96-well plate OT-2 liquid handler integrated live-cell and endpoint viability drug activity screen
Rolando DZ Lyles, M Julia Martinez, Benjamin Sherman, Kerry Burnstein
Abstract
This protocol details OT-2 liquid handler integrated live-cell and endpoint viability drug activity screening on a 96-well plate.
Before start
Before starting :
Select the drug to be tested and determine the max concentration to be tested.
-
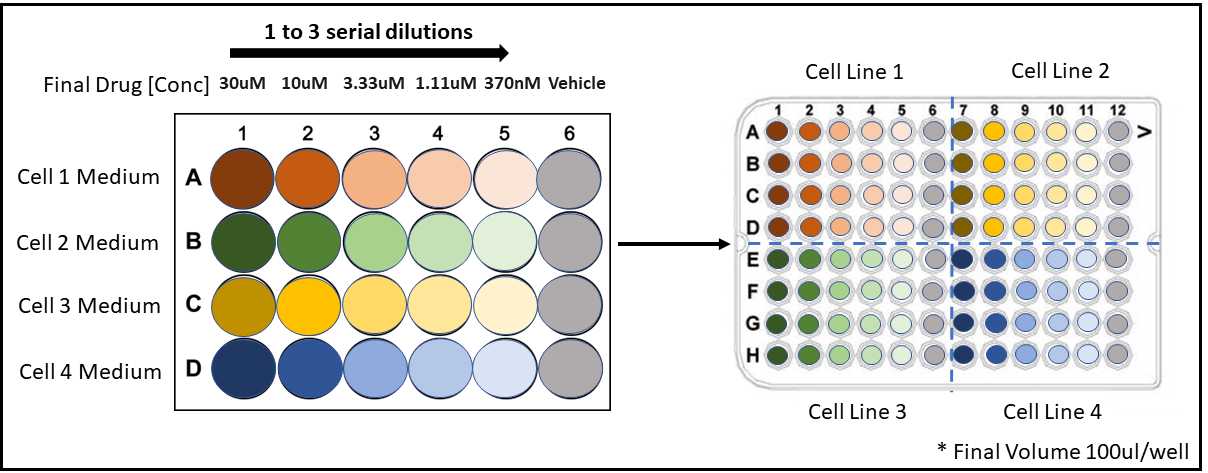
Starting with the defined max concentration, this protocol tests a single drug at 6 concentrations (including a vehicle control) at 1:3 serial dilutions.
-
Determine the cell line models which will be tested. Expand cell lines to ~50-60% confluence in any standard tissue t75-Flask or 15cm tissue culture dish.
-
Determine cell-specific seeding density and media condition to be tested.
Attachments
Steps
Day 1: Prepare cell lines for seeding into 96-well plates via Opentrons OT-2 liquid handler
In a sterile tissue culture hood, aspirate media from t75-flask.
Perform a wash with 5mL Phosphate Buffered Saline (PBS) (Hyclone, Cat# SH30028.02).
To detach cells from tissue culture flasks, add 3mL of trypsin (Corning, Cat# 25-052-cl) directly on to the cells. Gently tilt the flask until the surface is equally covered with trypsin then place cells into tissue culture incubator for 0h 3m 0s.
After incubation, gently tap the sides of the flask to insure complete detachment of adherent cells. Return to incubator for 0h 1m 0s increments if needed.
Add 3mL of trypsin inhibitor (Thermofisher, Cat# R007100) (or equal volume to trypsin used) to deactivate the trypsin enzyme, collect all the cell suspension, and then transfer into a sterile 15mL conical tube.
Pellet cells via centrifugation at 100-300x g.
After centrifugation, aspirate all supernatant while being careful to not disrupt the cell pellet.
Uniformly resuspend the cell pellet in 1mL of cell-specific culture media using a p1000 pipette.
Using the well-mixed cell suspension, accurately count cells via your preferred hemocytometry method to a final unit of [cells/mL].
