Amplicon sequencing on the MinION platform
Yoshiyuki Matsuo
Abstract
Amplicon sequencing on the MinION platform
Steps
Materials
KAPA2G Robust HotStart ReadyMix (2X) (KAPA BIOSYSTEMS, KK5701)* Inner primer (forward): 5'-TTTCTGTTGGTGCTGATATTGC - target-specific sequence -3' (User-supplied)
- Inner primer (reverse): 5'-ACTTGCCTGTCGCTCTATCTTC - target-specific sequence -3' (User-supplied)
- PCR Barcoding Kit (Oxford Nanopore Technologies, SQK-PBK004)
- AMPure XP (Beckman Coulter, A63880)
- 70% ethanol
- Elution buffer: 10 mM Tris-HCl pH 8.0 with 50 mM NaCl
- QuantiFluor ONE dsDNA System (Promega, E4871)
- Flow cell R9.4 (Oxford Nanopore Technologies, FLO-MIN106D)
- Flow Cell Priming Kit (Oxford Nanopore Technologies, EXP-FLP001)
Equipment
Thermal cycler* Gel electrophoresis device
- Magnetic rack
- Quantus Fluorometer (Promega, E6150)
- MinION sequencer (Oxford Nanopore Technologies)
Workflow
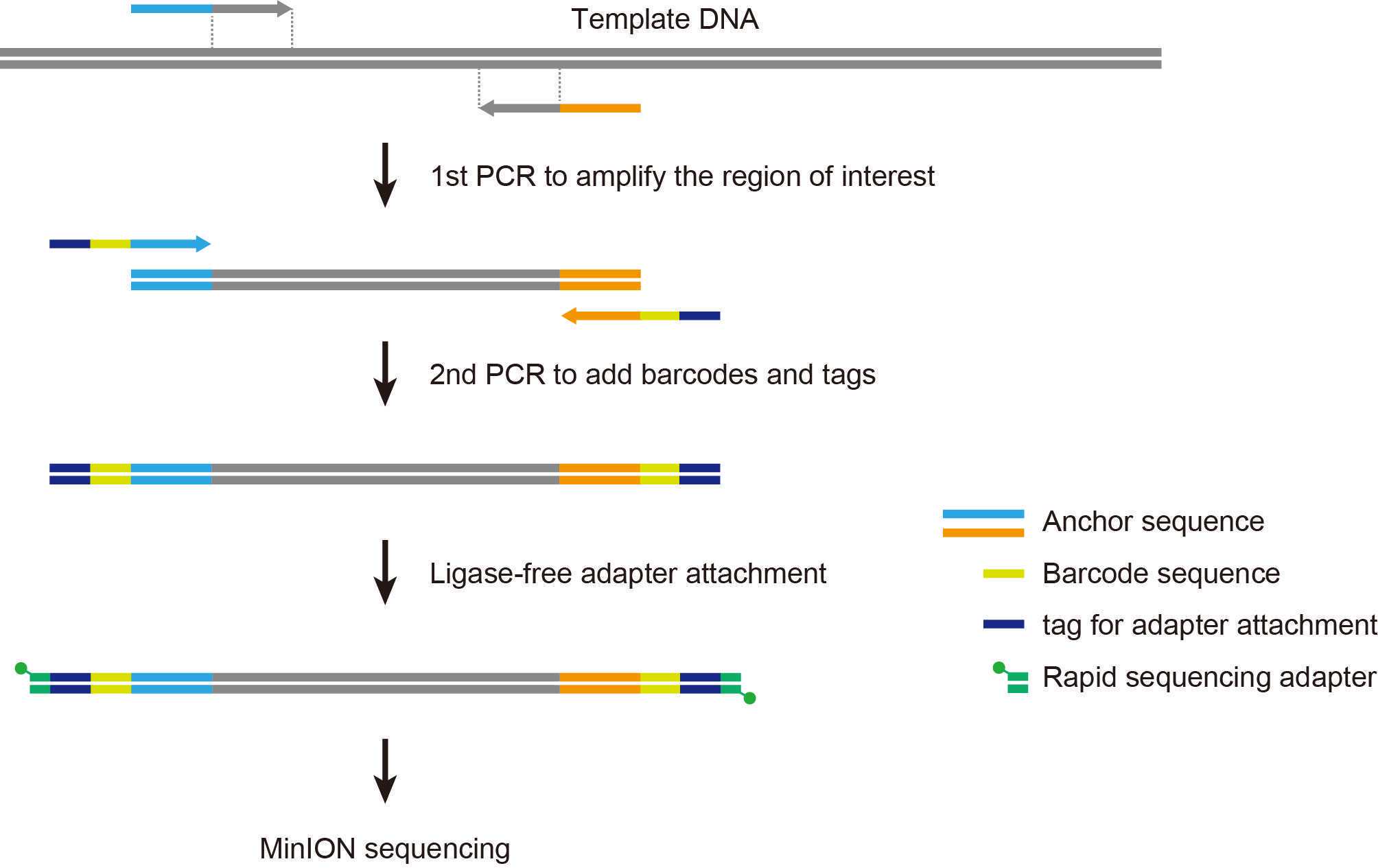
1st PCR with inner primers
Prepare the PCR master mix.
