Barcoded and targeted cDNA library preparation for Oxford Nanopore Technologies sequencing
Jonathan Mill, Aaron Jeffries, Rosemary A Bamford, Szi Kay Leung
Abstract
The NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (NEB) was adapted for the purpose of adding Oxford Nanopore Technologies (ONT) compatible barcodes during reverse transcription of RNA. This is a useful process for multiplexing low input samples for ONT transcriptome library preparation.
An optional step before ONT library preparation is the targeted enrichment of cDNA molecules using IDT hybridisation probes. Here we provide a protocol for this process based on the 'PacBio cDNA capture using IDT xGen Lockdown Probes' protocol.
Using this approach, up to 100 samples can be barcoded with individual ONT barcodes via reverse transcription. Samples can then be pooled together and a cDNA PCR amplification performed. This allows sufficient material for cDNA enrichment and/or ONT ligation sequencing library preparation.
Before start
Order the bespoke IDT primers and dilute to the correct concentration.
Steps
Bespoke barcoding of cDNA
This protocol is adapted from that provided with NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (NEB, E6420S). Input and component values have been modified from the original kit protocol.
Primer annealing for first strand synthesis
Prepare in a PCR tube on ice. A reaction mix of up to 7µL of total RNA (75ng ) is added to 1µL 2millimolar (mM) TSO-ONT primer, 1µL 10millimolar (mM) dNTPs (NEB) and made up to 9µL with nuclease-free H2O. Gently invert a few times and spin briefly.
Incubate the mixture in a thermocycler for 0h 5m 0s at 70°C with the heated lid at 105°C , then snap cool on ice for 0h 1m 0s .
Reverse Transcription (RT) and Template Switching
Meanwhile, prepare the reverse transcription mix which consists of 2.5µL NEBNext Single Cell RT buffer (lilac); 0.5µL NEBNext Template Switching Oligo (lilac); 1µL NEBNext Single Cell RT enzyme mix (lilac) and 1.5µL nuclease-free water. Mix by gentle inversion and spin briefly. Add 5.5µL of reverse transcription mix to each sample. Gently invert a few times and spin briefly.
Place the mix in a thermocycler (heated lid at 105°C ):
42°C for 1h 30m 0s
70°C for 0h 10m 0s
hold at 4°C .
cDNA amplification by PCR
Prepare the cDNA amplification mix which consists of 25µL NEBNext Single
Cell cDNA PCR Master Mix (orange);1µL NEBNext Single Cell cDNA PCR Primer (orange);0.25µL NEBNext Cell Lysis Buffer (10X) (white) and13.75µL nuclease-free water. Mix by gentle inversion and spin briefly. Add40µL of cDNA amplification mix to each sample. Gently invert a few times and spin briefly.
PCR cycling conditions:
Heated lid set to 105°C
Initial denaturation:
98°C for 0h 0m 45s
For 14 cycles:
98°C for 0h 0m 10s
65°C for 0h 12m 30s ,
72°C for 0h 3m 0s ,
Final extension:
72°C for 0h 5m 0s
hold at 4°C
Cleanup of Amplified cDNA
Bring Promega Pronex beads to Room temperature for 0h 30m 0s prior to use.
Spin tubes down briefly. 0.85X Promega Pronex beads are added. Gently invert a few times and spin briefly. Incubate at Room temperature on Hula mixer for 0h 5m 0s . Briefly spin tubes and place on magnet. Leave beads to settle for 0h 5m 0s . Perform 2x 80% EtOH washes. Amplified cDNA is eluted in 27µL 1X TE.
cDNA enrichment
We performed an adapted version of the 'PacBio cDNA Capture Using IDT xGen Lockdown Probes' protocol (Other Documentation - PacBio) and xGen Lockdown Panel.
Barcoded cDNA is equimolar pooled for a total of 1µg cDNA per capture reaction. Prepare in a 0.2mL PCR tube.
1µL of TSO blocker and 1µL of polyT blocker (poly(dT)30) oligonucleotides (both at 1millimolar (mM) ) were added to the pooled cDNA.
Dry the mixture using a DNA vacuum concentrator (set temperature to 45°C ). Do not overdry.
Add IDT 2X hybridisation buffer (8.5µL ), hybridisation buffer enhancer (2.7µL ) and nuclease free water (1.8µL ) to the dried-down sample. Replace the lid (do not cover the hole with tape). Mix the reaction by gently tapping the tube, followed by a quick spin.
