Enzymatic padlock probe preparation
Kun Zhang, Chien-Ju Chen, Kian Kalhor
Abstract
This protocol accompanies the manuscript Mapping Human Tissues with Highly Multiplexed RNA in situ Hybridization (https://doi.org/10.1101/2023.08.16.553610). It protocol outlines the steps for the enzymatic amplification and preparation of padlock probes from an oligo pool. This strategy has a lower upfront cost compared to individual synthesis of probes and thus permits the use of tens of thousands of padlock probes per probe set. Additionally, the oligo pool serves as an unlimited source of padlock probes, as it can be expanded by PCR amplification upon need.
Before start
The main starting material is an oligo pool with probe-set specific amplification primers on the 5' and 3' ends. In the example below, AP1V7 and AP2V7 are the primer pair that specifically amplify our probe set of interest.
Steps
oligo pool pre-amplification
Resuspend the oligo pool in water to a concentration of 7.02 ng/uL. Dilute the pool 100 times and run PCR to amplify the brain probe set.
| A | B |
|---|---|
| 100x diluted oligo pool | 5 |
| 10uM AP1V7U primer | 2 |
| 10uM AP2V7 primer | 2 |
| 2x KAPA SYBR FAST qPCR master mix | 25 |
| ultrapure water | 16 |
PCR program:
95°C , 0h 0m 30s -> (95°C , 0h 0m 30s ->55°C 0h 0m 45s ->72°C 0h 0m 45s )x17 cycles ->72°C , 0h 2m 0s -> 15°C , hold
Purify the PCR product by QIAquick PCR purification kit and elute each QIAquick column with 50µL
Use Qubit with HS dsDNA kit to quantify the concentration and dilute the PCR product to 10nM by adding ultrapure water. This is called "10nM oligo pool 1st amplicon" in the following section.
PCR mass production
Prepare PCR production mix in a 15mL tube and add load 100µL to each well of a 96-well plate
| A | B | C |
|---|---|---|
| 10nM oligo pool 1st amplicon | 0.2 | 20 |
| 2x KAPA SYBR FAST qPCR master mix | 50 | 5000 |
| 100uM AP1V7U primer | 0.4 | 40 |
| 100uM AP2V7 primer | 0.4 | 40 |
| ultrapure water | 49 | 4900 |
run PCR program to amplify the oligo pool
98°C , 0h 1m 0s -> (98°C , 0h 0m 30s ->55°C 0h 0m 45s ->72°C 0h 0m 45s )x14 cycles ->72°C , 0h 2m 0s -> 4°C , hold
Pool the PCR production mix from the 96-well plate and prepare the ethanol precipitation mix in 5mL-tubes. (One 96-well plate can split into 8 5mL-tubes)
| A | B |
|---|---|
| PCR product | 1200 |
| 100% ethanol | 3000 |
| glycoblue coprecipitant | 4 |
| 3M NaOAc | 120 |
Mix the tubes by shaking and incubate them in a -80°C freezer for 1h 0m 0s.
Centrifuge the tubes at 4,000rpm, 4°C for 0h 30m 0s and discard the supernatant
Add 800µL to each 5mL tube and transfer the solution into another 1.5mL tube. (8 5mL-tubes transfer into 8 1.5mL tubes).
Centrifuge at 14,000rpm, 4°C for 0h 5m 0s and discard the supernatant.
Air-dry the DNA pallets on ice bucket in AirClean hood for 0h 10m 0s
Rehydrate DNA pallets with 50µL
Pool the DNA solutions and use 8 QIAquick columns to purify.
Use Nanodrop to quantify the concentration.
Aliquot 3uL for TBU gel check
Lambda Exo digestion
Prepare Lambda Exo digestion mix and split it into PCR tubes (100µL per tube). Adjust the volume of DNA amplicon and ultrapure water so that the DNA amplicon per tube is around 5µg
| A | B | C |
|---|---|---|
| DNA amplicon | 25 | 400 |
| 10x Lambda Exo buffer | 10 | 160 |
| Lambda Exo (5U/uL) | 10 | 160 |
| ultrapure water | 55 | 880 |
Run PCR program: 37°C ,2h 0m 0s ->4°C ,hold
Use 16 ssDNA/RNA Zymo-spin columns to purify the Lambda Exo-digested products and elute the columns with 50µL
Use Nanodrop to quantify the concentration.
