y., Xing, G., Chen, M., Hao, Z., Zhang, C., Zhang, J., Ma, B., Liu, Z., Yuan, H., Liu, Z., Long, Q., Zhou, Y., Qi, J., Zhao, D., Gao, M., Pei, D., … Liu, X. (2020). Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nature Metabolism , 2(9), 882–892. https://doi.org/10.1038/s42255-020-0267-9
- Li, X. L., Yang, Y. Y., Zhang, B., Lin, X. T., Fu, X. X., An, Y., Zou, Y., Wang, J. - X., Wang, Z., & Yu, T. (2022). Lactate metabolism in human health and disease. Signal Transduction and Targeted Therapy , 7(1), 305. https://doi.org/10.1038/s41392-022-01151-3
- Li, Y., Sabari, B. R., Panchenko, T., Wen, H., Zhao, D., Guan, H., Wan, L., Huang, H., Tang, Z., Zhao, Y., Roeder, R. G., Shi, X., Allis, C. D., & Li, H. (2016). Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Molecular Cell , 62(2), 181–193. https://doi.org/10.1016/j.molcel.2016.03.028
- Liberti, M. V., & Locasale, J. W. (2016). The warburg effect: How does it benefit cancer cells? (vol 41, pg 211, 2016). Trends in Biochemical Sciences , 41(3), 287–287. https://doi.org/10.1016/j.tibs.2016.01.004
- Liberti, M. V., & Locasale, J. W. (2020). Histone Lactylation: A New Role for Glucose Metabolism. Trends in Biochemical Sciences , 45(3), 179–+. https://doi.org/10.1016/j.tibs.2019.12.004
- Lin, Y., Peng, W., Jiang, M., Lin, C., Lin, W., Zheng, Z., Li, M., & Fu, Q. (2018). Clinical, biochemical and genetic analysis of Chinese patients with isobutyryl-CoA dehydrogenase deficiency. Clinica Chimica Acta , 487, 133–138. https://doi.org/10.1016/j.cca.2018.09.033
- Liu, S., Yu, H., Liu, Y., Liu, X., Zhang, Y., Bu, C., Yuan, S., Chen, Z., Xie, G., Li, W., Xu, B., Yang, J., He, L., Jin, T., Xiong, Y., Sun, L., Liu, X., Han, C., Cheng, Z., … Shang, Y. (2017). Chromodomain protein CDYL acts as a Crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Molecular Cell , 67(5), 853–+. https://doi.org/10.1016/j.molcel.2017.07.011
- Liu, X., Wang, L., Zhao, K. H., Thompson, P. R., Hwang, Y., Marmorstein, R., & Cole, P. A. (2008). The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature , 451(7180), 846–850. https://doi.org/10.1038/nature06546
- Liu, X., Wei, W., Liu, Y., Yang, X., Wu, J., Zhang, Y., Zhang, Q., Shi, T., Du, J. X., Zhao, Y., Lei, M., Zhou, J. - Q., Li, J., & Wong, J. (2017). MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discovery , 3, 17016. https://doi.org/10.1038/celldisc.2017.16
- Liu, Y., Li, M., Fan, M., Song, Y., Yu, H., Zhi, X., Xiao, K., Lai, S., Zhang, J., Jin, X., Shang, Y., Liang, J., & Huang, Z. (2019). Chromodomain Y-like protein-mediated histone crotonylation regulates stress-induced depressive behaviors. Biological Psychiatry , 85(8), 635–649. https://doi.org/10.1016/j.biopsych.2018.11.025
- Loupatty, F. J., Clayton, P. T., Ruiter, J. P. N., Ofman, R., Ijlst, L., Brown, G. K., Thorburn, D. R., Harris, R. A., Duran, M., Desousa, C., Krywawych, S., Heales, S. J. R., & Wanders, R. J. A. (2007). Mutations in the gene encoding 3-hydroxyisobutyryl-CoA hydrolase results in progressive infantile neurodegeneration. American Journal of Human Genetics , 80(1), 195–199. https://doi.org/10.1086/510725
