DIAMOND+MEGAN: Fast and Easy Taxonomic and Functional Analysis of Short and Long Microbiome Sequences
Caner Bağcı, Caner Bağcı, Sascha Patz, Sascha Patz, Daniel H. Huson, Daniel H. Huson
functional binning
metagenome assembled genomes
microbiome sequencing
protein alignment
software
taxonomic binning
Abstract
One main approach to computational analysis of microbiome sequences is to first align against a reference database of annotated protein sequences (NCBI-nr) and then perform taxonomic and functional binning of the sequences based on the resulting alignments. For both short and long reads (or assembled contigs), alignment is performed using DIAMOND, whereas taxonomic and functional binning, followed by inter- active exploration and analysis, is performed using MEGAN. We provide two step-by-step descriptions of this approach: © 2021 The Authors.
This article was corrected on 20 July 2022. See the end of the full text for details.
Basic Protocol 1 : Taxonomic and functional analysis of short read microbiome sequences
Support Protocol 1 : Preprocessing
Basic Protocol 2 : taxonomic and functional analysis of assembled long read microbiome sequences
Support Protocol 2 : Taxonomic binning and CheckM
INTRODUCTION
One main approach to taxonomic and functional binning of microbiome shotgun sequences is based on protein homology (Glass, Wilkening, Wilke, Antonopoulos, & Meyer, 2010; Huson, Auch, Qi, & Schuster, 2007). In this approach, the sequences are first aligned against a reference database of protein sequences of known taxonomic and functional identity, and then the resulting alignments are used to assign the sequences to taxonomic and functional bins.
Why align against protein sequences? While analysis of microbiome sequences using DNA alignment against genomic references is feasible, there are a number of issues with this approach. First, currently, genomic reference databases cover only a small fraction of the diversity present in the environment (Wu et al., 2009). Second, the high level of redundancy of genomic sequences causes performance issues when query sequences display very large numbers of equally good alignments. Translated alignment ameliorates these issues to a degree because protein sequences are much more conserved than genomic sequences. Finally, a proper biological understanding of processes within a given microbiome requires detailed knowledge of the proteins present and their alignments to reference sequences of known function (Willmann et al., 2015).
The core computation of the approach presented here is the translated alignment of microbiome sequences against the NCBI-nr database (Benson, Karsch-Mizrachi, Lipman, Ostell, & Wheeler, 2005). In early microbiome studies (Huson et al., 2007; Poinar et al., 2006; Venter et al., 2004), BLASTX (Altschul et al., 1997) was used to align small numbers of reads (on the order of hundreds of thousands) against a small database (in 2007, NCBI-nr contained approximately 2 million sequences). For subsequent studies involving hundreds of millions of reads (Mackelprang et al., 2011; Qin et al., 2010), BLASTX was run at super-computer centers. Our lab developed DIAMOND (Buchfink, Xie, & Huson, 2015b) to replace BLASTX in such analyses, providing a 20,000-fold speedup over BLASTX on short sequencing reads, while maintaining sufficient sensitivity. DIAMOND is used as the main alignment engine in a number of analysis pipelines (Franzosa et al., 2018; Huson et al., 2016; Zhu et al., 2017).
Analysis of short read microbiome samples usually involves determining the highest scoring alignments of each read to a set of reference sequences, followed by assignment to taxonomic and functional bins, using heuristics such as the naïve LCA (lowest common ancestor) approach for taxonomic binning (Huson et al., 2007) and the best hit approach for functional assignment (Huson, Mitra, Weber, Ruscheweyh, & Schuster, 2011). Long reads or assembled contigs require modified algorithms during alignment and binning, and both DIAMOND and MEGAN provide long read modes to operate on long (erroneous) sequences (Arumugam et al., 2019; Huson et al., 2018). Microbiome studies can be performed using either short read technology (Bentley, 2006), see e.g., (Willmann et al., 2015) or long read sequencing technology (Jain, Olsen, Paten, & Akeson, 2016; Rhoads & Au, 2015), see e.g., (Arumugam et al., 2019).
While short read sequences have lengths measured in hundreds of bases, long read technologies produce reads that are tens of kilobases (kb) in length, on average. Hence, one might assume that assembly is more useful for short reads than for long reads. Paradoxically, for microbiome sequences, this is not the case. For sequencing reads obtained from a mixed community of organisms, assembly of short reads usually leads to disappointingly low average scaffold lengths (a couple of kb; Boisvert, Raymond, Godzaridis, Laviolette, & Corbeil, 2012), whereas the assembly of long reads can result in complete closed circular chromosomes (Arumugam et al., 2019). Thus, in this protocol, we follow two different strategies for taxonomic and functional classification of reads, one for short reads and one for long reads. For short reads, we follow a naïve read binning approach, whereas for long reads we first carry out an assembly and correction procedure and then perform the taxonomic and functional classification on the assembled contigs.
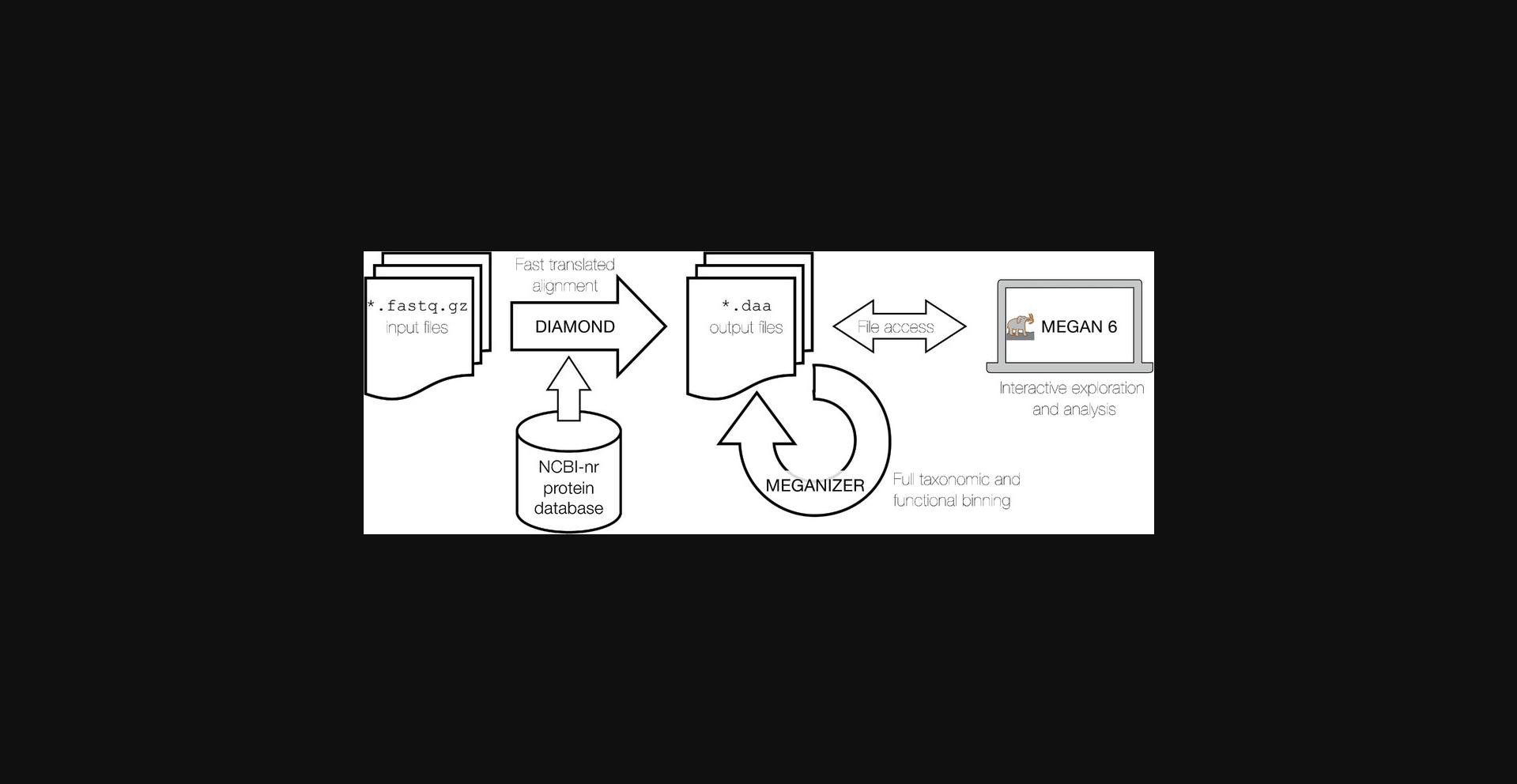
The core “DIAMOND+MEGAN” analysis of microbiome shotgun sequencing datasets (both short reads and assembled long reads) consists of three subsequent steps, namely:
- 1.Alignment of all reads against a protein reference database using DIAMOND
- 2.Taxonomic and functional analysis of the resulting alignments using a program called MEGANIZER (part of the MEGAN package), or MEGAN
- 3.Interactive exploration and analysis using MEGAN
Both DIAMOND and MEGANIZER can be run either in short read mode (by default) or long read mode, so as to accommodate the two different types of input. In addition, in a preprocessing step, both types of sequences can be subjected to quality control and, especially prudent in the case of long reads, sequence assembly.
We provide two protocols. In the first protocol, we will discuss how to apply the core DIAMOND+MEGAN microbiome analysis pipeline in the context of a typical short read project. We will illustrate this protocol using a small, published dataset (Hu, Pang, Huang, Zhang, & Zhang, 2018) of 11 gut microbiome samples. In Support Protocol 1, we will discuss optional preprocessing. A general workflow of this process can be seen in Figure 1.
In the second protocol, we will discuss application of the core pipeline to long read data. Here, assembly of the reads in an initial preprocessing step is highly recommended, and is illustrated using the Unicycler pipeline. We will illustrate the necessary steps using a published dataset (Arumugam et al., 2019) collected from an enrichment bioreactor seeded with waste-water treatment sludge.
STRATEGIC PLANNING
Necessary hardware
A typical microbiome shotgun sequencing project will involve multiple samples, each represented by millions of sequencing reads (Willmann et al., 2015). The initial computational analysis of such datasets should be performed on a server with a good number of cores, ∼24 or more, at least 64 GB of main memory, and sufficient disk space (multiple TB).
Once all samples have been processed, the resulting files can be downloaded onto a desktop or laptop computer for interactive exploration and analysis. This computer should be a recent model with a fast processor, at least 16 GB of memory and a large SSD disk.
Installing the software
For short read analysis, two programs are required. On a server, install DIAMOND and MEGAN.
- DIAMOND can be obtained from http://www.diamondsearch.org. DIAMOND is provided as a single binary file, and installation on a computer running Linux can be performed by simply downloading and unpacking the program. DIAMOND can also be installed in conda, using the command conda install -c bioconda diamond.
- Installers for MEGAN can be obtained from http://megan.husonlab.org. There are two different editions of MEGAN. The Community Edition (CE; Huson et al., 2016) is open-source and free to use. The Ultimate Edition (UE) contains additional functionality, including an up-to-date version of KEGG (Kanehisa & Goto, 2000), and is licensed through Computomics GmbH. For either edition, three different installers are provided, targeting Linux, MacOS, and Windows. The program is installed by downloading the appropriate installer and then launching it. By default, the installer presents a graphical user interface. For installation on a server that does not support this, use the command-line option -c to launch the installer in a non-graphical console mode. During installation, you will be asked to specify the amount of memory that MEGAN can use. We recommend specifying at least 16 GB, but more is better and the program will run faster with more memory.
- ∘ MEGAN should be installed both on the server used for the main computational processing and on the desktop or laptop used for interactive exploration and analysis.
- For long read processing, we require installation of the long read assembly tool Unicycler (Wick, Judd, Gorrie, & Holt, 2017), which uses miniasm (Li, 2016) and Racon (Vaser, Sović, Nagarajan, & Šikić, 2017), on a server. The program can be installed locally on a server using the commands:
git clone https://github.com/rrwick/Unicycler.git cd Unicycler make
∘ The program is then launched by typing: unicycler
∘ Alternatively, Unicycler can also be installed in conda, using the command:
conda install -c bioconda unicycler
- In addition, we will use the long read assembly correction tool medaka (ONT, 2020), which can be installed in conda using the command:
conda install -c bioconda medaka
Basic Protocol 1: ANALYSIS OF SHORT READ SHOTGUN SEQUENCES
Microbiome projects employing second-generation, short read sequencing technologies, such as provided by Illumina or Ion-Torrent, typically involve tens or hundreds of sequencing datasets, each containing millions of reads. Here we discuss how to apply the core analysis pipeline to such data.
We will illustrate the necessary steps using a published dataset (Hu et al., 2018; SRA accession: PRJNA490628) collected from gastric wash samples isolated from six patients with advanced gastric adenocarcinoma (GC) and from five patients with superficial gastritis (SG). The data can be obtained by following the Support Protocol 1, step 1, “Data acquisition.” Every sample is represented by two files of reads (forward and reverse reads), and thus the total dataset consists of 22 files of Illumina paired-end reads, each containing 16 million reads of length 150 bp, on average. In this protocol, we apply the preprocessing steps described in Support Protocol 1 to the example dataset. Whether preprocessing is necessary depends on the data quality, whether the data is host associated, and on the availability of sufficient computational resources.
Materials
Hardware
A server (typically Linux) with a good number of cores, 24 or more, with at least 64 GB of main memory and sufficient disk space (multiple TB)
Software
- DIAMOND (http://www.diamondsearch.org)
- MEGAN (http://megan.husonlab.org)
- Unicycler (https://github.com/rrwick/Unicycler#installation)
- Medaka (https://github.com/nanoporetech/medaka)
NOTE : DIAMOND is designed to align a large set of short read sequences against a large protein reference database (Buchfink et al., 2015b). This typically involves aligning hundreds of millions of reads against the NCBI-nr database (Benson et al., 2005) of non-redundant protein reference sequences, which currently contains approximately 180 million entries. The program is typically run on a Linux server, from the command line.
1.Build a DIAMOND index.
In preparation of using DIAMOND to align sequences, you must first build a DIAMOND index.
From the command line, download the latest NCBI-nr database as follows:
Then, run DIAMOND to build the DIAMOND index, as follows:
- diamond makedb --in nr.gz --db nr
This will create a file called nr.dmnd that contains the DIAMOND index.
2.Align short reads.
When running DIAMOND to align sequences, we have to provide a number of command-line parameters:
1.First, specify the command blastx, which requests alignment of translated reads against the reference database. 2.Use -d nr to determine the index file to use. This refers to the index file nr.dmnd, but must be specified without the file suffix. 3.Use -q ( input file.) to specify the input file. The file must be in FastA or FastQ format, and can be compressed (suffix .gz). 4.Use -o ( output-file ) to specify the output file. Use .daa as file suffix. 5.Use -f 100 to specify that the output be written in DAA (DIAMOND alignment archive) format, which is required for processing with MEGANIZER and MEGAN.
To run DIAMOND on an input file in compressed FastQ format, for example on the first sample of the example dataset after preprocessing, type the following:
- diamond blastx -d nr -q SRR7828855_merged.fastq -o SRR7828855_merged.daa -f 100
Again, use paths to programs, and to files that are not in your current directory.
3.Meganization.