After counting, calculate the volume of cells suspended in cell-specific culture media needed to prepare 10mL cells at the seeding density concentration range previously determined.
Preparing for cell seeding via Opentrons OT-2 Liquid handler
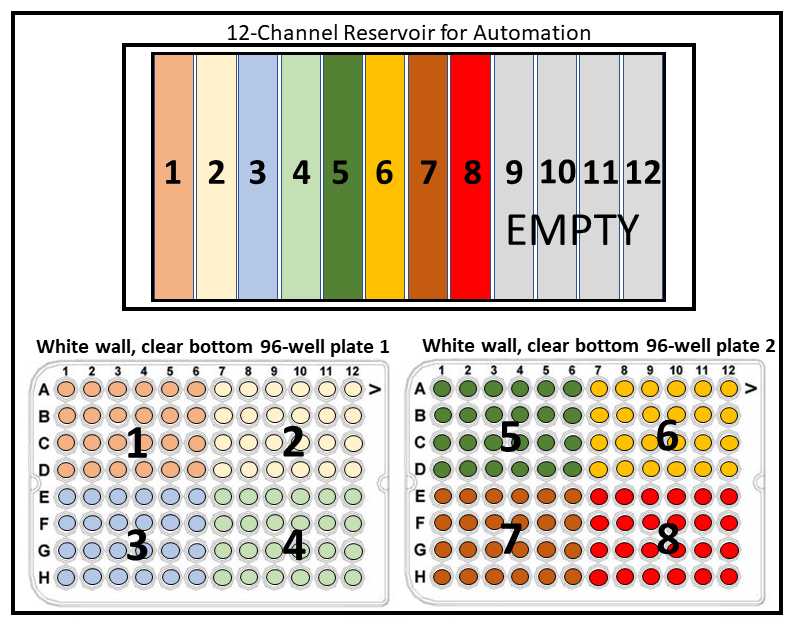
Alternate between a full row of tips (for mixing in the 12-channel reservoir) in one row and then 4 tips for the corresponding quadrant where the cells will seeded. This protocol seeds the cells in the upper quadrants first, and then returns to the cells in the lower quadrants from left to right. Two tip racks will be needed for this protocol. See image below:

Corresponding starting deck state compatible with OT-2 seeding protocol:
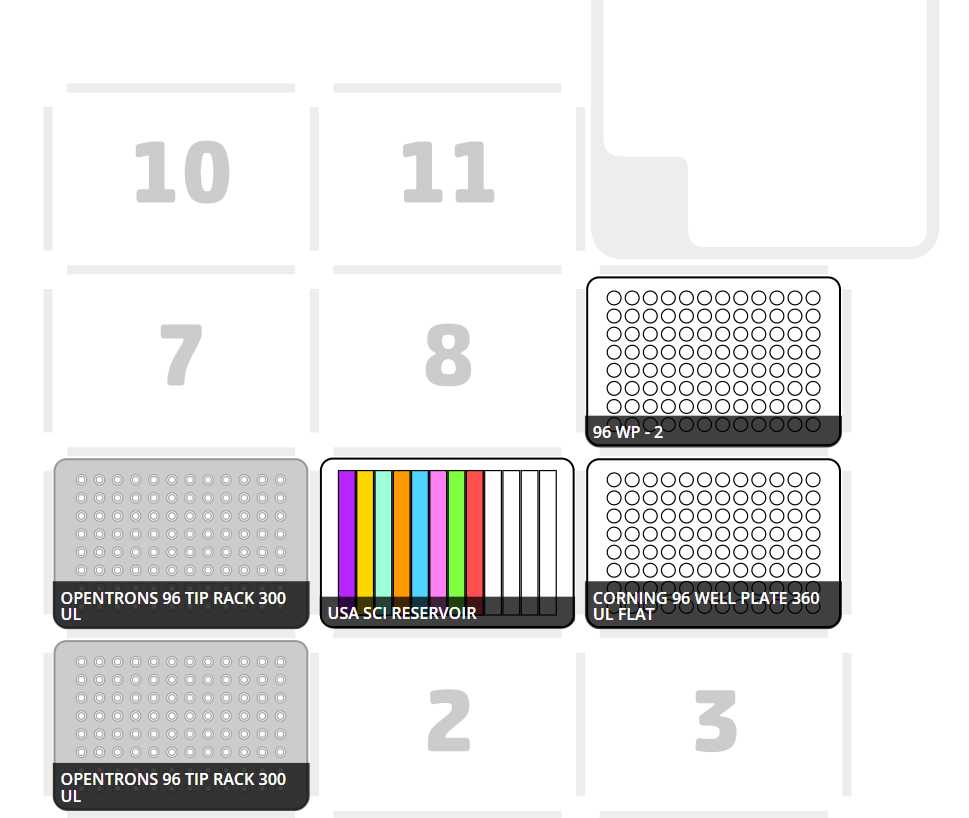
Transfer loaded 12-channel reservoir, pre-arranged tip racks, and two empty white walled/clear bottomed 96-well plates into the OT-2 deck and begin OT-2 procedure “Cell Seeding 8 Cell Lines.json”.