- Ma, K. W., & Ma, W. (2016). YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiology and Molecular Biology Reviews , 80(4), 1011–1027. https://doi.org/10.1128/MMBR.00032-16
- Marin, T. L., Gongol, B., Zhang, F., Martin, M., Johnson, D. A., Xiao, H., Wang, Y., Subramaniam, S., Chien, S., & Shyy, J. Y. - J. (2017). AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Science Signaling , 10(464), eaaf7478. https://doi.org/10.1126/scisignal.aaf7478
- Marsh, V. L., Peak-Chew, S. Y., & Bell, S. D. (2005). Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. Journal of Biological Chemistry , 280(22), 21122–21128. https://doi.org/10.1074/jbc.M501280200
- Meng, L., Chan, W. S., Huang, L., Liu, L., Chen, X., Zhang, W., Wang, F., Cheng, K., Sun, H., & Wong, K. C. (2022). Mini-review: Recent advances in post-translational modification site prediction based on deep learning. Computational and Structural Biotechnology Journal , 20, 3522–3532. https://doi.org/10.1016/j.csbj.2022.06.045
- Meng, X. X., Baine, J. M., Yan, T. C., & Wang, S. (2021). Comprehensive analysis of lysine lactylation in rice (Oryza sativa) grains. Journal of Agricultural and Food Chemistry , 69(29), 8287–8297. https://doi.org/10.1021/acs.jafc.1c00760
- Moreno-Yruela, C., Galleano, I., Madsen, A. S., & Olsen, C. A. (2018). Histone deacetylase 11 is an epsilon-N-myristoyllysine hydrolase. Cell Chemical Biology , 25(7), 849–856.e848. https://doi.org/10.1016/j.chembiol.2018.04.007
- Moreno-Yruela, C., Zhang, D., Wei, W., Baek, M., Liu, W. C., Gao, J. J., Danková, D., Nielsen, A. L., Bolding, J. E., Yang, L., Jameson, S. T., Wong, J., Olsen, C. A., & Zhao, Y. M. (2022). Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Science Advances , 8(3), https://doi.org/10.1126/sciadv.abi6696
- Nagarajan, P., Garcia, P. A. A., Iyer, C. C., Popova, L. V., Arnold, W. D., & Parthun, M. R. (2019). Early-onset aging and mitochondrial defects associated with loss of histone acetyltransferase 1 (Hat1). Aging Cell , 18(5), e12992. https://doi.org/10.1111/acel.12992
- Neumann, H., Peak-Chew, S. Y., & Chin, J. W. (2008). Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nature Chemical Biology , 4(4), 232–234. https://doi.org/10.1038/nchembio.73
- Nguyen, T. V., Andresen, B. S., Corydon, T. J., Ghisla, S., Razik, N. A. E., Mohsen, A. W. A., Cederbaum, S. D., Roe, D. S., Roe, C. R., Lench, N. J., & Vockley, J. (2002). Identification of isobutyryl-CoA dehydrogenase and its deficiency in humans. Molecular Genetics and Metabolism , 77(1-2), 68–79. https://doi.org/10.1016/S1096-7192(02)00152-X
- Nie, L. T., Shuai, L., Zhu, M. R., Liu, P., Xie, Z. F., Jiang, S. W., Jiang, H. - W., Li, J., Zhao, Y., Li, J. - Y., & Tan, M. J. (2017). The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Molecular & Cellular Proteomics, 16(7), 1324–1334. https://doi.org/10.1074/mcp.M117.067553
- Ntorla, A., & Burgoyne, J. R. (2021). The regulation and function of histone crotonylation. Frontiers in Cell and Developmental Biology , 9, 624914. https://doi.org/10.3389/fcell.2021.624914
- Ott, M., Schnolzer, M., Garnica, J., Fischle, W., Emiliani, S., Rackwitz, H. R., & Verdin, E. (1999). Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Current Biology , 9(24), 1489–1492.