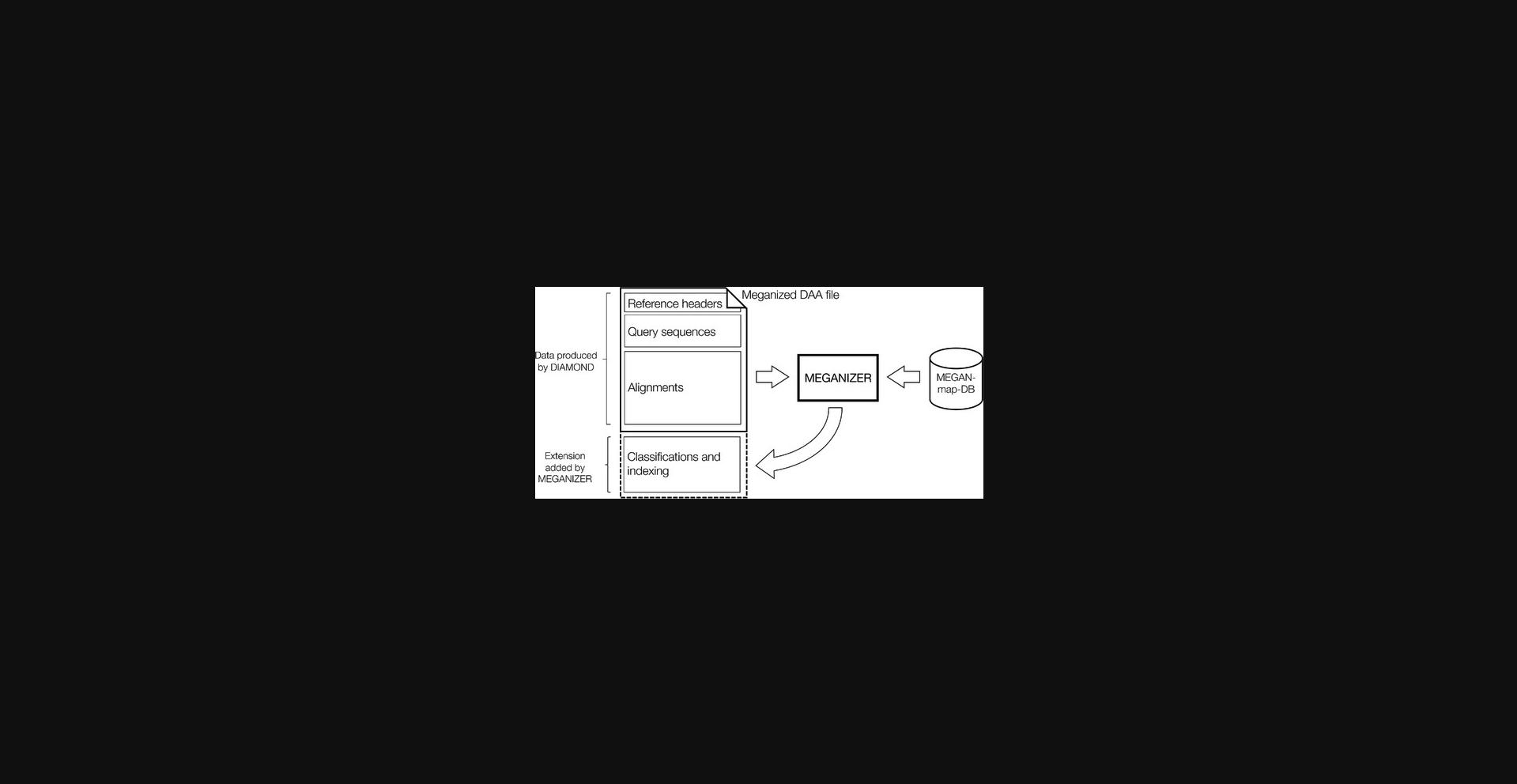
A DAA file computed by DIAMOND contains aligned sequences and their alignments. This data can be used to perform taxonomic and functional binning of the sequences.
The term meganization refers to the process of first analyzing all sequences and alignments in a DAA file, so as to perform taxonomic and functional binning or classification of the sequences, and then placing the result of this analysis in an additional block at the end of the DAA file (see Fig. 2). For short reads, taxonomic binning is performed using the naïve LCA algorithm (Huson et al., 2007), whereas functional binning is performed using the best-hit approach (Huson et al., 2011).
MEGAN supports a number of different classifications. Taxonomic classification is performed using the NCBI taxonomy and the GTDB taxonomy (Parks et al., 2020). Functional classification is currently performed using EC (Barrett, 1992), eggNOG (Powell et al., 2012), InterPro (Mitchell et al., 2015), or SEED (Overbeek et al., 2013). Functional classification using KEGG (Kanehisa & Goto, 2000) is available in the Ultimate Edition of MEGAN. Other classifications will be added in future releases. The mappings of reference protein accessions to classes in the supported classifications are provided in an SQLite database.
To prepare for meganization, download and unzip the database file from: https://software-ab.informatik.uni-tuebingen.de/download/megan6. The current version is megan-map-Jan2021.db.
A DAA file containing the alignments computed by DIAMOND in the previous step can be meganized either using a command-line program or through the graphical user interface of MEGAN. The command-line MEGANIZER program daa-meganizer is installed with MEGAN and is found in the megan/tools sub-directory of the MEGAN installation directory.
To run MEGANIZER on the input file in DAA format called SRR7828855_merged.daa, from the previous step, execute the following:
- daa-meganizer -i SRR78288555_merged.daa -mdb megan-map-Jan2021.db
Again, use paths to programs, and to files that are not in your current directory.
The MEGANIZER program offers a number of additional options, which can be listed by running:
- daa-meganizer -h
The daa-meganizer program can be run individually on each file in the dataset, similar to DIAMOND, or can also be run on a list of input files that are separated by spaces.
As an example of an additional feature, consider the task of contamination filtering. For human-associated microbiome samples, one might consider all reads with significant alignments to proteins from Metazoa (animals) as contamination. Or, as another example, in low biomass samples, laboratory contaminants might make up a significant fraction in the sample, and these can be identified using a tool like decontam (Davis, Proctor, Holmes, Relman, & Callahan, 2018).
For example, to perform meganization with contaminant filtering for all animals, first create a text file contaminants.txt that contains a list of NCBI taxon names or numerical ids, then pass this to the MEGANIZER program using the option -cf, as shown here:
echo "Metazoa" > contaminants.txt
daa-meganizer -i input.daa -mdb megan-map-Jan2021.db -cf contaminants.txt
With this feature turned on, in the case of short reads, any read that has a significant alignment to any contaminant taxon (recursively including all descendants) will be assigned to the Contaminants node in every classification. Contaminants can also be specified later, during an update of the analysis, as described below.
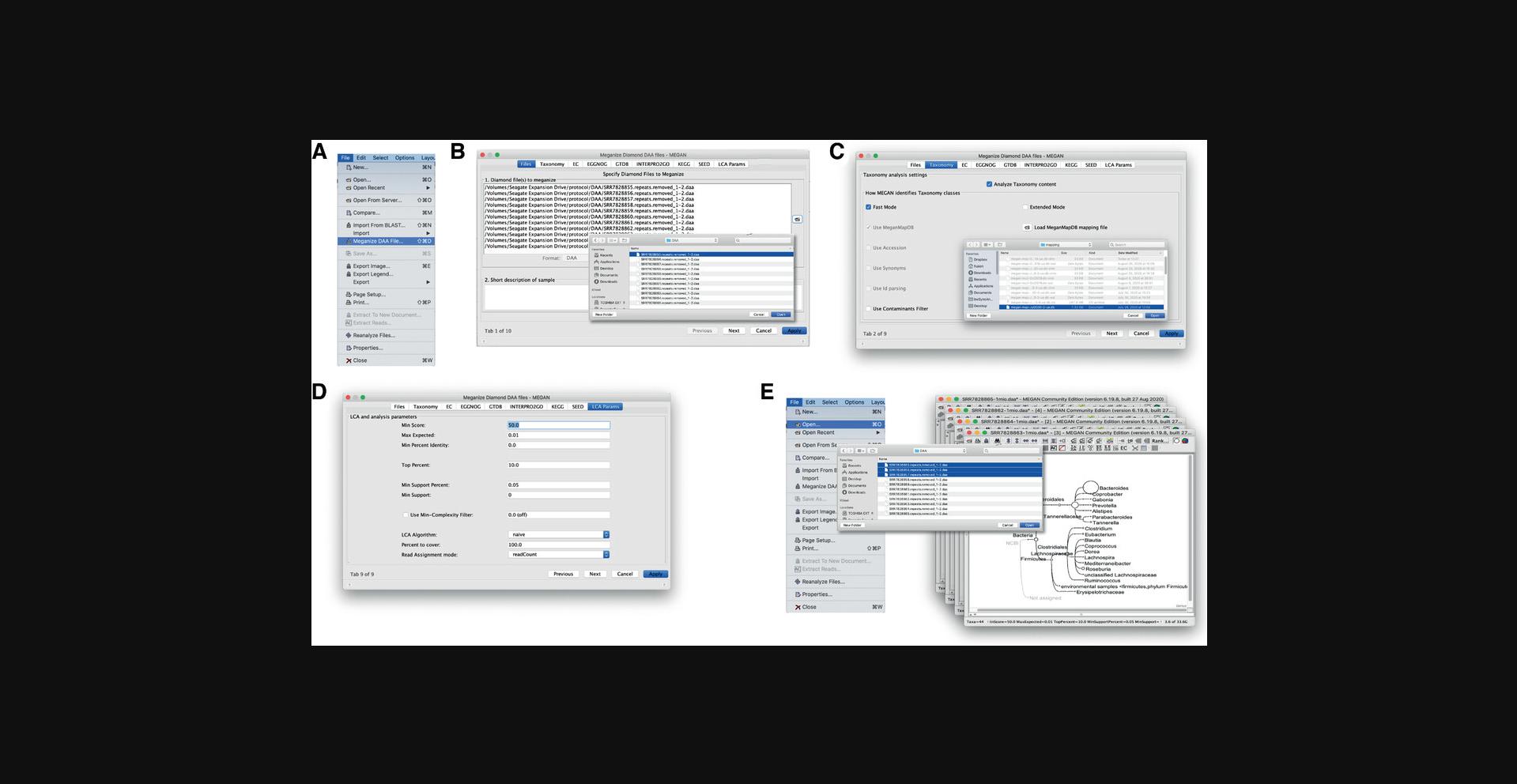
DAA files produced by DIAMOND can also be meganized using the graphical user interface (GUI) of the MEGAN program. To initiate this, launch MEGAN in GUI mode.
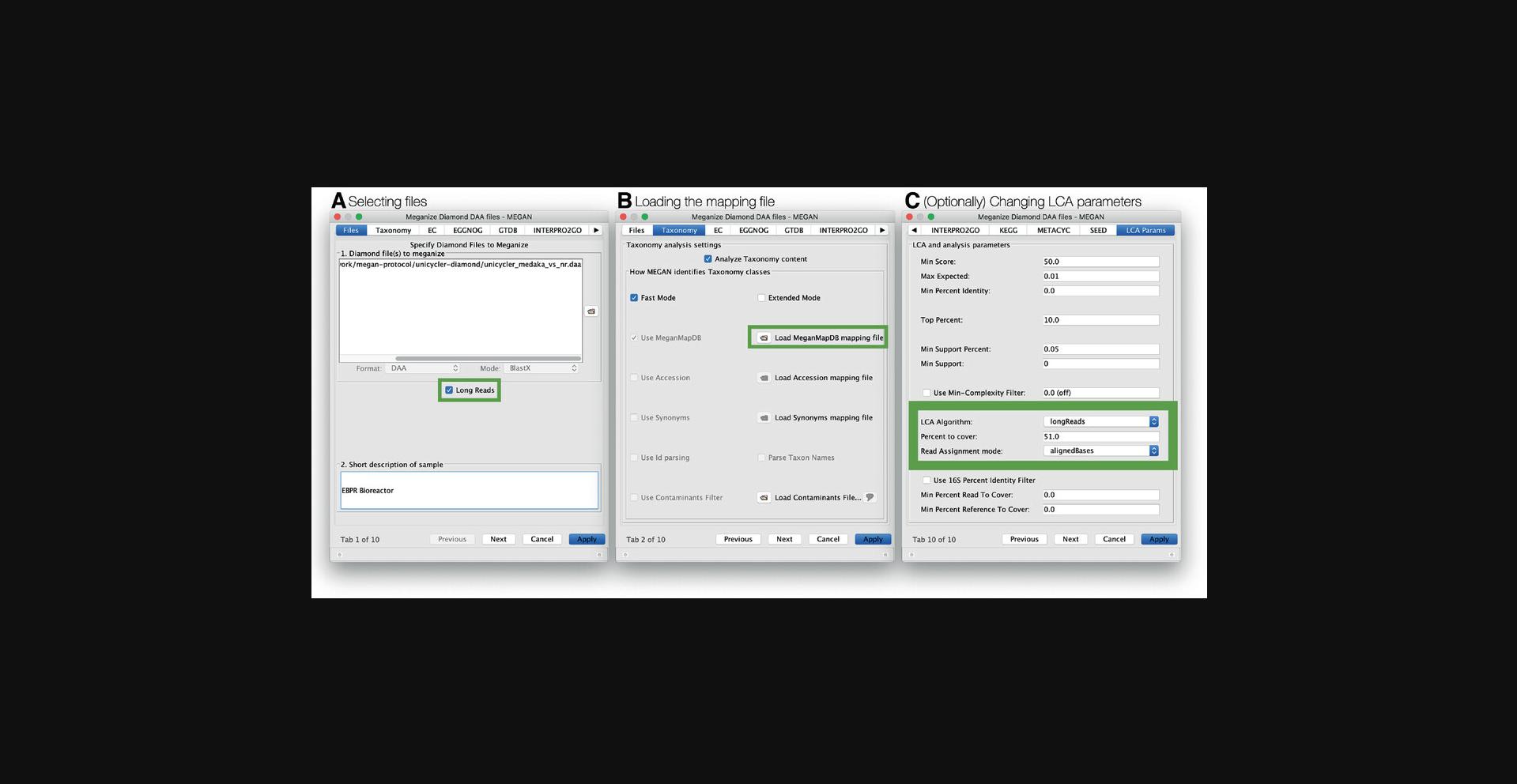
Once MEGAN is running, select the File → Meganize DAA File… menu item to open the meganization dialog (Fig. 3A). This dialog has multiple tabs. On the first tab, Files, enter all the DAA files that you want to meganize (Fig. 3B). These will be processed one-by-one. On the second tab, Taxonomy, use the button Load MeganMapDB mapping file to select the mapping file megan-map-Jan2021.db (Fig. 3C). By default, all classifications supported by the selected mapping file will be activated. Thus, there will generally be no need to explicitly turn on any of the classifications. The last tab, LCA Params, can be used to adjust the classification parameters (Fig. 3D).
To run meganization on all selected files, press the Apply button. When the computation is completed, the meganized DAA files can be opened in MEGAN using the File → Open… menu item (Fig. 3E).
Meganization does not change the initial content of a DAA file and a DAA file can be re-meganized any number of times. Meganization can take several hours on larger datasets, and so for large datasets, we recommend running the daa-meganizer command-line program on a server.
4.Open and update DAA files in MEGAN.
Usually, MEGAN is run interactively on a desktop or laptop. If your files were meganized on a server, then you must first copy them onto your local computer.
At startup, MEGAN displays a main taxonomy viewer and a message window, on which all commands are echoed and errors are reported. The File → Open… menu item can be used to open one or more meganized DAA files (Fig. 3E).
Once a file has been opened, the main taxonomy viewer displays the taxonomic classification of the sequences based on the NCBI taxonomy. Viewers for the other supported classifications can be opened as discussed below. The status bar at the bottom of the taxonomy viewer provides some information on the number of taxa displayed, the number of aligned and assigned reads, and the parameters used during meganization.
Note that the file chooser only allows selection of DAA files that have file extension .daa and that have been successfully meganized.
To re-meganize a DAA file, for example, to use a more recent mapping database, you can do so using the MEGANIZER program or MEGAN's meganization dialog, as described above.
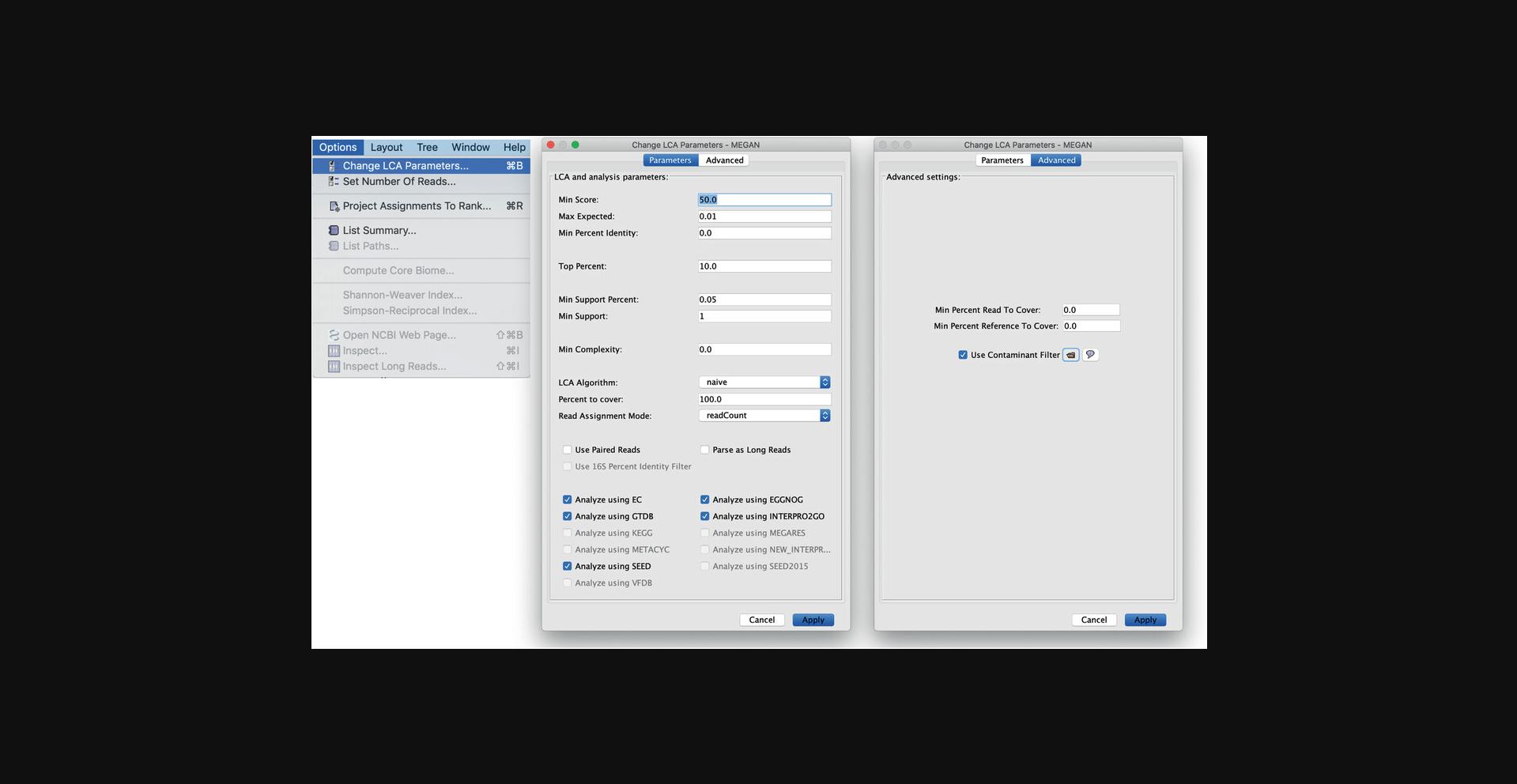
The parameters used for the initial taxonomic and functional analysis of a DAA file can be interactively modified, and then the analysis can be rerun using the Change LCA Parameters dialog, which is opened using the Options → Change LCA Parameters… menu item (see Fig. 4).