Place 96-well plates into standard tissue culture incubator .
Day 2: Prepare drug dilutions
Observe cells with a microscope at 10x magnification to verify that cells are settled and adhered to the inner surface of the 96-well plate.
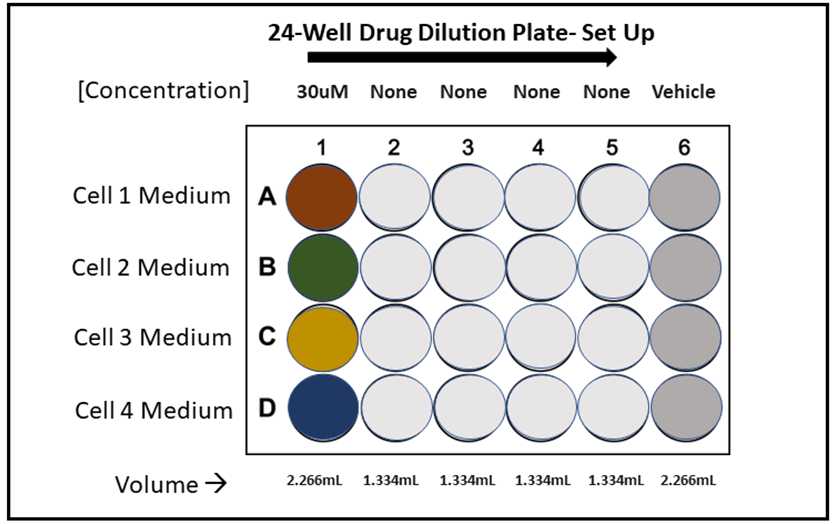
Prepare drug at pre-determined max concentration into pre-determined optimized media conditions. For this protocol, each cell line (in the optimized media conditions) will require 2.5µL of drug prepared at the pre-determined max concentration as well as 2.5µL of vehicle treated media at the same percentage of the max drug concentration.
Example: Example : If you are working with DrugX at a stock concentration of 50mM and desire a max concentration of 30µM in 2.5 mL of media use the formula:
where V1= Desired volume of DrugX at stock concentration to be added into optimized media conditions, C1= Concentration [µM] of DrugX at stock concentration, C2=Desired final concentration of DrugX (30micromolar (µM)), & V2= Desired final volume of new cell preparation (2.5mL). For the vehicle preparation, add equal volume as V1 of vehicle into media with optimized conditions.
In a sterile 24-well plate transfer 2.266mL of DrugX at max concentration in column 1 for each cell line using the cell-specific media conditions. Similarly, transfer 2.266mL of vehicle treated media into column 6. In the center columns 2-5, add 1.334mL of untreated media at optimized conditions for the corresponding cell lines (See plate map above). There should be one 24-well plate per white opaque 96-well plate (seeded the previous day).