- Palsson-McDermott, E. M., & O'Neill, L. A. J. (2013). The Warburg effect then and now: From cancer to inflammatory diseases. BioEssays , 35(11), 965–973. https://doi.org/10.1002/bies.201300084
- Pan, R. Y., He, L., Zhang, J., Liu, X. H., Liao, Y. J., Gao, J., Liao, Y., Yan, Y., Li, Q., Zhou, X., Cheng, J., Xing, Q., Guan, F., Zhang, J., Sun, L., & Yuan, Z. Q. (2022). Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer's disease. Cell Metabolism , 34(4), 634–+. https://doi.org/10.1016/j.cmet.2022.02.013
- Park, H. W., Park, E. H., Yun, H. M., & Rhim, H. (2011). Sodium benzoate-mediated cytotoxicity in mammalian cells. Journal of Food Biochemistry , 35(4), 1034–1046. https://doi.org/10.1111/j.1745-4514.2010.00432.x
- Parker, C. G., & Pratt, M. R. (2020). Click chemistry in proteomic investigations. Cell , 180(4), 605–632. https://doi.org/10.1016/j.cell.2020.01.025
- Pavlova, N. N., & Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metabolism , 23(1), 27–47. https://doi.org/10.1016/j.cmet.2015.12.006
- Peng, C., Lu, Z. K., Xie, Z. Y., Cheng, Z. Y., Chen, Y., Tan, M. J., Luo, H., Zhang, Y., He, W., Yang, K., Zwaans, B. M. M., Tishkoff, D., Ho, L., Lombard, D., He, T. - C., Dai, J., Verdin, E., Ye, Y., & Zhao, Y. M. (2011). The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & Cellular Proteomics, 10(12), M111.012658. https://doi.org/10.1074/mcp.M111.012658
- Peters, H., Buck, N., Wanders, R., Ruiter, J., Waterham, H., Koster, J., Yaplito-Lee, J., Ferdinandusse, S., & Pitt, J. (2014). ECHS1 mutations in Leigh disease: A new inborn error of metabolism affecting valine metabolism. Brain , 137, 2903–2908. https://doi.org/10.1093/brain/awu216
- Phillips, D. M. (1963). The presence of acetyl groups of histones. Biochemical Journal , 87, 258–263.
- Pongsavee, M. (2015). Effect of sodium benzoate preservative on micronucleus induction, chromosome break, and Ala40Thr superoxide dismutase gene mutation in lymphocytes. BioMed Research International , 2015, 103512. https://doi.org/10.1155/2015/103512
- Pougovkina, O., te Brinke, H., Wanders, R. J. A., Houten, S. M., & de Boer, V. C. J. (2014). Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. Journal of Inherited Metabolic Disease , 37(5), 709–714. https://doi.org/10.1007/s10545-014-9684-9
- Praphanproj, V., Boyadjiev, S. A., Waber, L. J., Brusilow, S. W., & Geraghty, M. T. (2000). Three cases of intravenous sodium benzoate and sodium phenylacetate toxicity occurring in the treatment of acute hyperammonaemia. Journal of Inherited Metabolic Disease , 23(2), 129–136. https://doi.org/10.1023/A:1005661631281
- Prescher, J. A., & Bertozzi, C. R. (2005). Chemistry in living systems. Nature Chemical Biology , 1(1), 13–21. https://doi.org/10.1038/nchembio0605-13
- Ran-Ressler, R. R., Bae, S., Lawrence, P., Wang, D. H., & Brenna, J. T. (2014). Branched-chain fatty acid content of foods and estimated intake in the USA. British Journal of Nutrition , 112(4), 565–572. https://doi.org/10.1017/S0007114514001081
- Ren, X., Zhou, Y., Xue, Z., Hao, N., Li, Y., Guo, X., Wang, D., Shi, X., & Li, H. (2021). Histone benzoylation serves as an epigenetic mark for DPF and YEATS family proteins. Nucleic Acids Research , 49(1), 114–126. https://doi.org/10.1093/nar/gkaa1130
- Robinson, W. G., Nagle, R., Bachhawat, B. K., Kupiecki, F. P., & Coon, M. J. (1957). Coenzyme-a thiol esters of isobutyric, methacrylic, and beta-hydroxyisobutyric acids as intermediates in the enzymatic degradation of valine. Journal of Biological Chemistry , 224(1), 1–11.