In more detail, the alignments considered during taxonomic and functional analysis can be filtered by minimal bit score (Min score), e-value (Max Expected), or minimal percentage identity (Min Percent Identity). In addition, the Top Percent parameter (default: 10%) filters all alignments whose bitscore does not lie within the specified percentage of the best score seen for a given read.
Two alternative parameters, Minimum Support Percent and. Min Support, determine the percent or number of assigned reads, respectively, that must be assigned to a taxon (and its descendants) so that the taxon appears in the taxonomic tree. For any taxon that does not meet this criterion, the read counts are passed up the tree (toward the root) until a taxon is reached that has a sufficiently high read count.
The LCA Algorithm parameter can be used to select between the naïve LCA, the “weighted LCA,” (Buchfink, Huson, & Xie, 2015a), or the “long reads LCA” (Huson et al., 2018). For the latter two algorithms, the Percent to cover parameter is used to set the percent of weight to cover, or percent of coding sequence to cover, respectively.
On the advanced tab of the dialog, one can select a contaminants file and thus rerun the analysis so as to assign contaminant sequences to the “contaminants” node.
In this protocol, we focus on how to meganize DAA files and then work with them in MEGAN. However, the program can also import a wide range of other formats, including alignments in BLAST, xml, tab-delimited, or text files, in SAM format, and classifications in csv text files and BIOM format. As with DAA files, this can be done both either using command-line tools, such as blast2rma or sam2rma, or directly in MEGAN.
5.Taxonomic inspection.
By default, upon loading a meganized DAA file into MEGAN, the program will open the main taxonomy viewer that provides an overview of the assignment of all sequences to taxa or nodes in the NCBI taxonomy (Fig. 5A). By default, each node or taxon is represented by a circle whose area is proportional to the number of reads assigned to the taxon. Selection of a node will display two numbers, labeled Assigned and Summed, reporting the number of reads assigned to the given node, or to the given node and any of its descendants, respectively, as illustrated for Pseudomonas in Figure 5B. Display of these values can also be turned on using the Tree → Show Number of Assigned and Tree → Show Number of Summarized menu items.
The taxonomy viewer displays a subtree of the NCBI taxonomy that contains all taxa to which a read has been assigned. The tree is drawn from left to right, from the root towards the leaves of the taxonomy. To facilitate viewing of the tree at different levels of detail, the user can “collapse” selected nodes so as to have them drawn as a leaf node, suppressing the subtree that lies below the selected node(s), using the Tree Collapse menu item, or expand them using Tree → Uncollapse. There are a number of other related menu items, such as Tree → Uncollapse All or Tree → Uncollapse Subtree, which uncollapses all nodes, or all nodes below any select nodes, respectively.
The user can also collapse all nodes at a given taxonomic rank, such as phylum, class, order, etc., using the Tree → Rank… menu item or corresponding toolbar item (Fig. 5B).
The GTDB taxonomy (Parks et al., 2020) encompasses both bacteria and archaea and is based on marker gene similarity. In addition to taxonomic analysis using the NCBI taxonomy, MEGAN also provides a binning of reads based on the GTDB taxonomy. This alternative taxonomic view can be opened using the Window → Open GTDB Viewer… menu item. The GTDB viewer provides the same functionality as MEGAN's main taxonomic viewer.
6.Taxonomic Charts.
MEGAN provides a number of different charts that can be used to summarize in-sample diversity or between-sample diversity (as discussed further below). To use a chart, first select all nodes of interest in the taxonomy viewer and then open the desired chart using Window → Charts… menu item or Show → Chart tool bar item (Fig. 5D).
7.Functional inspection.
During meganization, functional classification of reads is performed, and reads are assigned to functional classes using a number of different classifications systems, currently using EC (Barrett, 1992), eggNOG (Powell et al., 2012), InterPro (Mitchell et al., 2015), or SEED (Overbeek et al., 2013). Functional classification using KEGG (Kanehisa & Goto, 2000) is available in the Ultimate Edition of MEGAN. Other classifications will be added in future releases. Functional classification is performed using the “best hit” algorithm (Huson et al., 2011), and so reads are usually assigned to the leaves of each functional analysis.
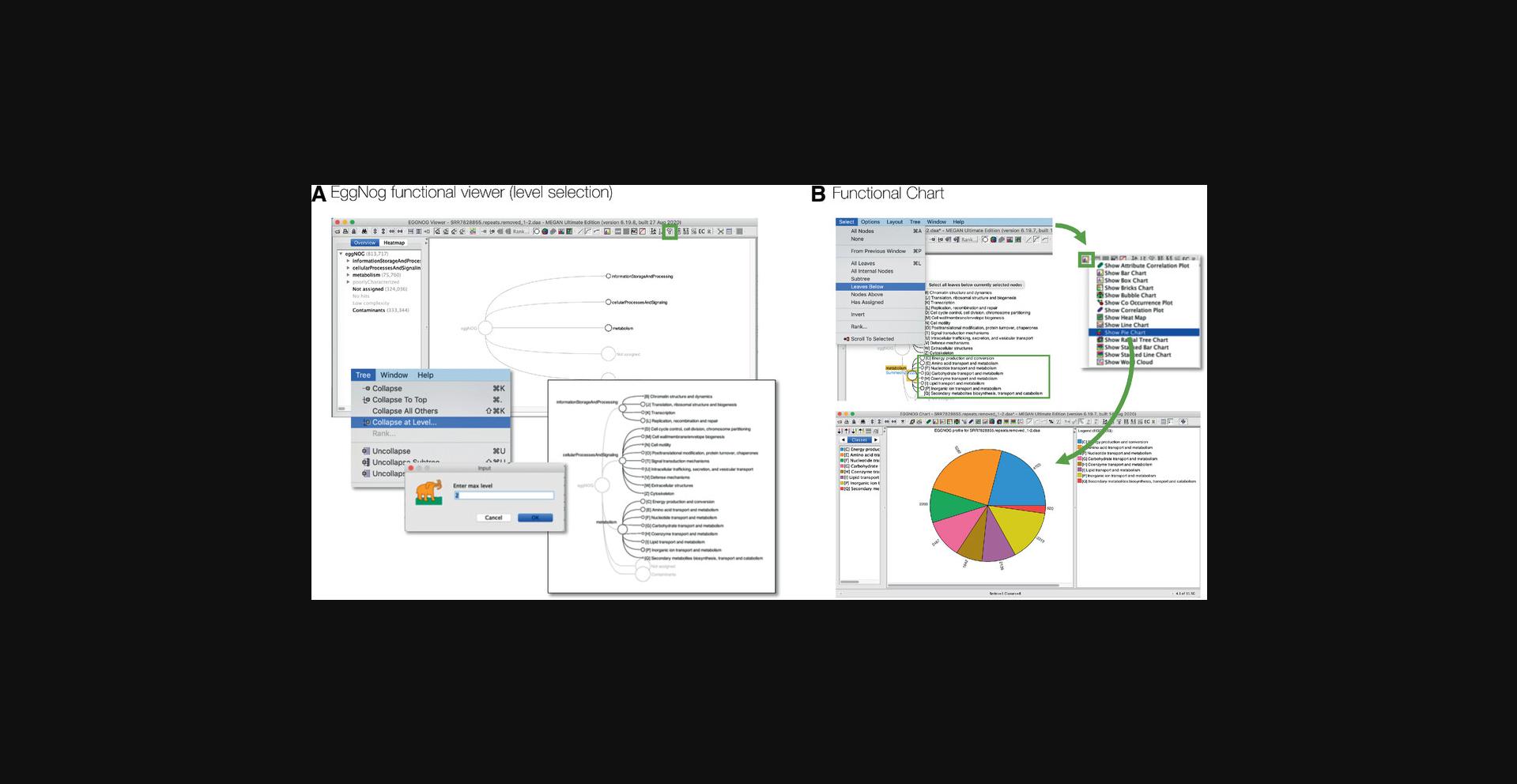
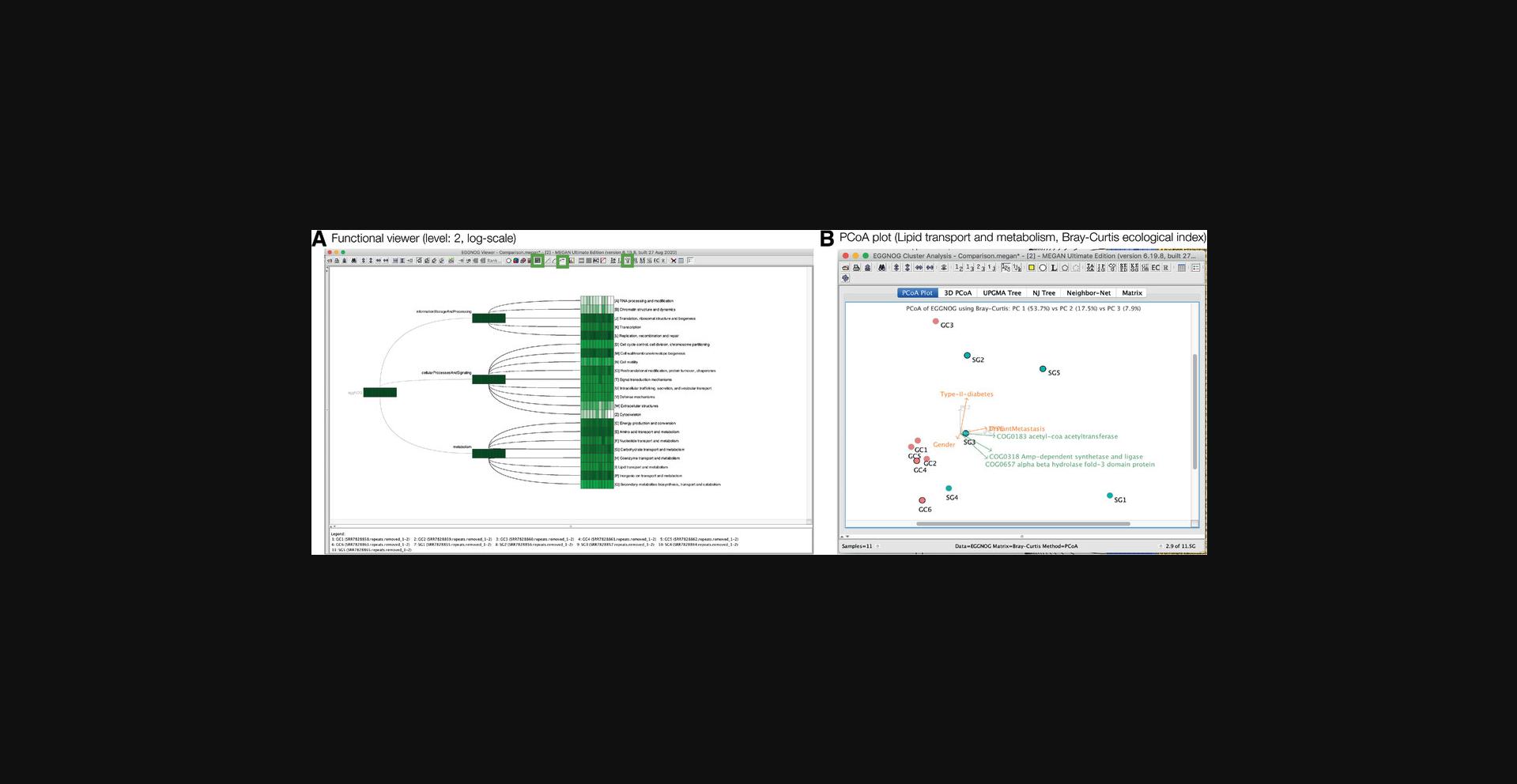
Each classification is represented by a rooted tree in MEGAN, which can be opened in a separate viewer, using the Window Open EGGNOG → Viewer… menu item to open the eggNOG viewer, for example, and then interactively explored in a manner similar to that described above for MEGAN's taxonomic viewers (Fig. 6).
In addition, the functional viewer provides a side bar on the left that contains two tabs, one presenting the full functional classification as a drop-down tree and the other containing a table listing the number of reads assigned to each leaf of the currently displayed tree (Fig. 6A).
Functional nodes can be collapsed and expanded, as discussed above for the taxonomic inspection. However, as functional classifications do not have taxonomic ranks, collapse to a specific rank is not supported. However, the user can use the Tree → Collapse at Level… menu item to collapse a functional classification at a given “level” or distance from the root (Fig. 6A, bottom).
MEGAN's charts can also be used to visualize functional assignments. To use a chart, first select all nodes of interest in the functional viewer and then open the desired chart using the Window → Charts… menu item or Show → Chart tool bar item (Fig. 6B). In the depicted example, all functional nodes at level 2 were selected and then displayed as a pie chart.
8.Read-level analysis (optional).
MEGAN allows the user to drill down to inspect individual reads and their alignments, explore the alignment of reads against a given reference sequence, and perform gene-centric assembly on specific genes or functional classes (Fig. 7).
The read inspector dialog can be used to view the assignment of reads to selected taxa or functional classes, and to inspect the reads and their alignments. To load taxonomic or functional nodes into the viewer, select them and then use choose Options → Inspect… menu item or corresponding node context menu item (Fig. 7A).
When performing a detailed analysis of specific genes in a microbiome sample, it may be desirable to investigate how reads align against specific reference sequences. To allow this, MEGAN provides an alignment viewer that can be opened by selecting the functional node of interest and then choosing the Window → Show Alignment… menu item or corresponding node context menu item (Fig. 7A).
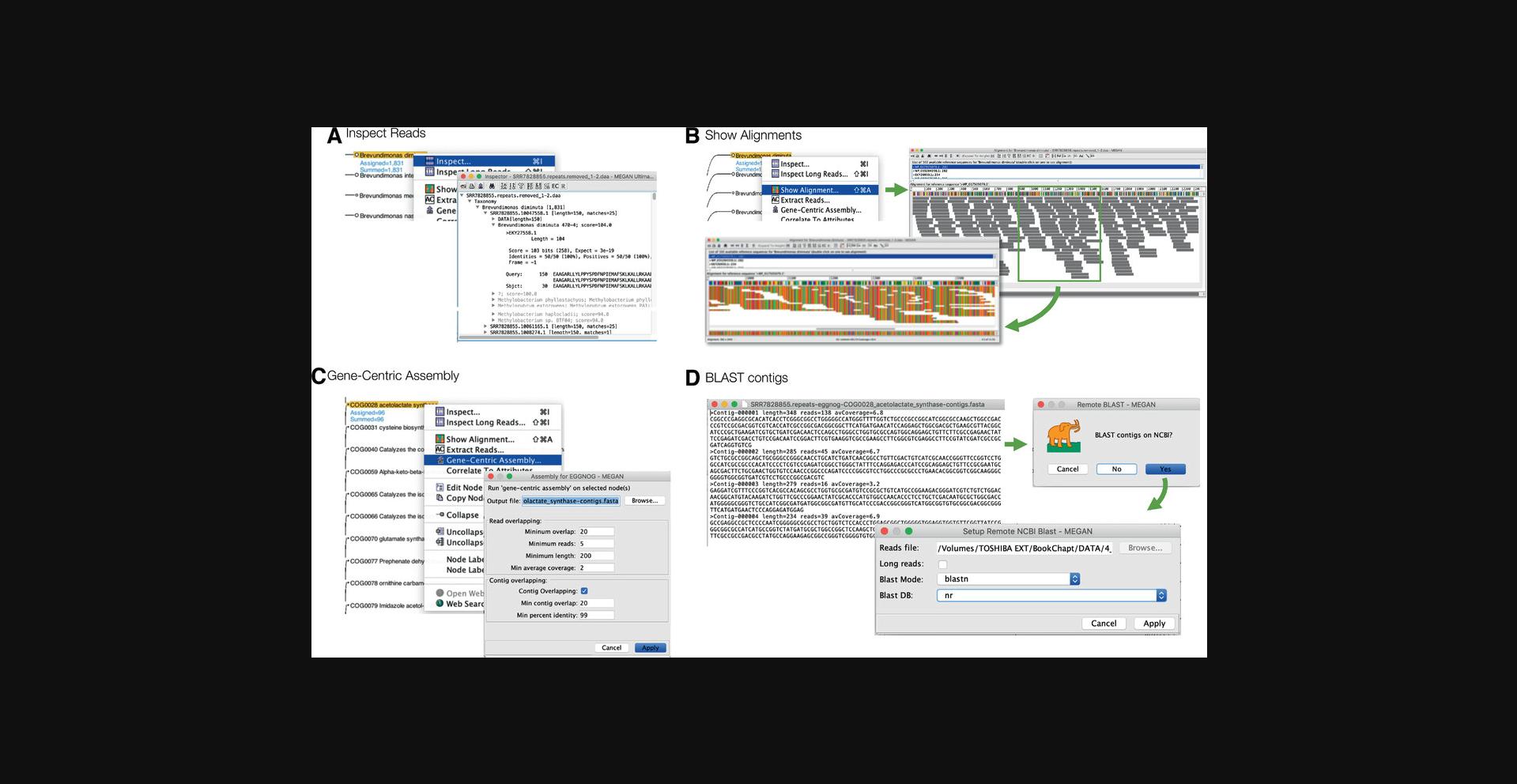
9.Gene-centric assembly (optional).
