CODEX Sample Preparation and Staining Experiment Protocol
Kavya.Anjani, Miguel Rivera, Zoltanlaszik
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
CODEX system is the combination of an (1) oligo-nucleotide based antibody labeling-detection technique, (2) a microfluidics instrument coupled with an inverted microscope capable of whole slide scanning, and an (3) ImageJ-based analysis platform. This experimental setup allows the automation of immunofluorescence signal detection across several staining cycles of a single tissue section
This protocol describes detailed methods used in CODEX sample preparation and staining at the Laszik Lab.
Steps
Tissue Sample Preparation Prior to CODEX Staining
Gather necessary materials
Coat coverslips with poly-lysine.
Gently place 20 coverslips on the bottom of the glass beaker and slowly swirl the beaker to fan out the coverslips.
Fill the beaker halfway with Milli-Q water.
Place two sets of paper towels on the benchtop.
Remove the coverslips from the beaker, then separate and lay each piece face down on the first set of paper towels.
Invert each coverslip to dry the reverse side on the second set of paper towels.
Once the coverslips dry completely, collect them in a plastic petri dish to store for up to 2 months.
Add 70mL of poly-lysine solution to the beaker, making sure to submerge all coverslips.
To prevent evaporation, use the saran wrap to cover the beaker and seal the wrap with a rubber band.
STOPPING POINT- Incubate coverslips in poly-lysine solution for a minimum of 12h 0m 0s and up to one week at Room temperature.
After incubation, dispose of the poly-lysine solution into a proper waste container.
Fill the beaker halfway with Milli-Q water and swirl contents to rinse coverslips.
Let the beaker and coverslips sit for 0h 1m 0s.
Dispose of the water into the sink.
Repeat steps 2.5-2.8 for a total of 7 washes.
Cut and embed frozen tissues on poly-lysine coated coverslips.
Place poly-lysine coated coverslips in the cryostat chamber to equilibrate.
Cut frozen tissue slices with 5 µm - 10 µm thickness.
Gently place tissue slice in the center of the coverslip.
Place a gloved finger on the underside of the coverslip behind the tissue slice to melt the OCT for better adherence. Do not exceed 2 seconds.
Store tissue slices separated at-80°C for up to 6 months prior to staining.
Antibody Conjugation with CODEX-tags for Custom Panel
Gather necessary materials.
Calculate and obtain 50µg of each antibody that will be conjugated.
Label a 50kDA MWCO filter unit (both the filter device and the collection tube) for each antibody that will be conjugated.
Add 500µL of Filter Block Solution to the filter device.
Spin down the filter unit at 12000x g.
Discard all liquid (from both the filter device and the collection tube).
Add 50µg of antibody to the filter device.
Spin down the filter unit at 12000x g.
Prepare the Antibody Reduction Master Mix while spinning.
Discard flow through.
Add 260µL of Antibody Reduction Master Mix to each filter device.
Vortex solution in filter device for0h 0m 2s - 0h 0m 3s.
Incubate tube for 0h 30m 0s at -80Room temperature.
Spin down the filter unit at 12000x g.
Discard flow-through.
Add 450µL of Antibody Conjugation Solution to each filter device.
Spin down the filter unit at 12000x g.
Prepare the CODEX antibody tag solution while spinning.
Label a new tube for each CODEX antibody tag (one tag per antibody).
Add 210µL of Antibody Conjugation Solution to each tube in step a. Set aside.
Add 10µL of Milli-Q water to resuspend each CODEX antibody tag (retrieved from -20 just prior). Mix by pipetting.
Add the entire resuspended CODEX antibody tag to step b and mix by pipetting.
Discard flow-through.
Add the CODEX antibody tag solution prepared in step 21 to the corresponding filter device.
Close the lid and vortex the filter unit for 2-3 sec.
Incubate tube for 2h 0m 0s at -80Room temperature.
After 2h 0m 0s, set aside 5µL of the purified solution for QC.
Spin down the filter unit at 12000x g.
Discard flow-through.
Add 450µL of Antibody Purification Solution to each filter device.
Spin down the filter unit at 12000x g.
Discard flow-through.
Repeat steps 29-31 two more times for a total of three times.
There will be solution left in the filter device.
Label a new collection tube for each filter device.
Add 100µL Antibody Storage Solution to each filter device.
Invert the filter device into its newly labeled collection tube.
Spin down the filter unit at3000x g.
Transfer solution in the collection tube to a labeled 2 mL screw-top tube and store at 4°C for up to 1 year.
Tissue Staining
Gather necessary materials.
Prepare a humidity chamber.
Place wet paper towels at the bottom of an empty pipet tip box.
Fill the box with enough water to cover the towels completely.
Recover the pipet tip tray and lid after rinsing.
Prepare Drierite absorbent beads.