- Ruiz-Andres, O., Sanchez-Nino, M. D., Cannata-Ortiz, P., Ruiz-Ortega, M., Egido, J., Ortiz, A., & Sanz, A. B. (2016). Histone lysine crotonylation during acute kidney injury in mice. Disease Models & Mechanisms, 9(6), 633–645. https://doi.org/10.1242/dmm.024455
- Ruthenburg, A. J., Li, H., Patel, D. J., & Allis, C. D. (2007). Multivalent engagement of chromatin modifications by linked binding modules. Nature Reviews Molecular Cell Biology , 8(12), 983–994. https://doi.org/10.1038/nrm2298
- Sabari, B. R., Tang, Z., Huang, H., Yong-Gonzalez, V., Molina, H., Kong, H. E., Dai, L., Shimada, M., Cross, J. R., Zhao, Y., Roeder, R. G., & Allis, C. D. (2015). Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Molecular Cell , 58(2), 203–215. https://doi.org/10.1016/j.molcel.2015.02.029
- Sabari, B. R., Zhang, D., Allis, C. D., & Zhao, Y. M. (2017). Metabolic regulation of gene expression through histone acylations. Nature Reviews Molecular Cell Biology , 18(2), 90–101. https://doi.org/10.1038/nrm.2016.140
- Sapountzi, V., & Cote, J. (2011). MYST-family histone acetyltransferases: Beyond chromatin. Cellular and Molecular Life Sciences , 68(7), 1147–1156. https://doi.org/10.1007/s00018-010-0599-9
- Serrano, L., Martinez-Redondo, P., Marazuela-Duque, A., Vazquez, B. N., Dooley, S. J., Voigt, P., Beck, D. B., Kane-Goldsmith, N., Tong, Q., Rabanal, R. M., Fondevila, D., Muñoz, P., Krüger, M., Tischfield, J. A., & Vaquero, A. (2013). The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes & Development, 27(6), 639–653. https://doi.org/10.1101/gad.211342.112
- Shang, S., Liu, J., & Hua, F. (2022). Protein acylation: Mechanisms, biological functions and therapeutic targets. Signal Transduction and Targeted Therapy , 7(1), 396. https://doi.org/10.1038/s41392-022-01245-y
- Simithy, J., Sidoli, S., Yuan, Z. F., Coradin, M., Bhanu, N. V., Marchione, D. M., Klein, B. J., Bazilevsky, G. A., McCullough, C. E., Magin, R. S., Kutateladze, T. G., Snyder, N. W., Marmorstein, R., & Garcia, B. A. (2017). Characterization of histone acylations links chromatin modifications with metabolism. Nature Communications , 8, 1141. https://doi.org/10.1038/s41467-017-01384-9
- Simon, R. P., Robaa, D., Alhalabi, Z., Sippl, W., & Jung, M. (2016). KATching-up on small molecule modulators of lysine acetyltransferases. Journal of Medicinal Chemistry , 59(4), 1249–1270. https://doi.org/10.1021/acs.jmedchem.5b01502
- Sletten, E. M., & Bertozzi, C. R. (2009). Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angewandte Chemie-International Edition , 48(38), 6974–6998. https://doi.org/10.1002/anie.200900942
- Smith, B. C., & Denu, J. M. (2007). Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. Journal of Biological Chemistry , 282(51), 37256–37265. https://doi.org/10.1074/jbc.M707878200
- Song, J., Han, Z., & Zheng, Y. G. (2022). Identification and profiling of histone acetyltransferase substrates by bioorthogonal labeling. Current Protocols , 2(7), e497. https://doi.org/10.1002/cpz1.497
- Song, J., & Zheng, Y. G. (2020). Bioorthogonal reporters for detecting and profiling protein acetylation and acylation. SLAS Discovery , 25(2), 148–162. https://doi.org/10.1177/2472555219887144