Genome assembly of short read microbiome shotgun data is challenging (Boisvert et al., 2012), and metagenome assembled genomes (MAGs) often consist of large collections of small contigs (Kang, Froula, Egan, & Wang, 2015). An alternative approach is to attempt to assemble individual gene families into contigs corresponding to genes from different genomes (Huson et al., 2017).
To apply gene-centric assembly in MEGAN, open the function viewer of interest and uncollapse all nodes (as described above). Further, select a leaf of interest, for example acetolactate synthase in the eggNOG viewer, and then use the File → Export → Gene-Centric Assembly… menu item or corresponding node context menu item to open the gene-centric assembly dialog (Fig. 7C). Press Apply to launch the assembly process.
Upon completion of gene-centric assembly, MEGAN will report basic assembly statistics, and the user will be presented with a dialog that can be used to launch a BLAST alignment of the resulting gene sequences on NCBI (Fig. 7D). The contigs and resulting alignments can be opened as a new MEGAN document in RMA format.
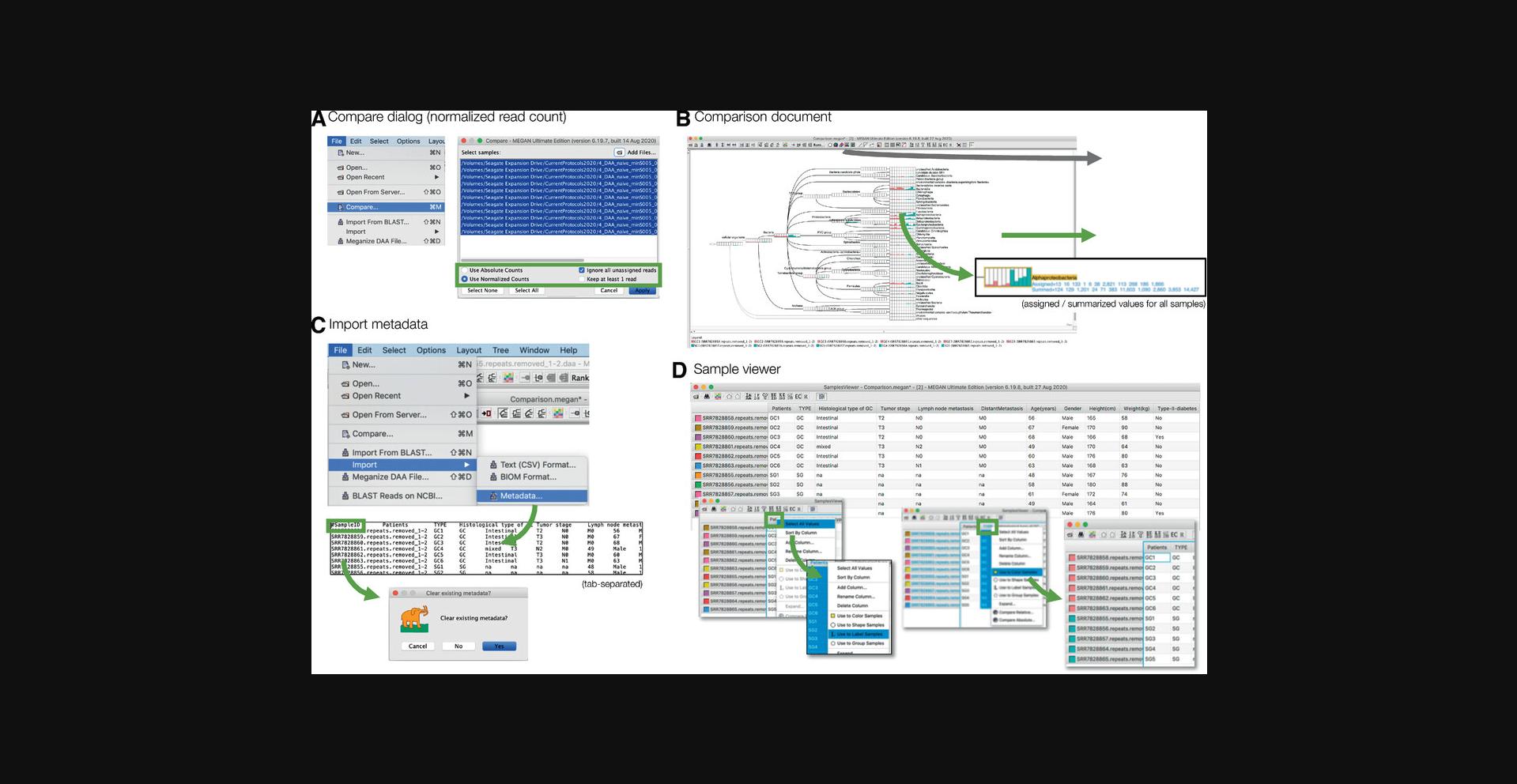
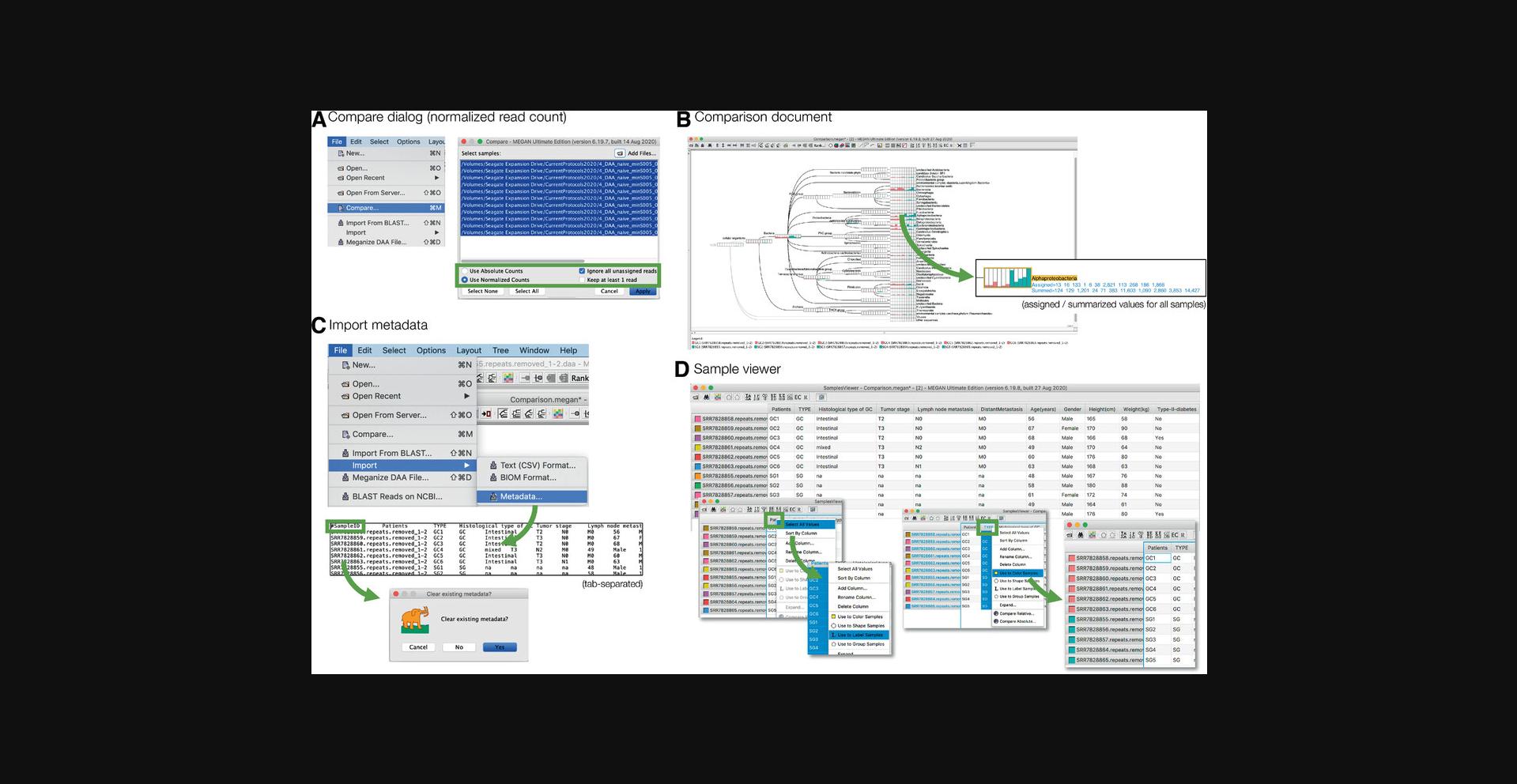
10.Comparison of DAA files.
Most microbiome projects involve multiple samples, and these must be analyzed in a comparative manner. To address this, MEGAN uses the concept of a comparison document that contains the result of the taxonomic and functional analysis of multiple individual samples, but not the corresponding reads and alignments (Fig. 8).
A comparison document is usually created from a set of meganized DAA files (e.g., for all samples SRR7828855-SRR7828865) using the compare dialog, which is opened by selecting the File → Compare… menu item (Fig. 8A).
The dialog provides buttons to choose whether the comparison is to be based on absolute counts or relative counts (normalized to the smallest sample size in the input), with the option to ignore all reads that are unassigned. Upon pressing Apply, a new comparison document will be created.
A newly computed comparison document can be saved to a file (extension .megan) using the File → Save As… menu item and reopened using the File → Open… menu item. The file is a small text file and thus can easily be shared, e.g., via e-mail.
A comparison document is displayed using the same taxonomy and functional viewers as for a single meganized DAA file (Fig. 8B).
Each sample in a microbiome project has associated metadata, reflecting technical parameters such as DNA extraction protocol, sequencing run, etc., and biological attributes, such as disease state, age, gender, etc., for human-associated samples. Metadata can be used both in visualization and statistical analysis.
Metadata should be prepared in a text file in tsv (tab-separated value) format. The first line starts with the special tag #SampleID followed by the names of all attributes (such as “Patient,” “Tumor type,” etc.). Then, there should be one row for each sample, with each row starting with the name of the sample (file name without path or suffix), then followed by the values for all the named attributes.
Metadata can be imported into a comparison document (but also into an individual meganized DAA file) using the File → Import → Metadata… menu item (Fig. 8C).
MEGAN provides a “Samples Viewer” that provides access to the imported metadata, and allows one to color, label, style, or group samples by attributes (Fig. 8D). The samples viewer is opened using the Window → Samples Viewer… menu item or the corresponding toolbar button.
For example, to color samples by a specific attribute, perform a context click on the table header for the given attribute, use the context menu to select all values, and then select the Use to Color Samples context menu item.
The samples viewer also provides methods for computing new comparison documents based on sub-selections or grouping of the samples. For example, the Options → Compute Core Biome… can be used to compute a new document containing those taxa that are present in large proportion of the samples at a high enough proportion or abundance.
For a comparison document, each taxonomic (or functional) node is displayed as a pie chart, coxcomb chart, bar chart, or heatmap, so as to indicate how many reads from each sample are assigned to the node (Fig. 8B). The displayed counts can be scaled linearly, by square root, or logarithmically. These aspects of the visualization are selected using the corresponding Layout menu items or tool bar items.
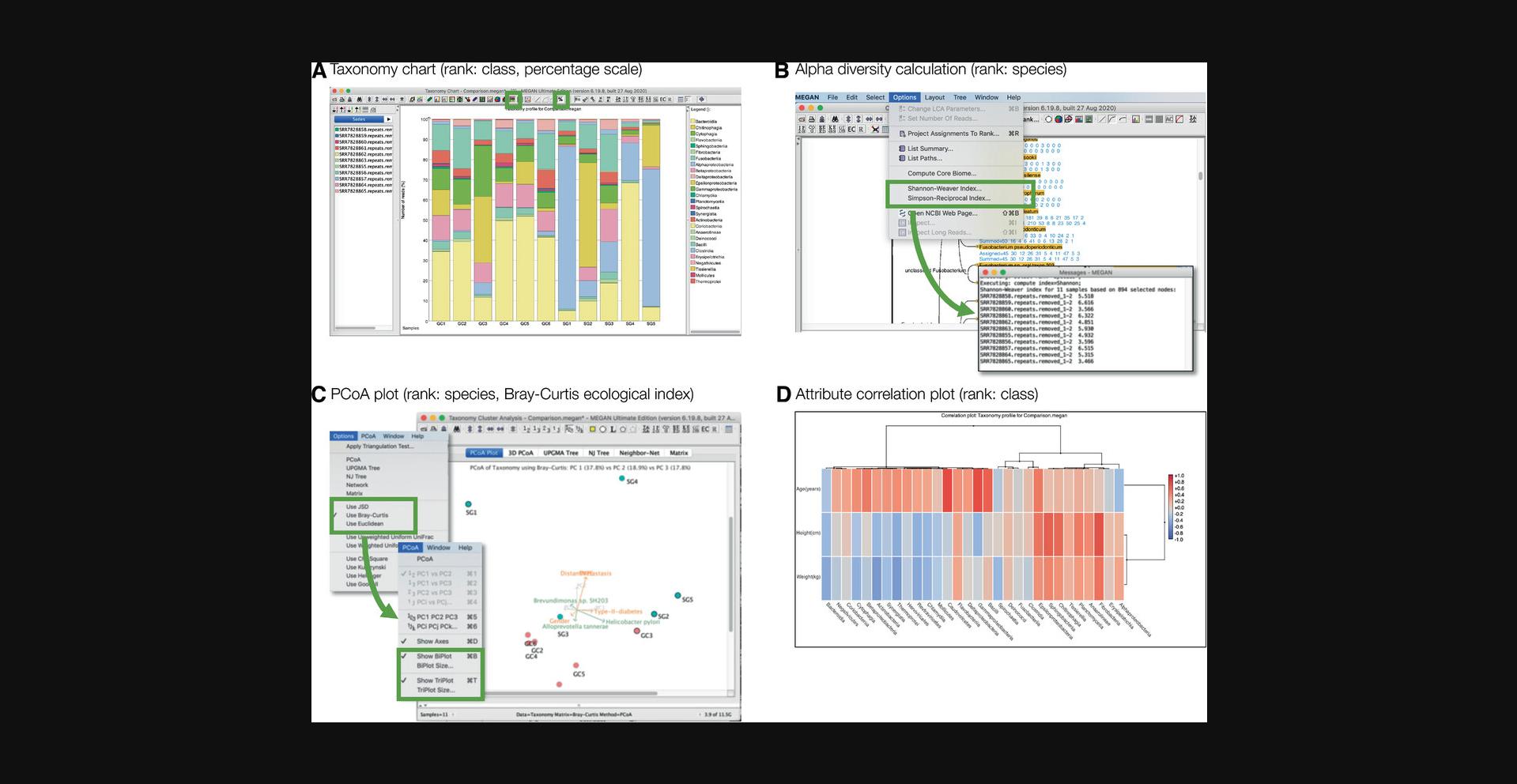
Taxonomic charts can be opened in the same way as described above. For example, in Figure 9A, we show a stacked bar chart displaying the percent of reads assigned to nodes of the taxonomic rank of class. In part D of the same figure, we show a plot that correlated a set of metadata attributes, subject age, height, and weight, with read counts assigned at the taxonomic rank of class.
Alpha diversity, or within-sample diversity, can be computed in MEGAN by selecting either the Options → Shannon-Weaver Index menu item or the Options → Simpson-Reciprocal Index menu item. Either measure will be calculated on the set of currently selected nodes, and the results will be written to the message window (Fig. 9B).
