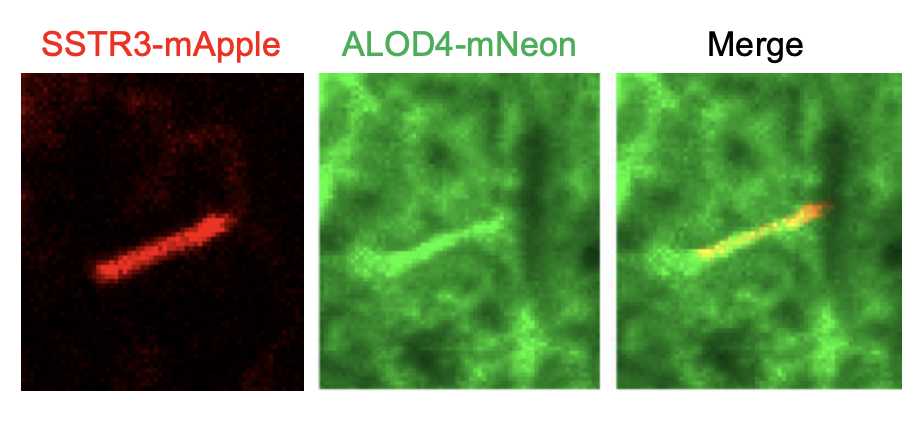
Detection of accessible cholesterol in primary cilia using purified His-ALOD4-mNeon in 3T3 Fibroblasts
Suzanne R Pfeffer, Ayan Adhikari, Sreeja V Nair
Abstract
There exist at least three different pools of cholesterol in the plasma membrane: the essential pool, the sphingomyelin-sequestered pool, and the accessible pool (Radhakrishnan et al, 2020). Ciliary Hedgehog signaling is regulated by the accessible pool of cholesterol (Kinnebrew et al, 2019), and His-mNeon-FLAG-ALOD4, a toxin-based probe, can be used to visualize this accessible pool. Here, we present a method for staining and measuring the amount of accessible cholesterol on cilia in 3T3 fibroblasts stably expressing Somatostatin Receptor (SSTR3)-mApple, a ciliary marker. The protocol described here is based upon previously established methods (Kinnebrew et al., 2019; Johnson and Radhakrishnan, 2021) and has also been used successfully to label cilia in RPE cells.
Steps
His-mNeon ALOD4 purification
Make a 50mL starter culture of BL21 DE3 Rosetta plysS cells expressing pRSET+ His-mNeon-FLAG-ALOD4, starting from a fresh colony picked from freshly transformed plates containing carbenicillin (100μg/ml) and chloramphenicol (34μg/ml). Grow with shaking in an Erlenmeyer flask at 37°C.
Day 2. Using a 6 liter flask, prepare 2 X 2 liters LB by mixing 50g LB powder with 2L water. Autoclave for 0h 20m 0s using a liquid cycle. Allow the media to cool to Room temperature
Add carbenicillin(100µg)and chloramphenicol (34µg) to each flask and shake at 37°C until an OD 0.5-0.6 is reached. This usually takes ~2.5 hr.
Transfer flask to an 18°C shaker and equilibrate temperature for 10-20 min.
Add IPTG to achieve a concentration of 0.3millimolar (mM)
Induce expression overnight with shaking at 180rpm at 18°C
Day 3. Transfer bacterial culture to 4, 1L centrifuge bottles.
Pellet bacteria by spinning at 4000 RPM for 20 min 4°C; discard supernatant.
Add 1 Roche protease inhibitor tablet to 50mL of lysis buffer, rotate at 4°C to dissolve.
Set up a ~200mL beaker with a stir bar at 4°C in the cold room.
Resuspend pelleted bacteria On ice or in cold room using ~10mL of lysis buffer for each bottle.
Vortex the bottles to help resuspend pellet; transfer resuspended pellets to the beaker with a stir bar.
Take 40mL of this suspension to the Emulsiflex cell breakage device and have ready, 4 x 65ml polycarbonate ultracentrifuge tubes on ice for the homogenate from the next step. Pass 10mL lysis buffer through the Emulsiflex once at 0-30K PSI to equilibrate the machine. Pass suspension through the Emulsiflex once at ~50K PSI to lyse cells. Collect directly into polycarbonate tubes On ice.
Balance tubes and then spin at 40,000 RPM for 0h 45m 0s in a 45Ti rotor at 4°Cto pellet cell debris. Collect supernatant and filter through a 500mL, 0.45µm Millipore filter. Collect the filtered supernatant in a 100mL glass beaker On ice.