- Soufi, B., Soares, N. C., Ravikumar, V., & Macek, B. (2012). Proteomics reveals evidence of cross-talk between protein modifications in bacteria: Focus on acetylation and phosphorylation. Current Opinion in Microbiology , 15(3), 357–363. https://doi.org/10.1016/j.mib.2012.05.003
- Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., & Escalante-Semerena, J. C. (2002). Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science , 298(5602), 2390–2392. https://doi.org/10.1126/science.1077650
- Starai, V. J., & Escalante-Semerena, J. C. (2004). Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. Journal of Molecular Biology , 340(5), 1005–1012. https://doi.org/10.1016/j.jmb.2004.05.010
- Sun, S. Y., Xu, X. X., Liang, L., Wang, X. L., Bai, X., Zhu, L. P., He, Q., Liang, H., Xin, X., Wang, L., Lou, C., Cao, X., Chen, X., Li, B., Wang, B., & Zhao, J. W. (2021). Lactic acid-producing probiotic saccharomyces cerevisiae attenuates ulcerative colitis via suppressing macrophage pyroptosis and modulating gut microbiota. Frontiers in Immunology , 12, 777665. https://doi.org/10.3389/fimmu.2021.777665
- Sun, Y. A., Chen, Y. C., & Peng, T. (2022). A bioorthogonal chemical reporter for the detection and identification of protein lactylation. Chemical Science , 13(20), 6019–6027. https://doi.org/10.1039/d2sc00918h
- Sunden-Cullberg, J., Norrby-Teglund, A., Rouhiainen, A., Rauvala, H., Herman, G., Tracey, K. J., Lee, M. L., Andersson, J., Tokics, L., & Treutiger, C. J. (2005). Persistent elevation of high mobility group box-1 protein (HMGB1) in patients with severe sepsis and septic shock. Critical Care Medicine , 33(3), 564–573. https://doi.org/10.1097/01.ccm.0000155991.88802.4d
- Szende, B., Juhasz, J., & Kendrey, G. (1967). [Electronmicroscopic studies on INH-induced liver changes and liver tumors in white rats]. Gegenbaurs Morphologisches Jahrbuch , 111(2), 216–224.
- Tan, D., Wei, W., Han, Z., Ren, X., Yan, C., Qi, S., Song, X., Zheng, Y. G., Wong, J., & Huang, H. (2022). HBO1 catalyzes lysine benzoylation in mammalian cells. iScience , 25(11), 105443. https://doi.org/10.1016/j.isci.2022.105443
- Tan, M., Luo, H., Lee, S., Jin, F., Yang, J. S., Montellier, E., Buchou, T., Cheng, Z., Rousseaux, S., Rajagopal, N., Lu, Z., Ye, Z., Zhu, Q., Wysocka, J., Ye, Y., Khochbin, S., Ren, B., & Zhao, Y. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell , 146(6), 1016–1028. https://doi.org/10.1016/j.cell.2011.08.008
- Tan, M., Peng, C., Anderson, K. A., Chhoy, P., Xie, Z., Dai, L., Park, J., Chen, Y., Huang, H., Zhang, Y., Ro, J., Wagner, G. R., Green, M. F., Madsen, A. S., Schmiesing, J., Peterson, B. S., Xu, G., Ilkayeva, O. R., Muehlbauer, M. J., … Zhao, Y. (2014). Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metabolism , 19(4), 605–617. https://doi.org/10.1016/j.cmet.2014.03.014
- Tian, H., Yang, J., Guo, A. - D., Ran, Y., Yang, Y. - Z., Yang, B., Huang, R., Liu, H., & Chen, X. - H. (2021). Genetically encoded benzoyllysines serve as versatile probes for interrogating histone benzoylation and interactions in living cells. Acs Chemical Biology , 16(11), 2560–2569. https://doi.org/10.1021/acschembio.1c00614
- Trefely, S., Lovell, C. D., Snyder, N. W., & Wellen, K. E. (2020). Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Molecular Metabolism , 38, 100941. https://doi.org/10.1016/j.molmet.2020.01.005