Beta diversity, or between-sample diversity, can be calculated using a number of different measures. To calculate such measures on a comparison document in MEGAN, open the cluster analysis viewer using the Window → Cluster analysis… menu item. MEGAN provides implementations of the Bray-Curtis ecological index (Bray & Curtis, 1957), Jensen-Shannon divergence (JSD), euclidean distances, and a number of other measures, which can be selected from the cluster analysis viewer's Options menu.
The measures are computed based on the currently selected nodes in the corresponding taxonomic (or functional) viewer, and the resulting distances are used to generate a PCoA (principal coordinate analysis) plot, a hierarchical clustering, an unrooted tree, an unrooted phylogenetic network, and a simple matrix, each displayed in a different tab.
The PCoA plot can be customized in a number of ways; one can select between two and three dimensions, select which principal components to display, and turn on both bi-plot and tri-plot vectors. For example, in Figure 9C, the bi-plot (shown in green) indicates that Helicobacter pylori is a main cause of differences between samples, whereas the tri-plot (show in orange) displays metadata-correlated variation.
Note that using euclidean distances and PCoA together is mathematically equivalent to performing a PCA (principal component analysis) calculation.
The naïve LCA and other similar algorithms used for taxonomic binning assign reads to taxa on different ranks of the NCBI taxonomy (Fig. 9D). As a consequence, the number of reads assigned at any given rank of the taxonomy might be much lower than the total number of assigned reads. This problem can be addressed, to a degree, using taxonomic projection, a simple algorithm that maps all read counts on a taxonomy to a selected rank. It operates by pushing all counts down the tree, from the root toward the selected rank, passing down counts to children in proportion to the number of reads assigned to a given child, or one of its descendants. This method is run using the Options → Project Assignments to Rank….
A functional comparison of multiple samples is very similar to the described taxonomic comparison (Fig. 10). The same visualizations, charts, and alpha- and beta-diversity are provided for all functional viewers.
11.Exporting data (optional).
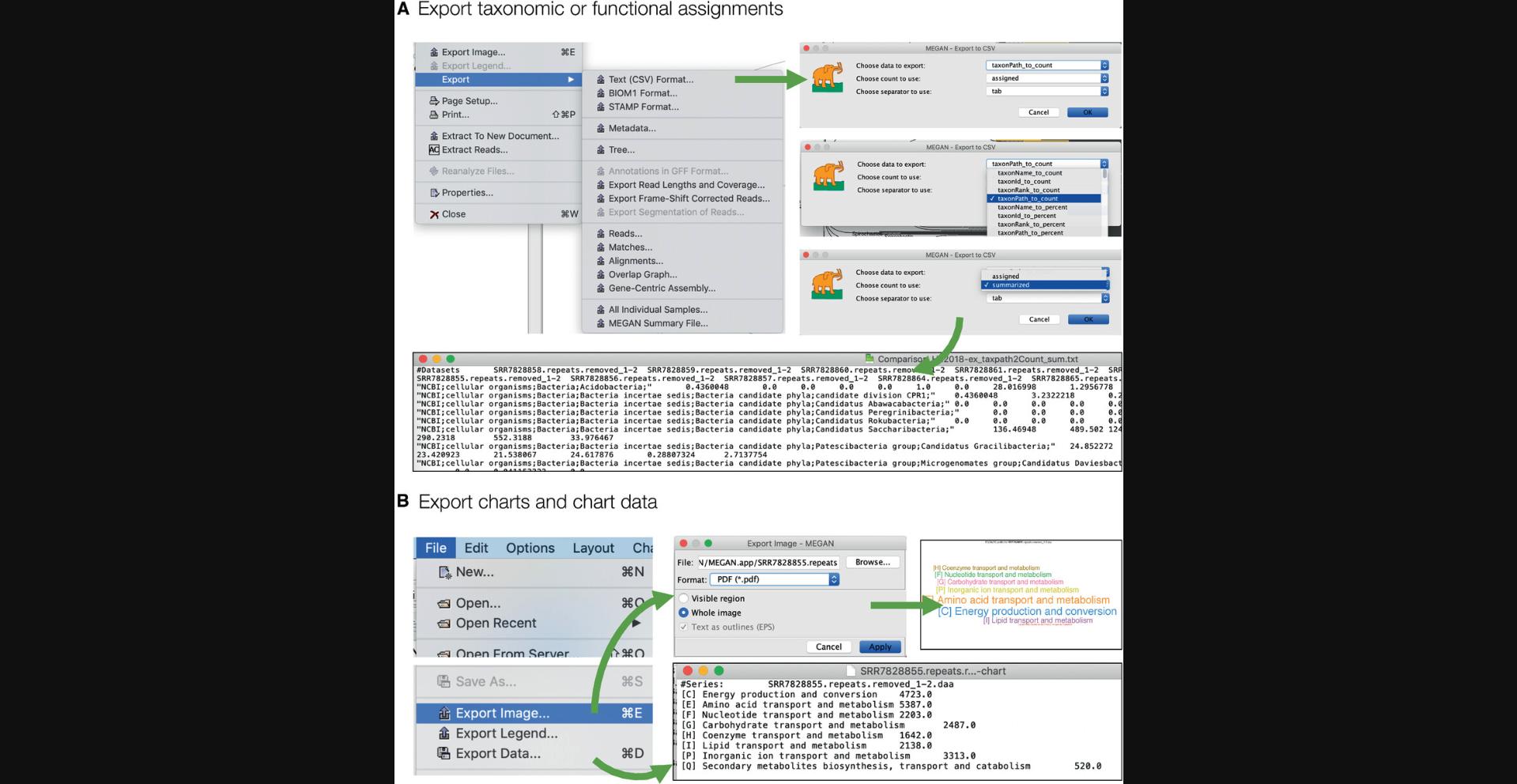
Data computed in MEGAN can be exported in a number of different ways, so as to allow additional analysis using other tools. The File → Export submenu provides 18 different menu items for exporting data, many of which support multiple formats. The taxonomic or functional classification of reads can be exported in a large number of different ways, in csv or tsv format. This is done by first selecting all nodes of interest in a taxonomic or functional viewer, and then selecting the File → Export → Text (CSV)….
In Figure 11A we show how to export the number of reads assigned to a particular taxon, the latter to be reported as a path from the root of the taxonomy down to the specific taxon.
Any chart can be exported as an image using the File → Export Image menu item, and using the File → Export Legend… menu item for its legend. The data displayed in a chart can be exported using the File → Export Data… menu item (Fig. 11B).
Support Protocol 1: PREPROCESSING
Before you can run the core DIAMOND+MEGAN analysis pipeline, there are a number of preprocessing steps to be considered. You may need to acquire public sequencing data. Usually, you will perform quality control, quality trimming, and contamination filtering on your input data.
1.Data acquisition.
The Sequence Read Archive (SRA) is a major source of microbiome sequencing datasets. The SRA Toolkit is a collection of tools that can be used to access the stored data in an efficient manner. For example, the program fastq-dump can be used to download sequences in fastq format. To download the data used in this protocol, we ran the command on all identifiers assigned to the referenced project (SRR7828855-SRR7828865):
- fastq-dump -I --split-files --gzip (identifier)
The --split-files option needs to be omitted for long read and single-end short read sequencing datasets, and the option --table SEQUENCE should usually be specified for long read datasets.
Details of all parameters can be found on the fastq-dump website.
2.Quality control.
The first step of any data analysis is to take a look at the data to check for obvious problems (the example dataset used in Basic Protocol 1 does not have any particular problems). FastQC is widely used form Illumina short reads. It can be run as follows:
- fastqc -f fastq -o (output directory) --extract -d (temporary directory) (input files)
The output is a collection of files in HTML format. These can be opened and viewed using a web browser and will provide an analysis of the quality of the sequencing data.
3.Quality trimming and contamination filtering.
It is not uncommon to analyze microbiome sequencing data without performing quality trimming and contamination control, especially if sequence assembly is not performed and the data is not host associated.
Because the example data is human-associated, here we illustrate this process using the tool KneadData ? (https://github.com/biobkery/kneaddata)? for quality trimming and host-associated contamination filtering. This first uses trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) (Bolger, Lohse, & Usadel, 2014) to trim poor-quality bases from the ends of the sequencing reads. It then uses bowtie2 (Langmead & Salzberg, 2012) to compare all reads against the human genome to remove human reads.
KneadData requires a bowtie2 index of the host genome (here, human genome) to be filtered. This is downloaded using the command:
- kneaddata_database --download human_genome bowtie2
Based on the information given in the respective publication, the tool was applied with the following parameters for the example dataset of Basic Protocol 1:
- kneaddata --input SRR7828855_1.fq --input SRR7828855_2.fq -db human-genome-db
- --output (output directory) -t 32 --trimmomatic-options="SLIDINGWINDOW:4:20 MINLEN:50 "
- --bowtie2-options=" -very-sensitive -dovetail " --output-prefix SRR7828855_kneaddata
The example dataset consists of overlapping read pairs, and so a tool may be applied to merge the pairs. Here we used the program fastq-join. It can be installed via conda using the command:
- conda install -c bioconda fastq-join
The program is executed on the output of KneadData for the example dataset as follows:
- fastq-join SRR7828855_kneaddata_1.fastq SRR7828855_kneaddata_2.fastq -o fastq-join/SRR7828855_%.fastq
The program fastq-join produces four output files, each replacing the “%” placeholder in the -o argument (fastq-join/SRR7828855_%.fastq for the above command, fastq-join being the directory where the files will be written). For example, the file SRR782885_join.fastq contains reads that were successfully merged, and SRR782885_un1.fastq and SRR782885_un2.fastq contain the reads that were not merged for the first and second read pairs, respectively. Before carrying out downstream analysis, we concatenate these three files in order to not lose any information, as follows:
- cat fastq-join/SRR7828855*.fastq > fastq-join/SRR7828855_merged.fastq
In Basic Protocol 1, we use only the files that are output of the cat command (quality trimmed, contamination filtered, overlapping read-pairs merged).
Basic Protocol 2: ANALYSIS OF ASSEMBLED LONG READ MICROBIOME SEQUENCES
This protocol describes the analysis of a typical single sample from a long read microbiome shotgun sequencing project, obtained using a third-generation sequencer, such as an ONT (Oxford Nanopore Technologies) MinION or a PacBio (Pacific Biosciences) sequencing device.
The analysis described here is built around the core pipeline depicted in Figure 1.For long reads, we add two steps. In a preprocessing step, all reads are assembled. The assembly of long read datasets, although more error-prone, results in some complete or near-complete, often circular chromosomes, and some contigs. The assembled, longer sequences make the task of accurate taxonomic binning easier, as well as making it possible to obtain a more complete and meaningful annotation of the genomes. In an optional post-processing step, contigs are subjected to protein-reference-based frame-shift correction, and then taxonomic bins are analyzed for completeness, contamination, and strain heterogeneity using a tool such as CheckM (Parks, Imelfort, Skennerton, Hugenholtz, & Tyson, 2014).
Long read sequencing data differ from short read sequencing data in terms of length, of course, but also in other aspects, including being more error-prone, even after assembly, and showing a high variability of sequence length.
Long reads and long read assemblies can contain many insertion and deletion errors, which cause problems when performing sequence alignment against the NCBI-nr protein reference database. To address this, we employ DIAMOND in frame-shift-aware alignment mode and use a long read version of the LCA algorithm for taxonomic binning in MEGAN (Huson et al., 2018). To address the length variability of long reads and contigs, instead of reporting the number of reads assigned to each taxonomic node, we report the number of aligned bases.
In this protocol, we will illustrate the necessary steps using a published dataset (Arumugam et al., 2019) that was collected from an enrichment bioreactor seeded with waste-water treatment sludge. The total dataset consists of ∼ 695, 000 long reads with an average length of 9 kb, totaling approximately 6 Gb of sequence.
To download the sequencing dataset, please follow Support Protocol 1 using the SRA accession SRR8305972.
1.Assembly.
De novo assembly of genomes from microbiome shotgun sequencing data is a long-standing, difficult problem. Short reads generated by second-generation sequencers are usually not informative enough to resolve repeats and conserved regions in genomes, resulting in disappointingly short contigs and scaffolds (Boisvert et al., 2012). Long reads obtained by third-generation sequencing technologies overcome the problem of resolving repeats and conserved regions in metagenomes, and allow the assembly of complete, circular microbial genomes (Arumugam et al., 2019; Moss, Maghini, & Bhatt, 2020).
In this protocol step, we describe the use of the long read assembly pipeline Unicycler (Wick et al., 2017). There are a number of other tools for the assembly of long read metagenomic datasets, such as Flye (Kolmogorov, Rayko, Yuan, Polevikov, & Pevzner, 2019a) and Canu (Koren et al., 2017), each with specific advantages and drawbacks (Wick & Holt, 2019).
Unicycler is a bacterial genome assembly pipeline that can run on long-reads-only datasets, as well as on hybrid (long read and short read) datasets. Here we describe the use of long-reads-only mode. In long-reads-only mode, the Unicycler pipeline consists of minimap2 (Li, 2018) and miniasm (Li, 2016) for assembly, racon (Vaser et al., 2017) for building consensus, and tBLASTn (Altschul et al., 1997) for detecting the origin of replication in circular genomes.
Unicycler requires only two parameters in long-reads-only mode: -l for the input reads (fastq file, can be gzipped) and -o for the output directory. The additional parameters we use are -t for setting the number of CPU threads, and --keep 3 to retain intermediate files generated by the pipeline, which can be useful for inspecting the assembly later on. It is run like this for the example dataset:
- unicycler -l SRR8305972.fastq.gz -o unicycler_asm -t (threads) --keep 3
Additionally, the assembly mode can be selected by the parameter --mode, which can take three values as input: conservative, normal, and bold, setting the trade-off between generating longer contigs and a higher risk of misassembly. We use the default setting, normal, in this protocol.
Unicycler places all output in the specified output directory, which is unicycler_asm in the example command. The final assembly is written to a file called assembly.fasta. Other produced files include assembly.gfa, which contains the assembly graph, and 001_string_graph.gfa, which contains the raw string graph. Such GFA files can be visualized and explored using the Bandage program (Wick, Schultz, Zobel, & Holt, 2015).
2.Correcting errors in draft assemblies.
Long-read-only assemblies of microbiome sequencing data can consist of long continuous, and even circular and complete sequences. However, the sequences will usually lack accuracy, which is due to systematic errors in sequencing, rather than random errors. Before continuing with the downstream analysis, these errors in the assembled contigs need to be corrected as much as possible. Racon (Vaser et al., 2017), a fast consensus algorithm, is one of the error-correction tools commonly used after assembling error-prone long read datasets. Unicycler, the assembly pipeline employed here, already performs three rounds of error-correction by Racon.
To improve the quality of the sequences further, we apply the tool medaka (ONT, 2020), which is a neural-network based correction algorithm designed for sequences obtained using an ONT device.
The medaka_consensus executable, which runs the pipeline of medaka, takes as input the draft assembly (-d) (assembly.fasta in the Unicycler output directory), the raw reads (-i), and a model name describing both the flow-cell used and the version of the employed base-calling algorithm (-m). It requires an output directory to be specified (-o). The software is run as follows for the example dataset:
- medaka_consensus -i SRR8305972.fastq.gz -d assembly.fasta -o unicycler_medaka -t (threads) -m r941_min_high_g330
If medaka needs to be run again (e.g., due to an error), the -f option can be specified to override the output directory; otherwise medaka will use the existing files in the output directory. The -b (batch size, default: 100) option can be used to control the memory usage of medaka, e.g., -b 1000, to increase its performance by using 10 times more memory than the default.
The available models can be listed with the command: medaka tools_list models and new model files can be downloaded using the command medaka tools download_models. The model names follow the notation: chemistry_device_accuracy_basecaller. For the model used for the example dataset (r941_min_high_g330), that is, R 9.4.1 chemistry, MinION, High accuracy base calling, and guppy version 3.3.0.
Medaka outputs the error-corrected assembly to a file called consensus.fasta in the specified output directory.
3.DIAMOND comparison.
Despite being error-corrected multiple times with several methods, long-read-only assemblies still contain errors, mostly insertions and deletions of single or a few bases. This causes problems for translated alignment algorithms such as BLAST×, because erroneous insertions and deletions cause frame-shifts that break alignments.
To address this, DIAMOND provides a frameshift-aware alignment mode (Buchfink et al., 2015b; Huson et al., 2018), which is activated by specifying a frameshift penalty using the -F option. We use a penalty of 15, which appears to strike a good balance between producing long alignments without excessive switching of frames.
By default, DIAMOND reports a set of high-scoring alignments for a given query sequence, regardless of their position along the query. Applied to long reads or contigs, this usually results a list of alignments that cover only a small, highly conserved region of the query. Hence, for the alignment of long reads and contigs, DIAMOND provides an alternative reporting mode, range culling , that reports alignments that are high-scoring in comparison only with other alignments that cover the same region of the query. This feature is activated using the flag --range-culling, and we also set the parameter --top 10 to instruct the program to reports all alignments whose bit-score lies within 10% of the best score of competing alignments.