Fill an empty pipet tip box with Drierite beads (about 1~2 cm deep).
Prepare 10mL of acetone in a 50mL beaker for each tissue.
Remove the tissue sections from -80C and place the coverslips on Drierite absorbent beads.
Let the tissue sections dry for 0h 2m 0s at 4Room temperature.
Place the tissue samples face-up in the beakers containing acetone.
Incubate for 0h 10m 0s.
Remove the coverslips from acetone and place them in the humidity chamber to dry for 0h 2m 0s at 4Room temperature.
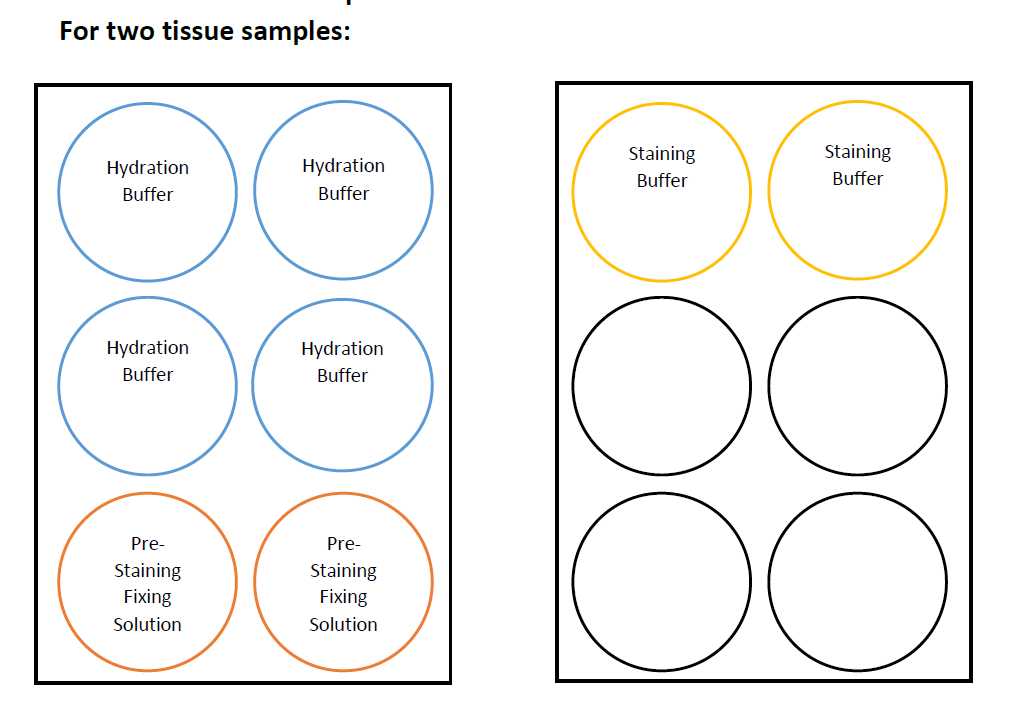
Hydrate tissue samples following the plate configuration.
Place the coverslips in the Hydration Buffer wells (dip 2-3 times before fully submerging the slide). Ensure tissue is facing up.
Incubate for 0h 2m 0s.
Place the coverslips in the next set of Hydration Buffer wells.
Incubate for0h 2m 0s.
Prepare Pre-Staining Fixing Solution while incubating tissue.
Place the coverslips in the Pre-Staining Fixing Solution wells.
Incubate for 0h 10m 0s at 4Room temperature.
Remove samples from Pre-Staining Fixing Solution and rinse the samples by dunking the coverslips 2-3 times each in the top and bottom Hydration Buffer wells.
Place the samples in the Staining Buffer wells and let equilibrate for 0h 20m 0s - 0h 30m 0s.
Prepare the antibody cocktail during equilibration.
To make CODEX Blocking Buffer for two samples, mix the following:
10µL Blocking Component N + 10µL Blocking Component G + 10µL Blocking Component J + 10µL Blocking Component S + 380µL Staining Buffer
Multiply the reagent volumes by the total number of samples that will be stained to get the total volume needed for the experiment.
Calculate the total antibody volume per antibody cocktail.
Total Ab volume = (# of antibodies) x (# of samples) x (Ab volume/sample)]
Calculate to CODEX Blocking Buffer needed per antibody cocktail.
CODEX Blocking Buffer volume needed to make 210 μL Ab Cocktail Solution/sample = CODEX Blocking Buffer volume - Total Ab volume
Combine the antibodies and the CODEX Blocking Buffer to make 210µL Antibody Cocktail Solution/tissue sample.
Remove the coverslips from Staining Buffer and place in the humidity chamber.
Quickly add 200µL of Antibody Cocktail Solution to each sample from the top corner of the coverslip.
Place lid back on the humidity chamber.
Incubate for 3h 0m 0s at 4Room temperature.