Attached File: “Drug Dilution and Transfer.json”
Duration: ~1h 5m 0s
Corresponding starting deck state compatible with OT-2 seeding protocol:

Transfer loaded 24-well plate, full tip racks, an empty standard clear 96-well plates into the OT-2 deck and begin OT-2 procedure “Drug Dilution and Transfer.json”.
After drug dilution and transfer into the template standard clear 96-well plate (~0h 43m 0s), the protocol pauses to allow the user to add the first 96-well plate at this step. Add plate then resume protocol.
After the first plate is completed, return treated white 96-well plate into an incubator, and repeat the drug dilution and treatment procedure with the second plate (~1h 5m 0s).
Afterwards, second plate into the incubator and proceed to the next step.
Day 2: Verify cell attachment to plates after drug treatments and begin IncuCyte ZOOM live-cell imaging
At this stage, the plate can be transferred to the preferred live-cell imaging platform. This protocol uses the IncuCyte Zoom platform. The plate will remain in the imaging platform for 72h 0m 0s .
Open IncuCyte Zoom software on computer desktop.
Click the “Properties” tab and label the plate as desired.
Click “Apply” on the bottom right corner to save changes and register the plate to the IncuCyte Zoom hardware.
Connect to device.
Under the “Task List” panel on the left-hand side, select “Schedule Scans”.
Click one of the “Empty” slots on the live representative plate map for the hardware then click “Add Vessel”.
Once prompted, search from the vessel (96-well plate) by catalog number. This protocol features the Greiner bio one cell culture microplate (#655098). Once selected proceed to setting up experimental parameters:
On the bottom panel on the left-hand side, select “Edit Scan Pattern” and select all wells and set the scan pattern to 4 images/well. Save this scan pattern.
In the Channel Selection section in the center, click “Phase” (no colored acquisition channel is need for this protocol).
In the “Scan Mode” section in on the top-right side, toggle to the scan pattern that was previously created and saved.
In the “Analysis Job Setup” section on the right-hand side, toggle the “Job Type” and select “Basic Analyzer.” Toggle the “Processing Definition” tab and select a pre-determined processing definition with masks optimized for your specific cell line. (If this hasn’t been created, the “DEMO Phase” processing definition can be used.
Transfer the 96-well plates into the same IncuCyte Zoom slot selected during the software setup and begin real-time image capture.
Day 5: End live-cell imaging procedure and perform CellTiter-glo endpoint viability assay
After 72-hours have passed, end the experiment on the live-cell imagining platform software then remove the 96-well plate and allow and allow it to equilibrate to Room temperature ~ 0h 20m 0s.
Open IncuCyte Zoom software on computer desktop.
Connect to device.
Under the “Task List” panel on the left-hand side, select “Schedule Scans”.
Click on the slot housing the plate being tested on the live representative plate map for the hardware then click “Remove Vessel”.
Click “Apply” on the bottom right corner to save changes.
Manually remove the corresponding 96-well plate to the IncuCyte Zoom hardware.
Cover the bottom of the 96-well plate with white opaque lab tape.

Transfer 10mL (per plate) of the thawed CellTiter-Glo reaction reagent into a standard 25-mL reagent reservoir.
Use an 8-channel p200 multi-channel pipette (or comparable multi-channel pipette) to transfer 100µL of CellTiter-Glo reaction reagent into each well of the 96-well plate.
Leave the plate cover off and transfer into a luminometer compatible with CellTiter-glo. Be mindful of the plate orientation and alignment to insure proper placement into the device. See example below:


Recommended parameters for the Luminometer are as follows:
- Shake in an orbital shaker for
0h 2m 0sat 500 cycles/minute with a 1mm shaking diameter (cell lysis). - Incubate for
0h 10m 0sin a dark environment (reaction). - Read luminescence of each well at an integration of 400ms (data acquisition).
Note
*It will take ~0h 13m 0s to read each plate.
Export data and remove plate from luminometer.