Place the mixture on a 95°C thermocycler for 0h 10m 0s to denature the cDNA.
After allowing the mixture to cool briefly for 0h 2m 0s at Room temperature. 4µL of xGen Lockdown Panel is added for a total volume of 17µL. Probes should never be added while at 95°C .
After 0h 5m 0s at Room temperature, briefly spin the tubes and incubate in a thermocycler at 65°C overnight (lid temperature 100°C).
Prepare the wash buffers and Dynabeads M-270 Streptavidin beads provided in the xGen Lockdown Hybridisation and Wash kit and use as per instructions below. The xGen Lockdown Hybridisation and Wash kit is optimal for 3 months at -20°C and poor yields may occur if the kit has expired.
Prepare wash buffers
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| Wash buffer I | 10X | 40 µL | 360 µL | 400 µL | 1X |
| Wash buffer II | 10X | 20 µL | 180 µL | 200 µL | 1X |
| Wash buffer III | 10X | 20 µL | 180 µL | 200 µL | 1X |
| Stringent wash buffer | 10X | 50 µL | 450 µL | 500 µL | 1X |
| Bead wash buffer | 2X | 250 µL | 250 µL | 500 µL | 1X |
These volumes are for a single sample - scale up for multiple samples.
Preheat the following wash buffers to 65°C in a heat block or water bath for at least 0h 15m 0s :
200µL of 1X wash buffer I
400µL of 1X stringent wash buffer
Other reagents can be kept at Room temperature
Prepare the capture beads
- Allow the Dynabeads M-270 Streptavidin to warm to room temperature for
0h 30m 0sprior to use. - Mix the beads thoroughly by vortexing for
0h 0m 15s - For a single sample, aliquot
100µLbeads into a1.5mLLoBind tube. Scale up volume for multiple samples. - Place the LoBind tube in a magnetic rack. When the supernatant is clear, remove and discard the supernatant being careful not to disturb the beads. Any remaining traces of liquid will be removed with subsequent wash steps. Give it
0h 5m 0sto settle. The Dynabeads are "filmy" and slow to collect to the side of the tube. - While the LoBind tube is in the magnetic rack, add
200µLof 1X bead wash buffer. For multiple samples, wash with200µLx X samples. - Remove the tube from the magnetic rack and vortex until the beads are in solution.
- Quickly spin and place the LoBind tube back in the magnetic rack to collect the beads to the side of the tube. Once clear, remove and discard the liquid.
- Repeat steps 5-7 for a total of two washes.
- Resuspend by vortexing the beads in
100µLof 1X bead wash buffer. For multiple samples, scale up accordingly. - Place the tube in the magnetic rack to collect beads to the side of the tube. Once clear, remove and discard the supernatant.
- The washed beads are now ready to bind the captured DNA. Proceed immediately to the next step. Do not allow the capture beads to dry. Small amounts of residual bead wash buffer will not interfere with binding of DNA to the capture beads.
Bind cDNA to the capture beads
Transfer the 17µL hybridised probe/sample mixture to the washed capture beads.
Mix by tapping the tube until the sample is homogenous.
Incubate in a heat block set to 65°C for 0h 45m 0s or transfer the mix to a PCR tube and incubate in a thermocycler (heated lid set to 75°C ). Hand mix periodically by gently tapping the tube to keep the beads in suspension, every 0h 10m 0s .
Wash the captured cDNA
- The 1X wash buffer I and 1X stringent wash buffer should have been pre-heated to
65°C. - After the above incubation, remove the tube from the heat block and add
100µLpre-heated 1X wash buffer I. - Mix thoroughly by tapping the tube until the sample is homogenous.
- If using a PCR tube, transfer the sample to a
1.5mLLoBind tube (careful of bubbles). - Place the tube in a magnetic rack to collect the beads to the side of the tube. Remove and discard the liquid once clear.
- Remove the tube from the magnetic rack and add
200µLof 1X stringent wash buffer heated to65°C. Mix by tapping the tube until the sample is homogenous. Work quickly so that the temperature does not drop. - Incubate at
65°Cfor0h 5m 0s. - Repeat steps 5-7 for a total of two washes using 1X stringent wash buffer heated to
65°C. - Place the tubes in the magnetic rack to collect the beads to the side of the tube. Remove and discard the liquid once clear.