- Ud-Din, A. I. M. S., Tikhomirova, A., & Roujeinikova, A. (2016). Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). International Journal of Molecular Sciences , 17(7), 1018. https://doi.org/10.3390/ijms17071018
- Varner, E. L., Trefely, S., Bartee, D., Von Krusenstiern, E., Izzo, L., Bekeova, C., O'connor, R. S., Seifert, E. L., Wellen, K. E., Meier, J. L., & Snyder, N. W. (2020). Quantification of lactoyl-CoA (lactyl-CoA) by liquid chromatography mass spectrometry in mammalian cells and tissues. Open Biology , 10(9), 200187. https://doi.org/10.1098/rsob.200187
- Verdin, E., & Ott, M. (2015). 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nature Reviews Molecular Cell Biology , 16(4), 258–264. https://doi.org/10.1038/nrm3931
- Vetting, M. W., LP, S. d. C., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L., & Blanchard, J. S. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Archives of Biochemistry and Biophysics , 433(1), 212–226. https://doi.org/10.1016/j.abb.2004.09.003
- Violante, S., Ijlst, L., Ruiter, J., Koster, J., van Lenthe, H., Duran, M., de Almeida, I. T., Wanders, R. J. A., Houten, S. M., & Ventura, F. V. (2013). Substrate specificity of human carnitine acetyltransferase: Implications for fatty acid and branched-chain amino acid metabolism. Biochimica Et Biophysica Acta-Molecular Basis of Disease , 1832(6), 773–779. https://doi.org/10.1016/j.bbadis.2013.02.012
- Voss, A. K., & Thomas, T. (2009). MYST family histone acetyltransferases take center stage in stem cells and development. BioEssays , 31(10), 1050–1061. https://doi.org/10.1002/bies.200900051
- Wagner, G. R., Bhatt, D. P., O'Connell, T. M., Thompson, J. W., Dubois, L. G., Backos, D. S., Yang, H., Mitchell, G. A., Ilkayeva, O. R., Stevens, R. D., Grimsrud, P. A., & Hirschey, M. D. (2017). A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metabolism , 25(4), 823–+. https://doi.org/10.1016/j.cmet.2017.03.006
- Wang, D., Yan, F., Wu, P., Ge, K., Li, M., Li, T., Gao, Y., Peng, C., … Chen, Y. (2022). Global profiling of regulatory elements in the histone benzoylation pathway. Nature Communications , 13(1), 1369. https://doi.org/10.1038/s41467-022-29057-2
- Wang, M., & Lin, H. N. (2021). Understanding the Function of Mammalian Sirtuins and Protein Lysine Acylation. Annual Review of Biochemistry , 90, 245–285. https://doi.org/10.1146/annurev-biochem-082520-125411
- Wang, Z. A., Whedon, S. D., Wu, M., Wang, S., Brown, E. A., Anmangandla, A., Regan, L., Lee, K., Du, J., Hong, J. Y., Fairall, L., Kay, T., Lin, H., Zhao, Y., Schwabe, J. W. R., & Cole, P. A. (2022). Histone H2B deacylation selectivity: Exploring Chromatin's dark matter with an engineered sortase. Journal of the American Chemical Society , 144(8), 3360–3364. https://doi.org/10.1021/jacs.1c13555
- Wei, W., Liu, X., Chen, J., Gao, S., Lu, L., Zhang, H., Ding, G., Wang, Z., Chen, Z., Shi, T., Li, J., Yu, J., & Wong, J. (2017). Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Research , 27(7), 898–915. https://doi.org/10.1038/cr.2017.68
- Xiao, H., Xuan, W., Shao, S., Liu, T., & Schultz, P. G. (2015). Genetic incorporation of epsilon-N-2-hydroxyisobutyryl-lysine into recombinant histones. Acs Chemical Biology , 10(7), 1599–1603. https://doi.org/10.1021/cb501055h