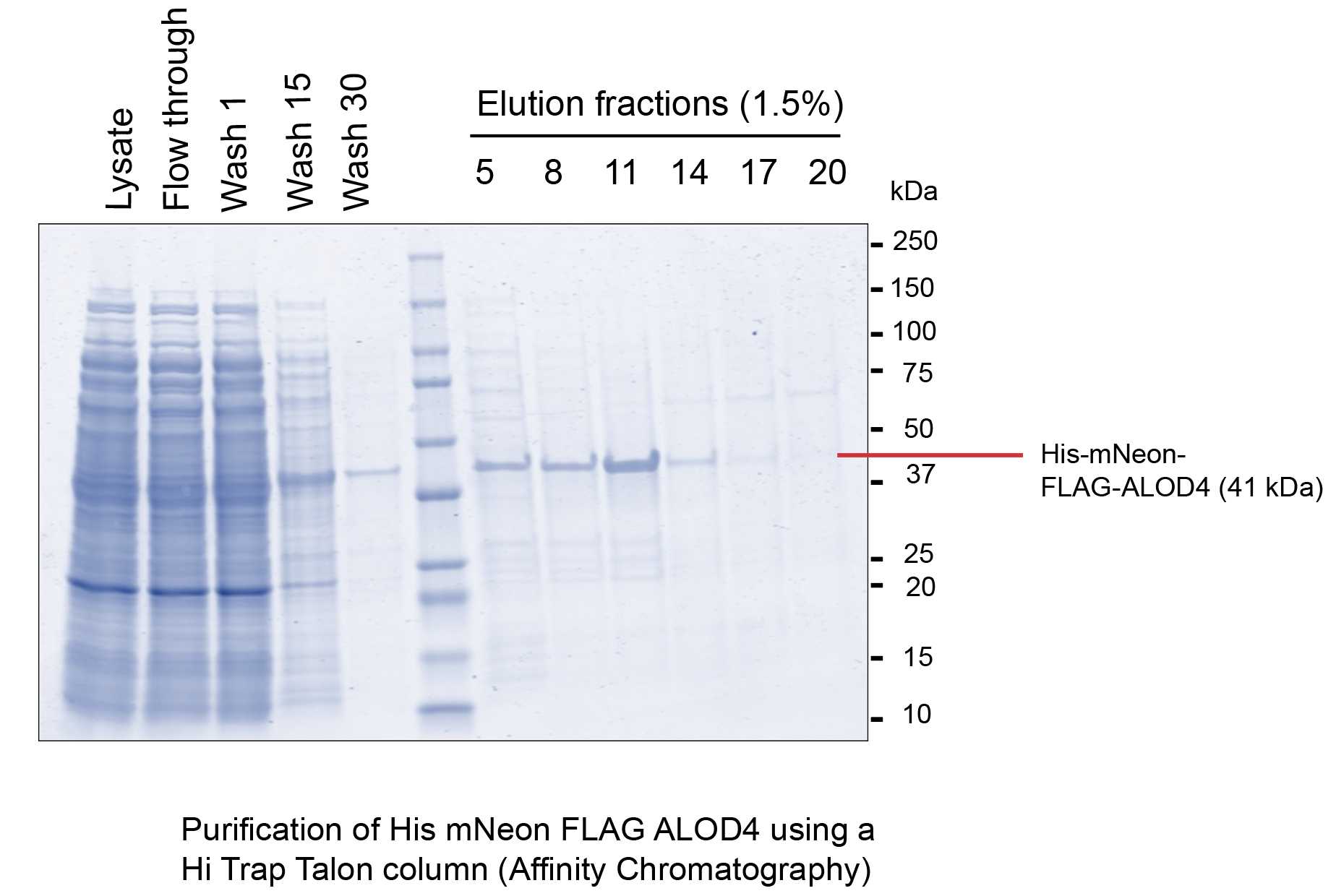
Take a 1mL HiTrap Talon column (Cytiva #28953766) and wash it with 10 column volumes (CV) of distilled, degassed water. Equilibrate column with 10CV lysis buffer (50mM HEPES pH 8, 500 mM NaCl, 5mM MgCl2, 0.5mM TCEP, 10% glycerol, EDTA-free protease tablet)
Run the lysate through the column. Collect the flow through.
Wash the column with 30CV wash buffer (50mM HEPES pH 8, 500 mM NaCl, 5mM MgCl2, 0.5mM TCEP, 10% glycerol, 20mM imidazole). Collect the wash fractions. Elute with an imidazole gradient of 100mM-500mM. Collect the elution fractions and analyze them by SDS PAGE.
Pool the fractions having the desired protein and buffer exchange the pool using a PD-10 desalting column (Cytiva #17085101) into the storage buffer (50mM HEPES pH 8, 150mM NaCl, 5mM MgCl2, 0.5mM TCEP, 10% glycerol).
Cilia Staining
Seed 0.3 X 106 cells onto collagen-coated coverslips. Once attached, starve the cells in media without serum or in low serum for 24h 0m 0s to induce ciliogenesis.
Set up an incubation chamber for ALOD4-mNeon staining: place a metal tray (covered with a layer of parafilm) on ice for 20-30 minutes before paraformaldehyde (PFA) fixation.
Aspirate media and fix cells in 4% PFA in 1X PBS for 0h 10m 0s at Room temperature.
Wash the cells three times in 1X PBS; transfer the coverslips to the incubation chamber and incubate them for 1h 0m 0s in 4μM ALOD4-mNeon diluted in 1% BSA 1X PBS, sterilized by passage through a 0.2 μm filter.
Wash the cells twice in 1X PBS; fix the cells again in 2% PFA diluted in 1X PBS for 5 minutes On ice. Wash the cells again twice in 1X PBS.
Optional: stain with DAPI (1:1000) for 4 minutes on ice. Wash the cells twice in 1X PBS and mount them onto clean glass slides using 4 µl Mowiol. Air-dry the coverslips and proceed to imaging.