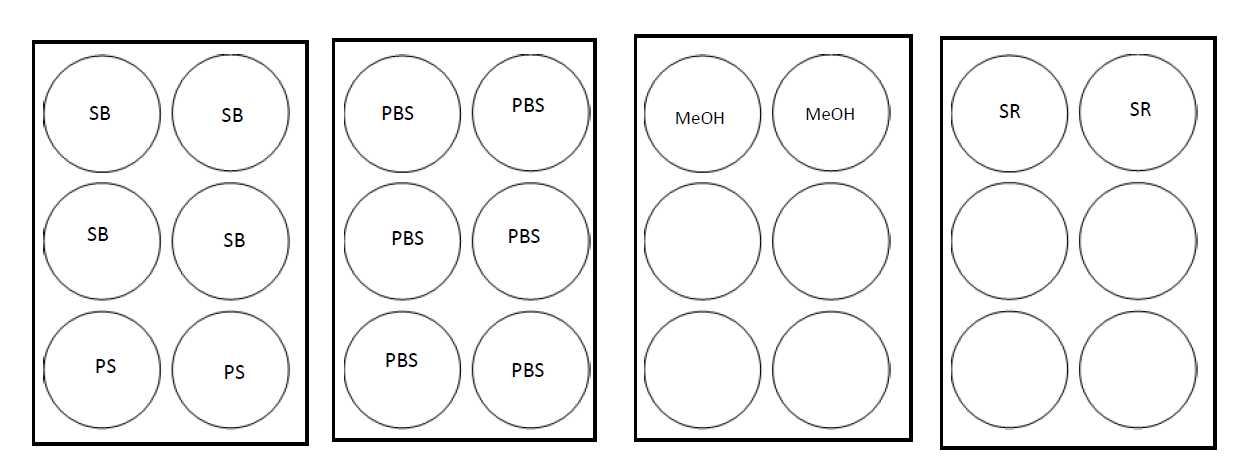
Wash the tissue samples following the plate configuration.
Place the coverslips in the Staining Buffer ( SB ) wells (dip 2-3 times before fully submerging the slide). Ensure tissue is facing up.
Remove the coverslips from 1x DPBS ( PBS ) and place in ice-cold methanol.
Incubate for 0h 5m 0s.
Bring plate containing 1x DPBS ( PBS ) to the ice bucket for quick transfer from methanol.
After incubation, rinse the samples by dunking the coverslips 2-3 times each in the 1x DPBS ( PBS ) wells for a total of 3 washes.
Incubate for 0h 2m 0s.
Place the coverslips in the next set of Staining Buffer ( SB ) wells.
Incubate for 0h 2m 0s.
Prepare Post-Staining Fixing Solution (PS) while incubating tissue.
For two samples, mix the following:
1mL 16% PFA + 9mL Storage Buffer
Multiply the PFA and buffer volumes by the total number of samples that will be stained to get the total volume needed from each reagent. Add 5mL of Post-Staining Fixing Solution to each well.
Place the coverslips in the Post-Staining Fixing Solution (PS) wells.
Incubate for 0h 10m 0s at 4Room temperature.
Prepare ice-cold methanol during incubation.
- Place a 6-well TC plate on ice.
- Pipet methanol up and down to equilibrate the serological pipet tip.
- Add 5 mL methanol to one well per sample.
Remove samples from Post-Staining Fixing Solution ( PS ) and rinse the samples by dunking the coverslips 2-3 times each in the 1x DPBS ( PBS ) wells for a total of 3 washes.
Rinse the humidity chamber if not previously washed.
Prepare the Final Fixative Solution. For 1-5 samples, mix the following:
1000µL 1x DPBS + 20µL CODEX Fixative Reagent
*20 μL of CODEX Fixative Reagent is one aliquot, thaw and spin down right before use.
Vortex gently to mix.
Remove coverslips from wells and place them in the humidity chamber.
Add 4Room temperature of Final Fixative Solution to the top corner of each sample.
Incubate for 0h 20m 0s.
Remove samples from the humidity chamber and rinse the samples by dunking the coverslips 2-3 times each in the 1x DPBS ( PBS ) wells for a total of 3 washes.
Place the coverslips in a new TC plate with 5mL Storage Buffer (SB) per sample in each well.
STOPPING POINT- use samples directly to run a CODEX experiment or store up to two weeks at 4C.
CODEX-tagged Dyes Cycle Prep
Gather necessary materials:
Determine the CODEX-tagged dyes combination for each running cycle. Each cycle may contain up to one HOECHST/DAPI dye, one FAM/FITC dye, one Cy3/TRITC dye, and one Cy5 dye.
Label an amber Eppendorf tube for each CODEX-tagged dyes combination.
Prepare the stock solution of CODEX Reporter Stock Solution.