- Xie, Z. Y., Dai, J. B. A., Dai, L. Z., Tan, M. J., Cheng, Z. Y., Wu, Y. M., Boeke, J. D., & Zhao, Y. M. (2012). Lysine succinylation and lysine malonylation in histones. Molecular & Cellular Proteomics, 11(5), 100–107. https://doi.org/10.1074/mcp.M111.015875
- Xie, Z., Zhang, D., Chung, D., Tang, Z., Huang, H., Dai, L., Qi, S., Li, J., Colak, G., Chen, Y., Xia, C., Peng, C., Ruan, H., Kirkey, M., Wang, D., Jensen, L. M., Kwon, O. K., Lee, S., Pletcher, S. D., … Zhao, Y. (2016). Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation. Molecular Cell , 62(2), 194–206. https://doi.org/10.1016/j.molcel.2016.03.036
- Xiong, J., He, J., Zhu, J., Pan, J., Liao, W., Ye, H., Wang, H., Song, Y., Du, Y., Cui, B., Xue, M., Zheng, W., Kong, X., Jiang, K., Ding, K., Lai, L., & Wang, Q. (2022). Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Molecular Cell , 82(9), 1660–1677.e1610. https://doi.org/10.1016/j.molcel.2022.02.033
- Xiong, X., Panchenko, T., Yang, S., Zhao, S., Yan, P., Zhang, W., Xie, W., Li, Y., Zhao, Y., Allis, C. D., & Li, H. (2016). Selective recognition of Histone crotonylation by double PHD fingers of MOZ and DPF2. Nature Chemical Biology , 12(12), 1111–+. https://doi.org/10.1038/Nchembio.2218
- Xu, G., Wang, J., Wu, Z., Qian, L., Dai, L., Wan, X., Tan, M., Zhao, Y., & Wu, Y. (2014). SAHA regulates histone acetylation, butyrylation, and protein expression in neuroblastoma. Journal of Proteome Research , 13(10), 4211–4219. https://doi.org/10.1021/pr500497e
- Yang, C., Mi, J. Q., Feng, Y., Ngo, L., Gao, T. L., Yan, L. L., & Zheng, Y. G. (2013). Labeling lysine acetyltransferase substrates with engineered enzymes and functionalized cofactor surrogates. Journal of the American Chemical Society , 135(21), 7791–7794. https://doi.org/10.1021/ja311636b
- Yang, D. W., Yin, J., Shan, L. Q., Yi, X. L., Zhang, W., & Ding, Y. B. (2022). Identification of lysine-lactylated substrates in gastric cancer cells. iScience , 25(7), 104630. https://doi.org/10.1016/j.isci.2022.104630
- Yang, H., Wang, H., & Andersson, U. (2020). Targeting inflammation driven by HMGB1. Frontiers in Immunology , 11, 484. https://doi.org/10.3389/fimmu.2020.00484
- Yang, K., Fan, M., Wang, X., Xu, J., Wang, Y., Tu, F., Gill, P. S., Ha, T., Liu, L., Williams, D. L., & Li, C. (2022). Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death and Differentiation , 29(1), 133–146. https://doi.org/10.1038/s41418-021-00841-9
- Yang, Y. Y., Ascano, J. M., & Hang, H. C. (2010). Bioorthogonal chemical reporters for monitoring protein acetylation. Journal of the American Chemical Society , 132(11), 3640–3641. https://doi.org/10.1021/ja908871t
- Yang, Z., He, M. Z., Austin, J., Pfleger, J., & Abdellatif, M. (2021). Histone H3K9 butyrylation is regulated by dietary fat and stress via an Acyl-CoA dehydrogenase short chain-dependent mechanism. Molecular Metabolism , 53, 101249. https://doi.org/10.1016/j.molmet.2021.101249
- Yao, Y., Bade, R., Li, G. T., Zhang, A. Q., Zhao, H. L., Fan, L. F., Zhu, R., & Yuan, J. (2022). Global-scale profiling of differential expressed lysine-lactylated proteins in the cerebral endothelium of cerebral ischemia-reperfusion injury rats. Cellular and Molecular Neurobiology , https://doi.org/10.1007/s10571-022-01277-6