Using the final polished assembly file as input file (consensus.fasta in the medaka output directory), we run DIAMOND as follows for the example dataset:
- diamond blastx -q consensus.fasta -d nr.dmnd -o SRR8305972_unicycler.daa -F 15 -f 100 --range-culling --top 10 -p (threads)
The output file must have the file extension .daa.
4.Taxonomic and functional binning.
To perform taxonomic binning of long reads or contigs, MEGAN provides an enhanced LCA approach called the interval-union LCA algorithm (Huson et al., 2018). This algorithm considers alignments along the whole sequence and assigns the sequence to the most specific taxon that covers a high percentage of the aligned regions of the sequence.
In more detail, the interval-union LCA algorithm works by first attempting to detect genes on the query sequence. This is done by grouping alignments based on their locations on the query sequence, and defining intervals where a gene starts and ends. The alignments are filtered locally within intervals, by their bit-score. The query sequence is then assigned to the taxon whose alignments cover at least the value of the Percent to cover parameter, or the taxon that is the lowest common ancestor of all taxa that are above this value.
To perform functional binning on each such interval, MEGAN sorts the alignments by bit score and assigns function of the first alignment in the sorted list that has a functional classification to the read. Thus, a query sequence can be classified into multiple functional categories, where each interval (each gene) is classified into a functional category.
In addition to using the interval-union LCA algorithm, when processing long reads or contigs, MEGAN reports the number of aligned bases, rather than number of reads, assigned to a given taxon or functional class.
A DAA file produced by DIAMOND with the appropriate parameters discussed in the DIAMOND comparison paragraph can be meganized either using command-line tool daa-meganizer or using the meganize dialog of MEGAN, very much as described above for short reads.
To meganize the DAA file from the previous section, use this command:
- daa-meganizer -i SRR8305972_unicycler.daa -mdb megan-map-Jan2021.db --longReads
Alternatively, DAA files for long reads can be meganized using the GUI. The meganize dialog in MEGAN is opened using the File → Meganize DAA File… menu item. After supplying the list of DAA files to be meganized, check the Long Reads box on the Files tab (Fig. 12A). This will activate the interval-union LCA algorithm and the reporting of aligned bases, rather than read count. You can verify this by inspecting the LCA tab. Here, the LCA algorithm will show longReads (this refers to the interval-intersection LCA) and the Read Assignment mode will show alignedBases (Fig. 12C). The Percent to cover will be set to 51%, by default. As described above for short reads, on the Taxonomy tab, select the mapping file (Fig. 12B).
Meganized DAA files can be inspected and analyzed the same way as in Basic Protocol 1 for short reads. Below we describe the functionality that is specific to long reads.
5.Long read inspection.
MEGAN provides a long read inspector that can be used to explore the alignments to reference proteins along a given long read or contig. To open this viewer for a specific taxon or functional class, select the node in the corresponding viewer and the choose the Inspect Long Reads… context menu item.
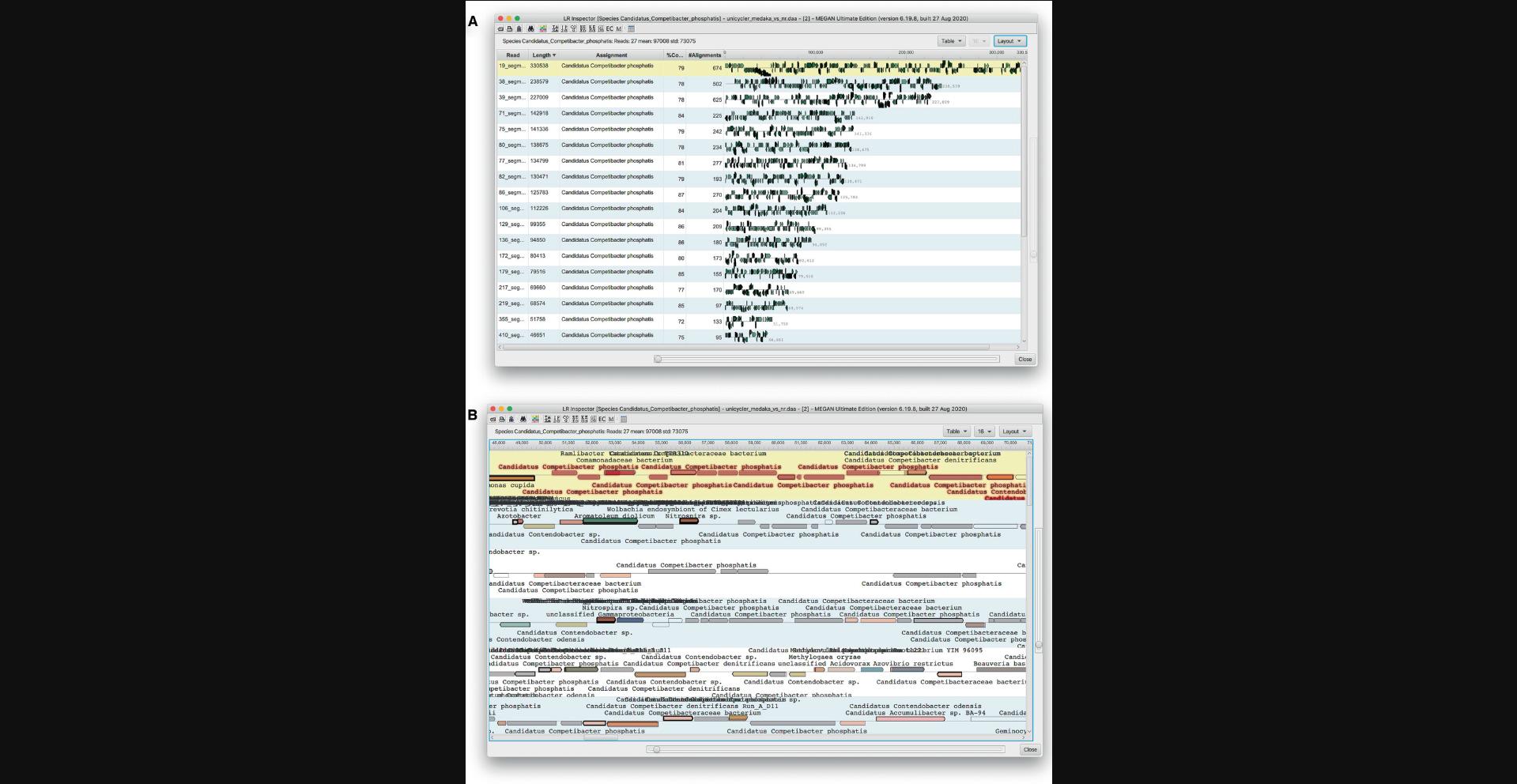
The long read inspector displays each sequence assigned to a given node in a separate row, listing the sequence name, length, taxonomic assignment, % coverage and number of alignments, together with an overview visualization of the sequence and its alignments to reference proteins (Fig. 13A). Alignments are represented by arrows in the direction of translation.
The visualization can be extended to show annotations of the aligned reference sequences, using the Layout drop-down menu item to select which classifications to use. The sliders on the right side and at the bottom of the window can be used to control the height of the rows and the zoom level of the layout of the alignments along the length of the sequence, respectively. Clicking on an arrow will select it, causing the corresponding alignments to be displayed in the message window. The context menu item Select Similar can be used to select all other arrows that represent alignments to the same taxon (Fig. 13B).
6.Exporting data.
All the methods for exporting data that are described above for short read microbiome sequences can also be applied to long read sequencing datasets. Here we describe two additional methods. The first can be used for short reads, too, whereas the second only makes sense for long read data.
MEGAN allows the user to export all reads to taxonomic or functional class-specific files. To use this feature, select all nodes of interest in a taxonomic or functional viewer and then use the File → Extract Reads… menu item to open a new dialog to specify the filename to use for output. During saving, any occurrences of the special placeholders %t or %i in the filename will be replaced by the name of the class, or the class integer id, respectively, thus ensuring that the reads assigned to each selected class are written to a different file.
As discussed above, even after multiple rounds of error correction, sequences assembled from long-reads-only datasets may contain many insertions and deletions, which leads to problems during translated alignment, thus significantly reducing the performance of tools like CheckM and Prokka (Seemann, 2014) on assembled long reads.
To address this, MEGAN implements a heuristic that aims at reducing the number of erroneous insertions and deletions by inserting an additional N at locations where the DIAMOND alignments suggest that a base is missing (causing a frame-shift of +1 in the alignments), or two additional Ns where the DIAMOND alignments indicate that the sequence contains a superfluous base (causing a frame-shift of −1 in the alignments) (Huson et al., 2018).
To save frame-shift-corrected sequences, select all nodes of interest in a taxonomic or functional viewer and then use the File → Export → Frame-Shift Corrected Reads… menu item to open a new dialog to specific the filename to use for output. Again, any occurrences of the special placeholders %t or %i in the output filename will be replaced by the name of the class, or the class integer id, respectively, during export.
Both export procedures described above can also be carried out on the command line, using the program read-extractor that can be found in MEGAN's tools sub-directory. The program takes as input a meganized DAA file (option -i), the name of an output file (option -o), which may contain occurrences of the special placeholders %t and %i that are replaced as discussed above, a classification name (option -c), such as Taxonomy or GTDB, and the flag -fsc, if frame-shift correction is desired.
The program is run like this for the example dataset:
- read-extractor -i SRR8305972_unicycler.daa -o %t_%i.fasta -c classification -fsc
Here, classification can be either Taxonomy or GTDB, and the output files will be written into the current directory with the format described above using the placeholders. The output can also be written into a directory, such as -o bins/%t_%i.fasta.gz. The -gz option can be used to compress the output files.
The alignment of reference proteins to long reads or contigs can be used to produce a crude annotation of the sequences, and this is implemented in MEGAN. To use this feature, select all nodes of interest in a taxonomic or functional viewer and then use the File → Export → Annotations in GFF Format… menu item to export the annotated sequences. The exported GFF file can then be visualized in an annotation viewer (Rutherford et al., 2000; Thorvaldsdóttir, Robinson, & Mesirov, 2013) or used in downstream analysis.
Support Protocol 2: TAXONOMIC BINNING AND CheckM
As discussed above, MEGAN places all reads into taxonomic bins, using both the NCBI taxonomy and the GTDB taxonomy. In either case, these bins are candidates for being “metagenome assembled genomes” (MAGs). These putative MAGs can be exported from MEGAN in FastA format using the above-described export of frame-shift corrected sequences. The quality of these bins can optionally be assessed using a tool such as CheckM, as discussed below.
1.Completeness and contamination analysis using CheckM.
To determine which of these output bins can be considered a high-quality MAG, we need to assess the level of completeness and level of contamination of each file.
The program CheckM (Parks et al., 2014) takes as input a collection of putative taxonomic bins and provides measures of completeness and contamination of each bin, based on the detection analysis of clade-specific, single-copy marker genes.
CheckM requires an input directory containing one FASTA file per bin, such as exported by MEGAN, as described above. The command to execute the standard pipeline is:
- checkm lineage_wf --tmpdir tmp -t (threads) --pplacer_threads (threads)
- -x fasta --tab_table -f SRR8305972_checkm.txt bins/ checkm_out/
Here, option --tmpdir specifies a temporary directory for use by the program, -t and --pplacer_threads sets the number of threads used by the program and during pplacer calculations, -x specifies the extension of the input files (e.g., fasta, fa, or fna), --tab_table makes the output tab-delimited, which is written into the file specified after -f, the directory called bins/ here is the input directory with all input files (e.g., the output of read-extractor from the Basic Protocol 2, step 6., “Exporting data”), whereas all intermediate and output files are written to the specified output directory (here checkm_out).
In Table 1, we present the CheckM results for all taxonomic bins exported from the GTDB taxonomy that show at least 50% completeness.
Highlighted in gray, for each taxonomic bin (column Bin Id), we list the reported percentage of completeness (Compl.), contamination (Cont.), and strain heterogeneity (SH). These values are inferred from lineage-specific marker genes. The Marker lineage column reports the most specific lineage CheckM could detect for the bin, whose marker genes were used for the analysis. The # Genomes column contains the number of genomes in the lineage used, while the # Markers and #Marker Sets columns contain number of marker genes and co-located marker gene sets for the lineage, respectively. The columns #0, #1, and #2 contain the number of genes found within the bin with the given number of copies. As CheckM uses single-copy marker genes, for a complete and uncontaminated genome, each marker gene should be found exactly once. Completeness and contamination are calculated based on the copy numbers of marker genes, whereas strain heterogeneity is an estimate of how much of the detected contamination is from a closely related organism.
| Bin ID | Marker lineage | #G | #M | #MS | #0 | #1 | #2 | Compl. | Cont. | SH |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteroidetes bacterium OLB12 | k Bacteria (UID2570) | 433 | 274 | 183 | 13 | 261 | 0 | 95.28 | 0 | 0 |
| Candidatus Accumulibacter | c Betaproteobacteria (UID3971) | 223 | 422 | 210 | 24 | 395 | 3 | 95.19 | 1.11 | 0 |
| Gammaproteobacteria | c Gammaproteobacteria (UID4266) | 1097 | 270 | 172 | 13 | 243 | 14 | 95.07 | 4.99 | 0 |
| Chlamydiia | k Bacteria (UID2982) | 88 | 230 | 148 | 12 | 215 | 3 | 94.6 | 1.58 | 0 |
| Rhodospirillaceae | o Rhodospirillales (UID3754) | 63 | 336 | 201 | 16 | 319 | 1 | 94.12 | 0.5 | 0 |
| Bacteroidetes bacterium OLB8 | p Bacteroidetes (UID2591) | 364 | 302 | 202 | 14 | 286 | 2 | 94.04 | 0.99 | 0 |
| Chlorobi bacterium OLB5 | k Bacteria (UID2570) | 433 | 273 | 183 | 25 | 246 | 2 | 90.87 | 0.66 | 0 |
| unclassified Thauera | f Rhodocyclaceae (UID3972) | 30 | 540 | 241 | 89 | 443 | 8 | 85.47 | 1.9 | 62.5 |
| Bacteroidetes | p Bacteroidetes (UID2591) | 364 | 303 | 203 | 43 | 238 | 22 | 83.16 | 5.61 | 4.55 |
| unclassified Nitrospira | k Bacteria (UID3187) | 2258 | 181 | 110 | 36 | 138 | 7 | 82.75 | 4.77 | 0 |
| Sphingobacteriales bacterium 44-15 | p Bacteroidetes (UID2591) | 364 | 302 | 203 | 85 | 211 | 5 | 74.54 | 2.46 | 12.5 |
| Chloroflexi bacterium | k Bacteria (UID1452) | 924 | 163 | 110 | 63 | 74 | 23 | 59.06 | 17.09 | 3.12 |
| Candidatus Competibacter phosphatis | k Bacteria (UID203) | 5449 | 104 | 58 | 69 | 32 | 3 | 53.45 | 4.31 | 33.33 |
| Chloroflexi | k Bacteria (UID1452) | 924 | 163 | 110 | 73 | 72 | 15 | 52.67 | 19.09 | 16.67 |
-
aA given bin, we report the percent completeness (Compl.), contamination (Cont.), and strain heterogeneity (SH) estimated by CheckM. In addition, we report the marker lineage used by CheckM to analyze the bin, and for that lineage, the number of genomes (#G), markers (#M), and marker sets (#MS), as well as the number of marker genes found 0, 1, or 2 times (columns #0, #1 or #2, respectively).
2.Alternative assembly using Flye.
Flye (Kolmogorov, Yuan, Lin, & Pevzner, 2019b; Kolmogorov et al., 2019a) is a long read assembly program, which also supports Nanopore-only assemblies of microbiome samples. It can be used as an alternative to the Unicycler pipeline. Flye performs better than Unicycler in terms of recovering plasmids and generating slightly more reliable assemblies, whereas Unicycler is better at achieving higher rates of complete, circular assemblies (Wick & Holt, 2019). Flye can be installed through conda with the command:
- conda install -c bioconda flye
For the example dataset from Basic Protocol 2, it can be run as:
- flye --nano-raw SRR8305972.fastq.gz -g 4m -o SRR8305972_flye_asm --plasmids --meta -t (threads)
The nano-raw option specifies the location of the raw base-called fastq file for Nanopore data. Alternatively, the --nano-corr option can be used for input reads that have undergone a preliminary error-correction step. The assembler also supports long reads generated by PacBio sequencers, using the options --pacbio-raw, --pacbio-corr, and --pacbio-hifi for raw, error-corrected, and HiFi reads, respectively. Flye expects an estimate of the genome size with the -g option (e.g., 4m); however, this option has very little effect in metagenome mode (it is nevertheless required, here we set it to 4m). The option -o specifies output directory. The flag --plasmids instructs Flye to look for plasmids in the dataset. The flag --meta turns on the metagenome assembly mode. The option -t sets the number of threads to use.
Flye outputs the assembled contigs in a file called assembly.fasta in the output directory, as well as the assembly graph in assembly_graph.gfa and assembly_graph.gv files.