Aliquot 3uL for TBU gel check
USER digestion
Prepare USER digestion mix and split into PCR tubes (80µL per tube). Adjust the volume of ssDNA and ultrapure water so that the ssDNA per tube is less than 2µg .
| A | B | C |
|---|---|---|
| ssDNA | 66.7 | 800 |
| USER (1U/uL) | 5 | 60 |
| 10x DpnII buffer | 8 | 96 |
| ultrapure water | 0.3 | 4 |
Run PCR program: 37°C ,3h 0m 0s ->4°C ,hold.
Aliquot 3uL for TBU gel check
DpnII digestion
Prepare DpnII oligo mix and add 15µL to each PCR tube
| A | B | C |
|---|---|---|
| 10x DpnII buffer | 2 | 24 |
| 100uM RE_DpnII_V7N primer | 5 | 60 |
| ultrapure water | 8 | 96 |
Run PCR program: 94°C ,0h 2m 0s -> 37°C ,0h 3m 0s -> 4°C , hold
Add 5µL to each PCR tube (total volume is 100µL per tube) and mix by pipetting.
Run PCR program:37°C ,5h 0m 0s ->4°C , hold
Use ssDNA/RNA Zymo-spin columns to purify each PCR tube and elute each column with 50µL
Use Nanodrop to quantify the concentration
Aliquot 3uL for TBU gel check
TBU gel check (optional)
Pre-run one 12-well TBU-gel for at least 0h 10m 0s
Prepare TBU gel check samples
| A | B | C | D |
|---|---|---|---|
| 10 bp DNA ladder | 1 | 4 | 5 |
| 1: PCR product | 1 | 4 | 5 |
| 2: Lambda Exo | 1 | 4 | 5 |
| 3: Lambda Exo +USER | 1 | 4 | 5 |
| 4: Lambda Exo +USER +DpnII | 1 | 4 | 5 |
Notes: If PCR product and Lambda Exo sample look overloaded, dilute the sample by 10-20 times before TBU gel check.
Incubate TBU gel check samples at 70°C for0h 5m 0s and immediately put on ice for 0h 5m 0s
Flush each well with TBE running buffer to remove excessive urea in the 12-well gel
Load the TBU gel check samples to the 12-well TBU gel
Run gel electrophoresis at 220-240 V for 0h 20m 0s
Example TBU gel for a padlock probe set with the final size of 150nt:
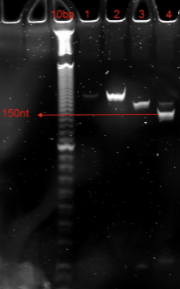
Size selection
Pre-run 1-well TBU gels for at least 0h 10m 0s
Prepare enzyme-digested probes: add 180µL to 180µL
Incubate the samples at 70°C for 0h 5m 0s and immediately put on ice for 0h 5m 0s
Flush each well with TBE buffer to remove excessive urea in each well.
Load samples to 1-well TBU gel.
Notes: Cap the amount of enzyme-digested probes per 1-well TBU gel at 2µg to avoid overloading and add the equivalent volume of 2x Novex buffer. The rest of the enzyme-digested probes can be stored at -20°C freezer for the next round of size selection.
Run gel electrophoresis at 220-240 V for 0h 20m 0s
Cut enzyme-digested probes at the right size and transfer the cut-out gels to 0.5mL tubes (with holes at the bottom) on top of 2mL tubes
Centrifuge the tubes at 15,000rpm for 0h 5m 0s to shred gels
Transfer the remaining gel in the 0.5mL tubes to the 2 mL tubes
Add 450µL to each 2mL tubes and mix the tubes by shaking
Freeze and thaw the solution in the -80°C freezer, and vortex the tubes in a 37°C incubator for 2h 0m 0s
Centrifuge the tubes at 15,000rpm, Room temperature for 0h 3m 0s
Transfer the clear supernatant to Nanosep columns and centrifuge at 15,000 rpm, Room temperature for 0h 3m 0s
Repeat Nanosep filtering once and transfer the flow-through to 1.5 mL tubes
Ethanol precipitation
Add 1mL , 1.4µL , 45µL to each tube and mix by shaking
Precipitate probes in a -80°C freezer for at least 1h 0m 0s
Centrifuge at 10,000 rpm, 4°C for 0h 30m 0s and discard the supernatant
Wash the pallets with 750µL and centrifuge at 14,000rpm at 4°C for 0h 5m 0s
Discard the supernatant and air-dry the pallets in AirClean hood for 0h 10m 0s
Rehydrate the pallets with 10µL per tube and pool the probe solution together
Use Qubit with ssDNA kit to quantify the concentration of probe solution