| A | B | C |
|---|---|---|
| Component | Volume | Final conc. |
| Template DNA | x µl | |
| 10 µM FW/RV primer mix | 0.5 µl | 0.2 µM each |
| 2X KAPA2G | 12.5 µl | 1X |
| Water | 12 - x µl | |
| Total | 25 µl |
Perform PCR.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 3 min | 1 |
| Denaturation | 95°C | 15 sec | 25-35 |
| Annealing | 55°C | 15 sec | |
| Extension | 72°C | 30 sec | |
| Hold | 4°C | ∞ | 1 |
The above is an example for amplifying the near-full length sequence of bacterial 16S rRNA genes (approximately 1,500 bp). The following inner primers are used, with 16S rRNA gene-specific sequences in bold letters.forward: 5'-TTTCTGTTGGTGCTGATATTGC AGRGTTYGATYMTGGCTCAG-3'reverse: 5'-ACTTGCCTGTCGCTCTATCTTC CGGYTACCTTGTTACGACTT-3'
Analyze the PCR product (2-5 µl) with gel electrophoresis to verify successful amplification.
2nd PCR with barcoded outer primers
Prepare the PCR master mix.
| A | B |
|---|---|
| Component | Volume |
| 1st PCR products | 1.0 µl |
| LWB 01-12 | 0.5 µl |
| 2X KAPA2G | 12.5 µl |
| Water | 11 µl |
| Total | 25 µl |
LWB 01-12: barcoded outer primers supplied in the PCR Barcoding Kit (SQK-PBK004)
Perform PCR.
| A | B | C | D |
|---|---|---|---|
| Step | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 3 min | 1 |
| Denaturation | 95°C | 15 sec | 8-10 |
| Annealing | 62°C | 15 sec | |
| Extension | 72°C | 30 sec | |
| Hold | 4°C | ∞ | 1 |
The above is an example for barcoding bacterial 16S rRNA gene amplicons (approximately 1500 bp).
[Optional] Analyze the PCR product (1 µl) with gel electrophoresis.
PCR cleanup
Resuspend the AMPure XP beads by vortexing.
For selecting fragments of over 500 bp, add 10 μl of AMPure XP to 20 µl of PCR product (0.5x ratio).
Mix by pipetting and incubate for 5 min at room temperature.
Place the tube on a magnetic rack for 2 min to magnetically separate out the beads.
Pipette off the supernatant.
Keeping on the magnetic rack, add 200 μl of 70% ethanol without disturbing the bead pellet, and discard the supernatant.
Repeat step 15 (wash the beads twice in total).
Spin down and place the tube back in the magnetic rack.
Pipette off any residual ethanol.
Air-dry for 1 min.
*Do not over dry the magnetic beads.
Remove the tube from the magnetic rack and resuspend the beads in 10 μl of elution buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl).
Incubate for 2 min at room temperature.
Place the tube on a magnetic rack for 2 min.
Transfer the eluate to a new tube.
DNA quantification
Warm QuantiFluor ONE dsDNA dye to room temperature.
Add 1 µl of eluted sample to 200 μl of QuantiFluor ONE dsDNA dye in 0.5 ml tube.
Mix thoroughly by vortexing.
Incubate for 5 min at room temperature, protected from light.
Measure fluorescence using the Quantus Fluorometer for quantifying DNA concentrations.
[Optional] Analyze the sample with gel electrophoresis.
Sequencing library preparation
Pool all barcoded amplicons to a total of 50-100 fmoles in 10 μl of 10 mM Tris-HCl pH 8.0 with 50 mM NaCl.
*For full-length 16S rRNA gene amplicons (approximately 1,500 bp), 50-100 fmoles of DNA equates to ~50-100 ng.
Add 1 μl of RAP and mix gently by pipetting.
*RAP is supplied in the PCR Barcoding Kit (SQK-PBK004).
Incubate for 5 min at room temperature.
Store the library on ice until ready to load.
Flow cell check
Open the MinION lid and insert the flow cell under the clip.
Perform flow cell QC.
Check the number of active pores available for the experiment.
Sample loading
Prepare flow cell priming mix by adding 30 µl of Flush Tether (FLT) to the tube of Flush Buffer (FB).
*FLT and FB are supplied in the Flow Cell Priming Kit (EXP-FLP001).
After opening the priming port, load 800 µl of the priming mix into the flow cell via the priming port and wait for 5 min.
Prepare the sequencing library for loading.
| A | B |
|---|---|
| Component | Volume |
| Library | 11 µl |
| Water | 4.5 µl |
| Sequencing Buffer (SQB) | 34 µl |
| Loading Beads (LB) | 25.5 µl |
| Total | 75 µl |
*SQB and LB are supplied in the PCR Barcoding Kit (SQK-PBK004).
Lift the spotON sample port cover and load 200 μl of the priming mix into the flow cell via the priming port (NOT the sample port).
Mix the prepared library gently by pipetting just prior to loading.
Load the library (75 µl) into the flow cell via the SpotON sample port in a dropwise fashion.
Replace the SpotON sample port cover and close the priming port.
Start the sequencing run.