- Add
200µLofRoom temperature1X wash buffer I. Hand mix by gently tapping the tube, followed by a quick spin. - Place the tube in the magnetic rack to collect the beads to the side of the tube. Remove and discard the liquid once clear. It will clear quickly but allow
0h 1m 0s. - Add
200µLofRoom temperature1X wash buffer II. Hand mix by gently tapping the tube, followed by a quick spin. - Place the tube in the magnetic rack to collect the beads to the side of the tube. Remove and discard the liquid once clear. Allow
0h 5m 0s. - Add
200µLofRoom temperature1X wash buffer III. Hand mix by gently tapping the tube. Quick spin. - Place the tube in the magnetic rack to collect the beads to the side of the tube. Remove and discard the liquid once clear. Allow
0h 5m 0s. - Remove the tubes from the magnetic rack and add
50µLof 1x TE. - Store the beads plus captured samples at
-20°Cor proceed to the next step. It is not necessary to separate the beads from the eluted DNA.
Amplification of captured DNA sample
| A | B |
|---|---|
| Water | 104.5 µL |
| 10x LA PCR buffer | 20 µL |
| 2.5 mM dNTPs | 16 µL |
| 12 uM TSO amp primer | 8.3 µL |
| Takara LA Taq DNA polymerase | 1.2 µL |
| Captured library | 50 µL |
| Total | 200 µL |
PCR reaction mix
Split the PCR mix into two tubes, 100µL each.
Amplify using the following PCR protocol:
| A | B | C |
|---|---|---|
| 1 | 95 °C | 2 mins |
| 2 | 95 °C | 20 s |
| 3 | 68 °C | 10 mins |
| 4 | Repeat steps 2-3 for a total of 11 cycles | |
| 5 | 72 °C | 10 mins |
| 6 | 4 °C | hold |
After amplification, pool the 100µL reactions.
Post amplification clean up
Bring AMPure beads to Room temperature for 0h 30m 0s prior to use.
Spin tubes down briefly. 1X AMPure beads are added. Gently invert a few times and spin briefly. Incubate at Room temperature for 0h 10m 0s . Briefly spin tubes and place on magnet. Leave beads to settle for 0h 5m 0s . Perform 2x 80% EtOH washes. Amplified cDNA is eluted in 27µL 1X TE.
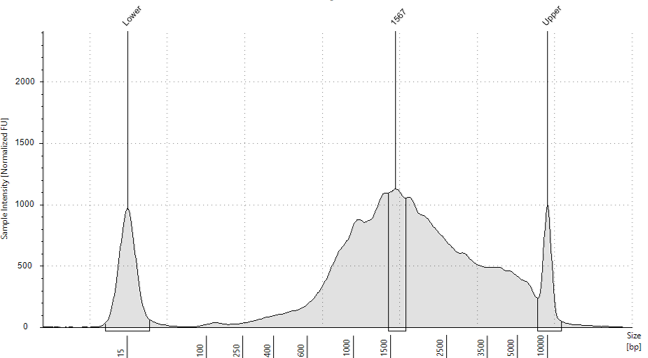
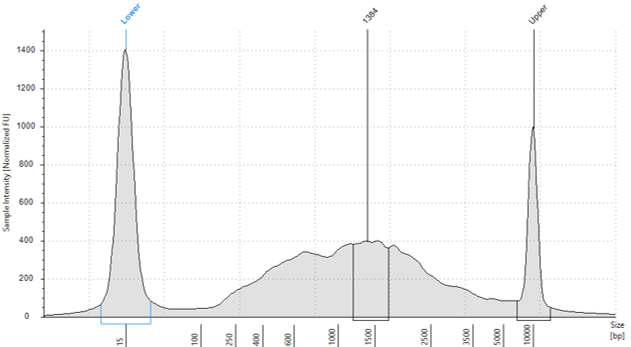
Quantification was determined using the Qubit DNA High sensitivity assay
(Invitrogen, UK) (1:5 dilution) and HS D5000 screentape (Agilent, UK) (1:5 dilution).
Library preparation and sequencing
Library preparation was performed using ONT’s ligation kit (SQK-LSK114). Follow supplier protocol, which also includes details for sequencing. Recommended library input is 100-200 fmol.