COMMENTARY
Background Information
MEGAN was originally developed in 2007 (Huson et al., 2007) as a taxonomic binning tool for second-generation metagenomic sequencing projects such as Poinar et al. (2006). This early version analyzed the output of a BLASTX comparison of all reads against the NCBI-nr database. MEGAN was later extended so as to also perform functional binning (Huson et al., 2011). To address the increasing size of metagenomic sequencing datasets and the NCBI-nr reference database, DIAMOND (Buchfink et al., 2015b) was developed as a high-throughput alternative to BLASTX, and the efficiency of MEGAN was much improved (Huson et al., 2016). More recently, to facilitate the analysis of long read metagenomic sequences and assemblies, both DIAMOND and MEGAN were extended so as to provide a dedicated long read mode (Arumugam et al., 2019; Huson et al., 2018).
Critical Parameters
When designing a microbiome sequencing project, the key questions are: how many samples should be collected, and at what time (or other) intervals? What sequencing depth is sufficient? Which sequencing technology should be used? The sampling strategy will depend on the strength and speed of the phenomena of interest. The required sequencing depth depends on whether the goal is only taxonomic profiling or high-level functional profiling, where one million reads might suffice, gene-centric assembly of specific genes of interest, where the number of reads required will depend on the relative abundance of the genes, or metagenome assembly and binning, where more reads are always beneficial, up to a point. Most microbiome projects to date have used short read sequencing technologies. Long read sequencing technologies are increasing being used, and are particularly of interest when the goal is to obtain chromosome-scale contigs of the most abundant organisms in a sample, for example to establish whether specific genes of interest appear together on some chromosome.
Troubleshooting
If you encounter problems while attempting to install or run DIAMOND, then please address them to the DIAMOND GitHub page at https://github.com/bbuchfink/diamond. Please address problems associated with running MEGAN to the MEGAN community webpage at http://megan.informatik.uni-tuebingen.de.
Acknowledgments
The authors acknowledge hardware support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC, and the German Research Foundation (DFG) through grant no. INST 37/935-1 FUGG. C.B. was supported by the German Research Foundation (D.F.G.) through grant no HU 566/12-1. Publication costs were supported by the Open Access Publishing Fund of University of Tübingen.
Author Contributions
Caner Bağcı : Formal analysis; writing-original draft; writing-review & editing. Sascha Patz : Formal analysis, writing-original draft; writing-review & editing. Daniel H. Huson : Conceptualization; Software; Supervision; writing-original draft; writing-review & editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing not applicable–no new data generated.
Literature Cited
- Altschul, S., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research , 25, 3389–3402. doi: 10.1093/nar/25.17.3389.
- Arumugam, K., Bagci, C., Bessarab, I., Beier, S., Buchfink, B., Gorska, A., … Williams, R. B. (2019). Annotated bacterial chromosomes from frame-shift-corrected long read metagenomic data. Microbiome , 7(61).
- Barrett, A. J. (1992). Enzyme nomenclature. recommendations 1992. European Journal of Biochemistry , 232(1), 1–1. doi: 10.1111/j.1432-1033.1995.tb20774.x.
- Benson, D., Karsch-Mizrachi, I., Lipman, D., Ostell, J., & Wheeler, D. (2005). Genbank. Nucleic Acids Research , 1(33), D34–D38.
- Bentley, D. (2006). Whole-genome re-sequencing. Current Opinion in Genetics and Development , 16, 545–552.
- Boisvert, S., Raymond, F., Godzaridis, E., Laviolette, F., & Corbeil, J. (2012). Ray meta: Scalable de novo metagenome assembly and profiling. Genome Biology , 13(12), R122. doi: 10.1186/gb-2012-13-12-r122.
- Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics , 30(15), 2114–2120. doi: 10.1093/bioinformatics/btu170.
- Bray, R. J., & Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs , 27, 325–349. doi: 10.2307/1942268.
- Buchfink, B., Huson, D. H., & Xie, C. (2015a). Metascope-fast and accurate identification of microbes in metagenomic sequencing data. Technical Report arXiv:1511.08753, arXiv.
- Buchfink, B., Xie, C., & Huson, D. (2015b). Fast and sensitive protein alignment using DIAMOND. Nature Methods , 12, 59–60. doi: 10.1038/nmeth.3176.
- Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., & Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome , 6(1): 226. doi: 10.1186/s40168-018-0605-2.
- Franzosa, E. A., McIver, L. J., Rahnavard, G., Thompson, L. R., Schirmer, M., Weingart, G., … Huttenhower, C. (2018). Species-level functional profiling of metagenomes and metatranscriptomes. Nature Methods , 15(11), 962–968. doi: 10.1038/s41592-018-0176-y.
- Glass, E. M., Wilkening, J., Wilke, A., Antonopoulos, D., & Meyer, F. (2010). Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harbor Protocols , 2010(1), pdb.prot5368+. doi: 10.1101/pdb.prot5368.
- Hu, Y.-L., Pang, W., Huang, Y., Zhang, Y., & Zhang, C.-J. (2018). The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Frontiers in Cellular and Infection Microbiology , 8, 433. doi: 10.3389/fcimb.2018.00433.
- Huson, D., Beier, S., Flade, I., G´orska, A., El-Hadidi, M., Mitra, S., … Tappu, R. (2016). MEGAN Community Edition-interactive exploration and analysis of large-scale microbiome sequencing data. PloS Computational Biology , 12(6), e1004957. doi: 10.1371/journal.pcbi.1004957.
- Huson, D. H., Albrecht, B., Bağcı, C., Bessarab, I., Górska, A., Jolic, D., & Williams, R. B. H. (2018). MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biology Direct , 13(1), 6. doi: 10.1186/s13062-018-0208-7.
- Huson, D. H., Auch, A. F., Qi, J., & Schuster, S. C. (2007). MEGAN analysis of metagenomic data. Genome Research , 17(3), 377–386. doi: 10.1101/gr.5969107.
- Huson, D. H., Mitra, S., Weber, N., Ruscheweyh, H.-J., & Schuster, S. C. (2011). Integrative analysis of environmental sequences using MEGAN 4. Genome Research , 21, 1552–1560. doi: 10.1101/gr.120618.111.
- Huson, D. H., Tappu, R., Bazinet, A. L., Xie, C., Cummings, M. P., Nieselt, K., & Williams, R. (2017). Fast and simple protein-alignment-guided assembly of orthologous gene families from microbiome sequencing reads. Microbiome , 5, 11.
- Jain, M., Olsen, H. E., Paten, B., & Akeson, M. (2016). The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biology , 17, 239. doi: 10.1186/s13059-016-1103-0.
- Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research , 28(1), 27–30. doi: 10.1093/nar/28.1.27.
- Kang, D. D., Froula, J., Egan, R., & Wang, Z. (2015). MetaBAT, an efficient tool for accurately recon- structing single genomes from complex microbial communities. PeerJ , 3, e1165. doi: 10.7717/peerj.1165.
- Kolmogorov, M., Rayko, M., Yuan, J., Polevikov, E., & Pevzner, P. (2019a). metaFlye: Scalable long-read metagenome assembly using repeat graphs. bioRxiv , 637637.
- Kolmogorov, M., Yuan, J., Lin, Y., & Pevzner, P. A. (2019b). Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology , 37(5), 540–546. doi: 10.1038/s41587-019-0072-8.
- Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., & Phillippy, A. M. (2017). Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Research , doi: 10.1101/gr.215087.116.
- Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods , 9(4), 357–359. doi: 10.1038/nmeth.1923.
- Li, H. (2016). Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics , 32(14), 2103–2110. doi: 10.1093/bioinformatics/btw152.
- Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics , 34(18), 3094–3100. doi: 10.1093/bioinformatics/bty191.
- Mackelprang, R., Waldrop, M., DeAngelis, K., David, M., Chavarria, K., Blazewicz, S., … Jansson, J. (2011). Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature , 480(7377), 368–371. doi: 10.1038/nature10576.
- Mitchell, A., Chang, H.-Y., Daugherty, L., Fraser, M., Hunter, S., Lopez, R., … Finn, R. D. (2015). The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Research , 43(D1), D213–D221. doi: 10.1093/nar/gku1243.
- Moss, E. L., Maghini, D. G., & Bhatt, A. S. (2020). Complete, closed bacterial genomes from microbiomes using Nanopore sequencing. Nature Biotechnology , 38(6), 701–707.
- Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., … Stevens, R. (2013). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Research , 42(Database issue), D206–D214.
- Oxford Nanopore Technologies (ONT). (2020). Medaka: Sequence correction tool provided by ONT research. Available at: https://github.com/nanoporetech?language=python.
- Parks, D., Imelfort, M., Skennerton, C., Hugenholtz, P., & Tyson, G. (2014). Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Research , 25, 1043–1055. doi: 10.1101/gr.186072.114.
- Parks, D. H., Chuvochina, M., Chaumeil, P.-A., Rinke, C., Mussig, A. J., & Hugenholtz, P. (2020). A complete domain-to-species taxonomy for bacteria and archaea. Nature Biotechnology , 38(9), 1079–1086.
- Poinar, H. N., Schwarz, C., Qi, J., Shapiro, B., Macphee, R. D. E., Buigues, B., … Schuster, S. C. (2006). Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science , 311(5759), 392–394. doi: 10.1126/science.1123360.
- Powell, S., Szklarczyk, D., Trachana, K., Roth, A., Kuhn, M., Muller, J., … Bork, P. (2012). eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Research , 40(Database Issue), 284–289. doi: 10.1093/nar/gkr1060.
- Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., … Wang, J. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature , 464(7285), 59–65. doi: 10.1038/nature08821.
- Rhoads, A., & Au, K. F. (2015). PacBio sequencing and its applications. Genomics, Proteomics and Bioinformatics , 13(5), 278–289.
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., & Barrell, B. (2000). Artemis: Sequence visualization and annotation. Bioinformatics , 16(10), 944–945. doi: 10.1093/bioinformatics/16.10.944.
- Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics , 30(14), 2068–2069. doi: 10.1093/bioinformatics/btu153.
- Thorvaldsdóttir, H., Robinson, J. T., & Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Briefings in Bioinformatics , 14(2), 178–192. doi: 10.1093/bib/bbs017.
- Vaser, R., Sović, I., Nagarajan, N., & Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Research , 27(5), 737–746. doi: 10.1101/gr.214270.116.
- Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., … Smith, H. O. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science , 304(5667), 66–74. doi: 10.1126/science.1093857.
- Wick, R. R., & Holt, K. E. (2019). Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Research , 8, 2138. doi: 10.12688/f1000research.21782.1.
- Wick, R. R., Judd, L. M., Gorrie, C. L., & Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Computational Biology , 13(6), 1–22. doi: 10.1371/journal.pcbi.1005595.
- Wick, R. R., Schultz, M. B., Zobel, J., & Holt, K. E. (2015). Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics , 31(20), 3350–3352. doi: 10.1093/bioinformatics/btv383.
- Willmann, M., Bezdan, D., Zapata, L., Susak, H., Vogel, W., Schröppel, K., … Peter, S. (2015). Analysis of a long-term outbreak of XDR Pseudomonas aeruginosa: A molecular epidemiological study. Journal of Antimicrobial Chemotherapy , 70, 1322–1330. doi: 10.1093/jac/dku546.
- Wu, D., Hugenholtz, P., Mavromatis, K., Pukall, R., Dalin, E., Ivanova, N. N., … Eisen, J. A. (2009). A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature , 462(7276), 1056–1060. doi: 10.1038/nature08656.
- Zhu, C., Miller, M., Marpaka, S., Vaysberg, P., Rühlemann, M. C., Wu, G., … Bromberg, Y. (2017). Functional sequencing read annotation for high precision microbiome analysis. Nucleic Acids Research , 46(4), e23–e23. doi: 10.1093/nar/gkx1209.
Corrections
In this publication, author's conflict of interest and data availability statement have been added.
The current version online now includes this information and may be considered the authoritative version of record.
Citing Literature
Number of times cited according to CrossRef: 61
- Manh Hung Hoang, Vu Hoang, Van-Vinh Le, Using Deep Learning for the Taxonomic Classification of Microbial Sequences, Journal of Technical Education Science, 10.54644/jte.2024.1521, 19 , 1, (8-14), (2024).
- Leandro Patiño, Andrea Denisse Benítez, Andrés Carrazco-Montalvo, Mary Regato-Arrata, Genomics for Arbovirus Surveillance: Considerations for Routine Use in Public Health Laboratories, Viruses, 10.3390/v16081242, 16 , 8, (1242), (2024).
- Saif Faraj, Ivana R. Sequeira-Bisson, Louise Lu, Jennifer L. Miles-Chan, Michael Hoggard, Daniel Barnett, Amber Parry-Strong, Meika Foster, Jeremy D. Krebs, Sally D. Poppitt, Michael W. Taylor, Akarsh Mathrani, Effect of a Higher-Protein Nut versus Higher-Carbohydrate Cereal Enriched Diet on the Gut Microbiomes of Chinese Participants with Overweight and Normoglycaemia or Prediabetes in the Tū Ora Study, Nutrients, 10.3390/nu16121971, 16 , 12, (1971), (2024).
- Brian P. Bourke, Robert J. Dusek, Koray Ergunay, Yvonne-Marie Linton, Sergei V. Drovetski, Viral pathogen detection in U.S. game-farm mallard (Anas platyrhynchos) flags spillover risk to wild birds, Frontiers in Veterinary Science, 10.3389/fvets.2024.1396552, 11 , (2024).
- Seth Commichaux, Tu Luan, Harihara Subrahmaniam Muralidharan, Mihai Pop, Database size positively correlates with the loss of species-level taxonomic resolution for the 16S rRNA and other prokaryotic marker genes, PLOS Computational Biology, 10.1371/journal.pcbi.1012343, 20 , 8, (e1012343), (2024).
- Faizan Saleem, Enze Li, Thomas A. Edge, Kevin L. Tran, Herb E. Schellhorn, Identification of potential microbial risk factors associated with fecal indicator exceedances at recreational beaches, Environmental Microbiome, 10.1186/s40793-024-00547-8, 19 , 1, (2024).
- Yabing Li, Brijen Miyani, Russell A. Faust, Randy E. David, Irene Xagoraraki, A broad wastewater screening and clinical data surveillance for virus-related diseases in the metropolitan Detroit area in Michigan, Human Genomics, 10.1186/s40246-024-00581-0, 18 , 1, (2024).
- Adrian Wallner, Livio Antonielli, Ouiza Mesguida, Patrice Rey, Stéphane Compant, Genomic diversity in Paenibacillus polymyxa: unveiling distinct species groups and functional variability, BMC Genomics, 10.1186/s12864-024-10610-w, 25 , 1, (2024).
- Jonathon B. Van Gray, Paul Ayayee, Examining the impacts of salt specificity on freshwater microbial community and functional potential following salinization, Environmental Microbiology, 10.1111/1462-2920.16628, 26 , 5, (2024).
- Harry T. Child, Lucy Wierzbicki, Gabrielle R. Joslin, Richard K. Tennant, Comparative evaluation of soil DNA extraction kits for long read metagenomic sequencing, Access Microbiology, 10.1099/acmi.0.000868.v3, 6 , 9, (2024).
- Elias Broman, Markus Olsson, Adele Maciute, Daniel Donald, Christoph Humborg, Alf Norkko, Tom Jilbert, Stefano Bonaglia, Francisco J A Nascimento, Biotic interactions between benthic infauna and aerobic methanotrophs mediate methane fluxes from coastal sediments, The ISME Journal, 10.1093/ismejo/wrae013, 18 , 1, (2024).
- Alexander Van Uffelen, Andrés Posadas, Nancy H. C. Roosens, Kathleen Marchal, Sigrid C. J. De Keersmaecker, Kevin Vanneste, Benchmarking bacterial taxonomic classification using nanopore metagenomics data of several mock communities, Scientific Data, 10.1038/s41597-024-03672-8, 11 , 1, (2024).
- Yabing Li, Brijen Miyani, Kevin L. Childs, Shin-Han Shiu, Irene Xagoraraki, Effect of wastewater collection and concentration methods on assessment of viral diversity, Science of The Total Environment, 10.1016/j.scitotenv.2023.168128, 908 , (168128), (2024).
- Brijen Miyani, Yabing Li, Heidy Peidro Guzman, Ruben Kenny Briceno, Sabrina Vieyra, Rene Hinojosa, Irene Xagoraraki, Bioinformatics-based screening tool identifies a wide variety of human and zoonotic viruses in Trujillo-Peru wastewater, One Health, 10.1016/j.onehlt.2024.100756, 18 , (100756), (2024).
- Ya-nan Wang, Qingzhao Wang, Yahui Li, Bingpeng Wang, Huawei Wang, Yingjie Sun, Jun Wu, Weihua Li, Rongxing Bian, Liejun Chen, Insights into bioaerosol contamination in the process of mineralized refuse mining: Microbial aerosolization behavior and potential pathogenicity, Journal of Hazardous Materials, 10.1016/j.jhazmat.2024.136142, (136142), (2024).