- Yi, F., Zhang, Y., Wang, Z., Wang, Z., Li, Z., Zhou, T., Xu, H., Liu, J., Jiang, B., Li, X., Wang, L., Bai, N., Guo, Q., Guan, Y., Feng, Y., Mao, Z., Fan, G., Zhang, S., Wang, C., … Cao, L. (2021). The deacetylation-phosphorylation regulation of SIRT2-SMC1A axis as a mechanism of antimitotic catastrophe in early tumorigenesis. Science Advances , 7(9), eabe5518. https://doi.org/10.1126/sciadv.abe5518
- Yilmaz, S., Unal, F., & Yuzbasioglu, D. (2009). The in vitro genotoxicity of benzoic acid in human peripheral blood lymphocytes. Cytotechnology , 60(1-3), 55. https://doi.org/10.1007/s10616-009-9214-z
- Yu, J., Chai, P., Xie, M., Ge, S., Ruan, J., Fan, X., & Jia, R. (2021). Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome biology , 22(1), 85. https://doi.org/10.1186/s13059-021-02308-z
- Yuliwulandari, R., Susilowati, R. W., Wicaksono, B. D., Viyati, K., Prayuni, K., Razari, I., Kristin, E., Syafrizal, Subagyo, Sri Diana, E., Setiawati, S., Ariyani, A., Mahasirimongkol, S., Yanai, H., Mushiroda, T., & Tokunaga, K. (2016). NAT2 variants are associated with drug-induced liver injury caused by anti-tuberculosis drugs in Indonesian patients with tuberculosis. Journal of Human Genetics , 61(6), 533–537. https://doi.org/10.1038/jhg.2016.10
- Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y., Liu, W., Kim, S., Lee, S., Perez-Neut, M., Ding, J., Czyz, D., Hu, R., Ye, Z., He, M., Zheng, Y. G., Shuman, H. A., Dai, L., Ren, B., … Zhao, Y. (2019). Metabolic regulation of gene expression by histone lactylation. Nature , 574(7779), 575–580. https://doi.org/10.1038/s41586-019-1678-1
- Zhang, K., Chen, Y., Mang, Z. H., & Zhao, Y. M. (2009). Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. Journal of Proteome Research , 8(2), 900–906. https://doi.org/10.1021/pr8005155
- Zhang, Q., Zeng, L., Zhao, C. C., Ju, Y., Konuma, T., & Zhou, M. M. (2016). Structural insights into histone crotonyl-lysine recognition by the AF9 YEATS domain. Structure (London, England) , 24(9), 1606–1612. https://doi.org/10.1016/j.str.2016.05.023
- Zhao, D., Guan, H., Zhao, S., Mi, W., Wen, H., Li, Y., Zhao, Y., Allis, C. D., Shi, X., & Li, H. (2016). YEATS2 is a selective histone crotonylation reader. Cell Research , 26(5), 629–632. https://doi.org/10.1038/cr.2016.49
- Zhao, D., Li, Y. Y., Xiong, X. Z., Chen, Z. L., & Li, H. T. (2017). YEATS domain-a histone acylation reader in health and disease. Journal of Molecular Biology , 429(13), 1994–2002. https://doi.org/10.1016/j.jmb.2017.03.010
- Zhao, K., Chai, X., & Marmorstein, R. (2004). Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. Journal of Molecular Biology , 337(3), 731–741. https://doi.org/10.1016/j.jmb.2004.01.060
- Zhou, T. T., Xu, H. W., Cheng, X., He, Y. Q., Ren, Q., Li, D. Z., Xie, Y., Gao, C., Zhang, Y., Sun, X., Xu, Y., & Huang, W. (2022). Sodium butyrate attenuates diabetic kidney disease partially via histone butyrylation modification. Mediators of Inflammation , 2022, 7643322. https://doi.org/10.1155/2022/7643322
- Zhu, Z., Han, Z., Halabelian, L., Yang, X., Ding, J., Zhang, N., Ngo, L., Song, J., Zeng, H., He, M., Zhao, Y., Arrowsmith, C. H., Luo, M., Bartlett, M. G., & Zheng, Y. G. (2021). Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Research , 49(1), 177–189. https://doi.org/10.1093/nar/gkaa1176