For 5 wells (cycles) per plate, mix the following:
150µL 10x CODEX Buffer + 125µL Assay Reagent + 5µL Nuclear Stain
1220µLNuclease-free water
Double or triple the volume of each reagent if running 10 or 15 cycles per plate. Mix solution by gently inverting.
Add CODEX Reporter Stock Solution to each labeled amber Eppendorf tube.
| A | B | C | D |
|---|---|---|---|
| 3 CODEX-tagged dyes | 2 CODEX-tagged dyes | 1 CODEX-tagged dye | |
| CODEX Reporter Stock Solution Volume per tube | 235 μL | 240 μL | 245 μL |
Thaw each CODEX-tagged dye immediately before use.
Quickly spin down the CODEX-tagged dyes.
Add 5µL of each CODEX-tagged dye to its corresponding tube.
Mix contents of each tube by gently pipetting.
Transfer 245µL of solution from each tube to its designated well in the 96-well plate.
Prepare a strip of foil seal and cover all wells containing CODEX-tagged dyes.
STOPPING POINT- use dyes directly to run a CODEX experiment or store up to two weeks at 4C.
Running the CODEX instrument
Gather necessary materials.
Turn on the CODEX Instrument.
Remove the antibody-stained tissue section(s) adhered to the Poly-lysine coated coverslip and the sealed, pre-loaded 96-well plate containing the Reporter Master Mix solutions from 4C. Allow them to equilibrate to 4Room temperature for at least 0h 15m 0s.
Prepare 1X CODEX Assay Buffers in 600-2000 mL glass beakers.
| A | B | C | D | E | F | G | H | I |
|---|---|---|---|---|---|---|---|---|
| 5 CODEX cycles | 8 CODEX cycles | 10 CODEX cycles | 12 CODEX cycles | 15 CODEX cycles | 18 CODEX cycles | 20 CODEX cycles | 25 CODEX cycles | |
| 10X CODEX Buffer | 31 mL | 44 mL | 53.3 mL | 63.3 mL | 78.5 mL | 93.5 mL | 103.5 mL | 128.5 mL |
| Milli-Q water | 281 mL | 394 mL | 480.5 mL | 570.5 mL | 705.5 mL | 840.5 mL | 930.5 mL | 1155.5 mL |
| Total | 312 mL | 438 mL | 534 mL | 634 mL | 784 mL | 934 mL | 1034 mL | 1284 mL |
| DMSO | 236 mL | 318 mL | 386 mL | 454 mL | 556 mL | 658 mL | 726 mL | 896 mL |
1X CODEX Buffer
Set aside 7mL of 1x CODEX Buffer in a 15mL conical tube for later use.
Place generated 1x CODEX Buffer in the CODEX Buffer Bottle, labeled Bottle One and DMSO in the amber bottle, labeled Bottle Two.
Load the stage insert.
Untwist the knobs at the top of the stage insert to remove the lid with the fluidic lines.
Quickly place the second gasket on top of the sample coverslip.
Quickly place the lid on top of the stage insert.
Secure the lid by turning the knobs.
Dispense 700µL of 1X CODEX Buffer on the corner of the sample.
Remove any salts formed on the coverslip from drying using a kimwipe with Milli-Q water. Use a dry kimwipe to remove residual liquid.
Remove the old coverslip using the forceps, push the corners of the gaskets towards the center to release the seal between the coverslip and the gasket.
Gently remove the coverslip using the forceps.
If the coverslip breaks, remove all glass pieces and rinse with water.
Soak fresh gaskets in 1X CODEX Buffer or in Storage Buffer for a few seconds.
Place the first gasket inside the squared well at the center of the stage insert.
Gently tap with forceps to ensure it adheres to the stage insert surface.
Gently place the tissue sample on top of the gasket, making sure it is perfectly inserted and the tissue is facing upwards.
Quickly and gently tap the coverslip edges with the forceps to ensure it adheres to the gasket.
Stain the CODEX sample with HOECHST.
Thaw CODEX Nuclear Staining Solution.
Add 700µL of 1x CODEX Buffer.
Have the 7mL of 1x CODEX Buffer set aside previously within reach.
Prepare a 2mL Eppendorf tube for the Nuclear Stain Solution.
Remove the solution within the sample well.
Add 700µL of Nuclear Stain Solution to the sample.
Incubate for 0h 5m 0s at 4Room temperature. Cover it from light.
Remove the solution within the sample well.
Add 700 μL of 1x CODEX Buffer.
Repeat steps 83.7-83.8 for a total of 5 washes.
Launch the CODEX Instrument Management Software and enter cycle, marker id and exposure time information. Minimize the window.
Place the plate with the CODEX-tagged dyes in its designated place in the instrument.
Clean any dried buffer on the bottom of the coverslip with a wet kimwipe and dry off any residual liquid.
Turn on the Keyence scanner and load the cassette into the scanner.