- Yu He, Yongzhe Li, Yue Pan, Jianying Shang, Weimin Sun, Meng Wang, Hao Fan, Robert A. Sanford, Na Wei, Shuming Peng, Daihong Xie, Weiguang Zhang, Shulin Chen, Yong Liu, Zhou Jiang, Yongguang Jiang, Yidan Hu, Shuyi Li, Na Hu, Yiran Dong, Liang Shi, Intimate microbe-water-mineral interactions mediate alkalization in the pyroxene-rich iron ore mines in Panxi area, Southwest China, Journal of Hazardous Materials, 10.1016/j.jhazmat.2024.136127, 480 , (136127), (2024).
- Ying Liu, Maozhen Han, Fengmin Li, Na Zhang, Shaoyong Lu, Xiaohui Liu, Fengchang Wu, Performance and mechanism of SMX removal by an electrolysis-integrated ecological floating bed at low temperatures: A new perspective of plant activity, iron plaque, and microbial functions, Journal of Hazardous Materials, 10.1016/j.jhazmat.2023.132802, 463 , (132802), (2024).
- Lauren F. Messer, Ruddy Wattiez, Sabine Matallana-Surget, A closer look at plastic colonisation: Prokaryotic dynamics in established versus newly synthesised marine plastispheres and their planktonic state, Environmental Pollution, 10.1016/j.envpol.2024.124479, 358 , (124479), (2024).
- Brian P. Bourke, Jader de Oliveira, Koray Ergunay, Yvonne-Marie Linton, Leveraging transcriptome sequence read archives for virus detection in wild and colony populations of triatomines (Hemiptera: Reduviidae: Triatominae), Archives of Virology, 10.1007/s00705-024-06130-3, 169 , 10, (2024).
- Hoseong Choi, Yeonhwa Jo, Hyunjung Chung, Soo Yeon Choi, Sang-Min Kim, Jin-Sung Hong, Bong Choon Lee, Won Kyong Cho, Investigating Variability in Viral Presence and Abundance across Soybean Seed Development Stages Using Transcriptome Analysis, Plants, 10.3390/plants12183257, 12 , 18, (3257), (2023).
- Fengcheng Song, Yishu Xu, Peng Peng, Hongxu Li, Ranxi Zheng, Haolin Zhang, Yingying Han, Qiang Weng, Zhengrong Yuan, Seasonal Changes in the Structure and Function of Gut Microbiota in the Muskrat (Ondatra zibethicus), Metabolites, 10.3390/metabo13020248, 13 , 2, (248), (2023).
- Nadim Ahmad, Manfred Ritz, Anjuli Calchera, Jürgen Otte, Imke Schmitt, Thomas Brueck, Norbert Mehlmer, Biosynthetic Potential of Hypogymnia Holobionts: Insights into Secondary Metabolite Pathways, Journal of Fungi, 10.3390/jof9050546, 9 , 5, (546), (2023).
- Rubén Martín-Blázquez, Mohammed Bakkali, Mercedes Ruiz-Estévez, Manuel A. Garrido-Ramos, Comparison between the Gametophyte and the Sporophyte Transcriptomes of the Endangered Fern Vandenboschia speciosa, Genes, 10.3390/genes14010166, 14 , 1, (166), (2023).
- Nguyen Nam, Hoang Do, Kieu Loan Trinh, Nae Lee, Metagenomics: An Effective Approach for Exploring Microbial Diversity and Functions, Foods, 10.3390/foods12112140, 12 , 11, (2140), (2023).
- Marat Kumar, Kobey Karamendin, Zhanara Mazhibayeva, Yermukhammet Kassymbekov, Temirlan Sabyrzhan, Kuanysh Isbekov, Saule Assylbekova, Aidyn Kydyrmanov, Evaluation of the Viral Diversity of Artemia Cysts from Saline Lakes in Kazakhstan Using Viral Metagenomics Analysis, Fishes, 10.3390/fishes8100487, 8 , 10, (487), (2023).
- Vahid J. Javaran, Abdonaser Poursalavati, Pierre Lemoyne, Dave T. Ste-Croix, Peter Moffett, Mamadou L. Fall, NanoViromics: long-read sequencing of dsRNA for plant virus and viroid rapid detection, Frontiers in Microbiology, 10.3389/fmicb.2023.1192781, 14 , (2023).
- Ruijie Xu, Sreekumari Rajeev, Liliana C. M. Salvador, The selection of software and database for metagenomics sequence analysis impacts the outcome of microbial profiling and pathogen detection, PLOS ONE, 10.1371/journal.pone.0284031, 18 , 4, (e0284031), (2023).
- Hong Li, Hao Wang, Huimin Ju, Jinquan Lv, Shixing Yang, Wen Zhang, Hongyan Lu, Comparison of gut viral communities in children under 5 years old and newborns, Virology Journal, 10.1186/s12985-023-02013-2, 20 , 1, (2023).
- K. Swapna Kumari, Sangita Dixit, Mahendra Gaur, Dibyajyoti Uttameswar Behera, Patitapabana Das, Enketeswara Subudhi, Shakti Rath, Comparative oral metagenome insight into acute and chronic root canal infections, Journal of Bio-X Research, 10.1097/JBR.0000000000000147, 06 , 04, (117-123), (2023).
- Anurag Chaturvedi, Xiaojing Li, Vignesh Dhandapani, Hollie Marshall, Stephen Kissane, Maria Cuenca-Cambronero, Giovanni Asole, Ferriol Calvet, Marina Ruiz-Romero, Paolo Marangio, Roderic Guigó, Daria Rago, Leda Mirbahai, Niamh Eastwood, John K Colbourne, Jiarui Zhou, Eamonn Mallon, Luisa Orsini, The hologenome of Daphnia magna reveals possible DNA methylation and microbiome-mediated evolution of the host genome , Nucleic Acids Research, 10.1093/nar/gkad685, 51 , 18, (9785-9803), (2023).
- Martijn J T N Timmermans, Hari Prabha, Stephen Kett, Wolbachia and Spiroplasma endosymbionts in the Anurida maritima (Collembola) species group , Evolutionary Journal of the Linnean Society, 10.1093/evolinnean/kzad001, 2 , 1, (2023).
- Anupam Gautam, Wenhuan Zeng, Daniel H Huson, MeganServer: facilitating interactive access to metagenomic data on a server, Bioinformatics, 10.1093/bioinformatics/btad105, 39 , 3, (2023).
- Rajveer Singh, Shivani Chandel, Arijit Ghosh, Anupam Gautam, Daniel H. Huson, V. Ravichandiran, Dipanjan Ghosh, Easy efficient HDR-based targeted knock-in in Saccharomyces cerevisiae genome using CRISPR-Cas9 system , Bioengineered, 10.1080/21655979.2022.2162667, 13 , 6, (14857-14871), (2023).
- Zijun Yang, Hao Wang, Shixing Yang, Xiaochun Wang, Quan Shen, Likai Ji, Jian Zeng, Wen Zhang, Haiyan Gong, Tongling Shan, Virome diversity of ticks feeding on domestic mammals in China, Virologica Sinica, 10.1016/j.virs.2023.02.001, 38 , 2, (208-221), (2023).
- Teija Ojala, Aino-Elina Häkkinen, Esko Kankuri, Matti Kankainen, Current concepts, advances, and challenges in deciphering the human microbiota with metatranscriptomics, Trends in Genetics, 10.1016/j.tig.2023.05.004, 39 , 9, (686-702), (2023).
- Beidi Chen, Jiabao Cao, Wei Liu, Yuqing Zhang, Yudong Liu, Min Wang, Fei Xiao, Jie Ma, Jun Wang, Xuan Zhang, Disturbed gut virome with potent interferonogenic property in systemic lupus erythematosus, Science Bulletin, 10.1016/j.scib.2023.01.021, 68 , 3, (295-304), (2023).
- Engy Mahmoud, Amro Hanora, Salah Abdalla, Ali A. Abdelrahman Ahmed, Samira Zakeer, Shotgun metagenomic analysis of bacterial symbionts associated with “Chromodoris quadricolor” mantle, Marine Genomics, 10.1016/j.margen.2023.101030, 69 , (101030), (2023).
- Adrián Alvarez-Molina, José F. Cobo-Díaz, Elena A. Alexa, Fiona Crispie, Miguel Prieto, Mercedes López, Paul D. Cotter, Avelino Alvarez-Ordóñez, Sequencing-based analysis of the microbiomes of Spanish food processing facilities reveals environment-specific variation in the dominant taxa and antibiotic resistance genes, Food Research International, 10.1016/j.foodres.2023.113442, 173 , (113442), (2023).
- Peng Lu, Heli Shi, Jiemeng Tao, Jingjing Jin, Sujie Wang, Qingxia Zheng, Pingping Liu, Bikun Xiang, Qiansi Chen, Yalong Xu, Zefeng Li, Jun Tan, Peijian Cao, Metagenomic insights into the changes in the rhizosphere microbial community caused by the root-knot nematode Meloidogyne incognita in tobacco, Environmental Research, 10.1016/j.envres.2022.114848, 216 , (114848), (2023).
- Yu He, Jie Pan, Dongmei Huang, Robert A. Sanford, Shuming Peng, Na Wei, Weimin Sun, Liang Shi, Zhou Jiang, Yongguang Jiang, Yidan Hu, Shuyi Li, Yongzhe Li, Meng Li, Yiran Dong, Distinct microbial structure and metabolic potential shaped by significant environmental gradient impacted by ferrous slag weathering, Environment International, 10.1016/j.envint.2023.108067, 178 , (108067), (2023).
- Bhagya Hathurusinghe, D.K.N.G. Pushpakumara, Pradeepa C.G. Bandaranayake, Unveiling the possible floral visitors and invisible pollination networks from Deep RNA-seq Profile, Ecological Genetics and Genomics, 10.1016/j.egg.2023.100178, 28 , (100178), (2023).
- Elias Broman, Mohanad Abdelgadir, Stefano Bonaglia, Sara C. Forsberg, Johan Wikström, Jonas S. Gunnarsson, Francisco J. A. Nascimento, Sara Sjöling, Long-Term Pollution Does Not Inhibit Denitrification and DNRA by Adapted Benthic Microbial Communities, Microbial Ecology, 10.1007/s00248-023-02241-7, 86 , 4, (2357-2372), (2023).
- Hong Thanh Pham, Van Hoai Tran, Van Vinh Le, imMeta: An Incremental Sub-graph Merging for Feature Extraction in Metagenomic Binning, Intelligence of Things: Technologies and Applications, 10.1007/978-3-031-46573-4_20, (214-223), (2023).
- Youngchul Kim, Bioinformatic and Statistical Analysis of Microbiome Data, Statistical Genomics, 10.1007/978-1-0716-2986-4_10, (183-229), (2023).
- Nadim Ahmad, Manfred Ritz, Anjuli Calchera, Jürgen Otte, Imke Schmitt, Thomas Brueck, Norbert Mehlmer, Biosynthetic gene cluster synteny: Orthologous polyketide synthases in Hypogymnia physodes, Hypogymnia tubulosa, and Parmelia sulcata, MicrobiologyOpen, 10.1002/mbo3.1386, 12 , 5, (2023).
- Elias Broman, Rinti Barua, Daniel Donald, Florian Roth, Christoph Humborg, Alf Norkko, Tom Jilbert, Stefano Bonaglia, Francisco J. A. Nascimento, No evidence of light inhibition on aerobic methanotrophs in coastal sediments using eDNA and eRNA, Environmental DNA, 10.1002/edn3.441, 5 , 4, (766-781), (2023).
- Alexander Lopez‐Roblero, David José Martínez Cano, Elia Diego‐García, Griselda Karina Guillén‐Navarro, Pavel Iša, Eugenia Zarza, Metagenomic analysis of plant viruses in tropical fresh and wastewater, Environmental DNA, 10.1002/edn3.416, 6 , 1, (2023).
- Melissa T. R. Hawkins, Mary Faith C. Flores, Michael McGowen, Arlo Hinckley, A comparative analysis of extraction protocol performance on degraded mammalian museum specimens, Frontiers in Ecology and Evolution, 10.3389/fevo.2022.984056, 10 , (2022).
- Kelly A. Speer, Melissa T. R. Hawkins, Mary Faith C. Flores, Michael R. McGowen, Robert C. Fleischer, Jesús E. Maldonado, Michael G. Campana, Carly R. Muletz-Wolz, A comparative study of RNA yields from museum specimens, including an optimized protocol for extracting RNA from formalin-fixed specimens, Frontiers in Ecology and Evolution, 10.3389/fevo.2022.953131, 10 , (2022).
- Jonathan Greenbaum, Xu Lin, Kuan-Jui Su, Rui Gong, Hui Shen, Jie Shen, Hong-Mei Xiao, Hong-Wen Deng, Integration of the Human Gut Microbiome and Serum Metabolome Reveals Novel Biological Factors Involved in the Regulation of Bone Mineral Density, Frontiers in Cellular and Infection Microbiology, 10.3389/fcimb.2022.853499, 12 , (2022).
- Thomas Grevesse, Céline Guéguen, Vera E. Onana, David A. Walsh, Degradation pathways for organic matter of terrestrial origin are widespread and expressed in Arctic Ocean microbiomes, Microbiome, 10.1186/s40168-022-01417-6, 10 , 1, (2022).
- Elias Broman, Dandan Izabel-Shen, Alejandro Rodríguez-Gijón, Stefano Bonaglia, Sarahi L. Garcia, Francisco J. A. Nascimento, Microbial functional genes are driven by gradients in sediment stoichiometry, oxygen, and salinity across the Baltic benthic ecosystem, Microbiome, 10.1186/s40168-022-01321-z, 10 , 1, (2022).
- Daniel Maddock, Dawn Arnold, Sandra Denman, Carrie Brady, Description of a novel species of Leclercia, Leclercia tamurae sp. nov. and proposal of a novel genus Silvania gen. nov. containing two novel species Silvania hatchlandensis sp. nov. and Silvania confinis sp. nov. isolated from the rhizosphere of oak, BMC Microbiology, 10.1186/s12866-022-02711-x, 22 , 1, (2022).
- Anupam Gautam, Hendrik Felderhoff, Caner Bağci, Daniel H. Huson, Using AnnoTree to Get More Assignments, Faster, in DIAMOND+MEGAN Microbiome Analysis, mSystems, 10.1128/msystems.01408-21, 7 , 1, (2022).
- Lotte J.U. Pronk, Marnix H. Medema, Whokaryote: distinguishing eukaryotic and prokaryotic contigs in metagenomes based on gene structure, Microbial Genomics, 10.1099/mgen.0.000823, 8 , 5, (2022).
- Induja Chandrakumar, Nick P. G. Gauthier, Cassidy Nelson, Michael B. Bonsall, Kerstin Locher, Marthe Charles, Clayton MacDonald, Mel Krajden, Amee R. Manges, Samuel D. Chorlton, BugSplit enables genome-resolved metagenomics through highly accurate taxonomic binning of metagenomic assemblies, Communications Biology, 10.1038/s42003-022-03114-4, 5 , 1, (2022).
- Lev G. Nemchinov, Brian M. Irish, Samuel Grinstead, Jonathan Shao, Paulo Vieira, Diversity of the virome associated with alfalfa (Medicago sativa L.) in the U.S. Pacific Northwest, Scientific Reports, 10.1038/s41598-022-12802-4, 12 , 1, (2022).
- Kazuyoshi Gotoh, Yoshihiko Sakaguchi, Haru Kato, Hayato Osaki, Yasutaka Jodai, Mitsutaka Wakuda, Akira Také, Shunji Hayashi, Eri Morita, Takehiko Sugie, Yoichiro Ito, Naoki Ohmiya, Fecal microbiota transplantation as therapy for recurrent Clostridioides difficile infection is associated with amelioration of delirium and accompanied by changes in fecal microbiota and the metabolome, Anaerobe, 10.1016/j.anaerobe.2021.102502, 73 , (102502), (2022).
- Hao Lin, Qingqing Guo, Zhiyong Wen, Songlin Tan, Jie Chen, Lijian Lin, Pengcheng Chen, Jianquan He, Jianbo Wen, Ye Chen, The multiple effects of fecal microbiota transplantation on diarrhea-predominant irritable bowel syndrome (IBS-D) patients with anxiety and depression behaviors, Microbial Cell Factories, 10.1186/s12934-021-01720-1, 20 , 1, (2021).
- Frank Maixner, Christian Mitterer, Heidi Y. Jäger, Mohamed S. Sarhan, Guido Valverde, Sebastian Lücker, Dario Piombino‐Mascali, Ildikó Szikossy, Erika Molnár, György Pálfi, Ildikó Pap, Giovanna Cipollini, Albert Zink, Linear polyacrylamide is highly efficient in precipitating and purifying environmental and ancient DNA, Methods in Ecology and Evolution, 10.1111/2041-210X.13772, 13 , 3, (653-667), (2021).
- Mosè Manni, Matthew R. Berkeley, Mathieu Seppey, Evgeny M. Zdobnov, BUSCO: Assessing Genomic Data Quality and Beyond, Current Protocols, 10.1002/cpz1.323, 1 , 12, (2021).

