Purification of mammalian telomeric DNA for single-molecule analysis
Giulia Mazzucco, Armela Huda, Martina Galli, Elia Zanella, Ylli Doksani




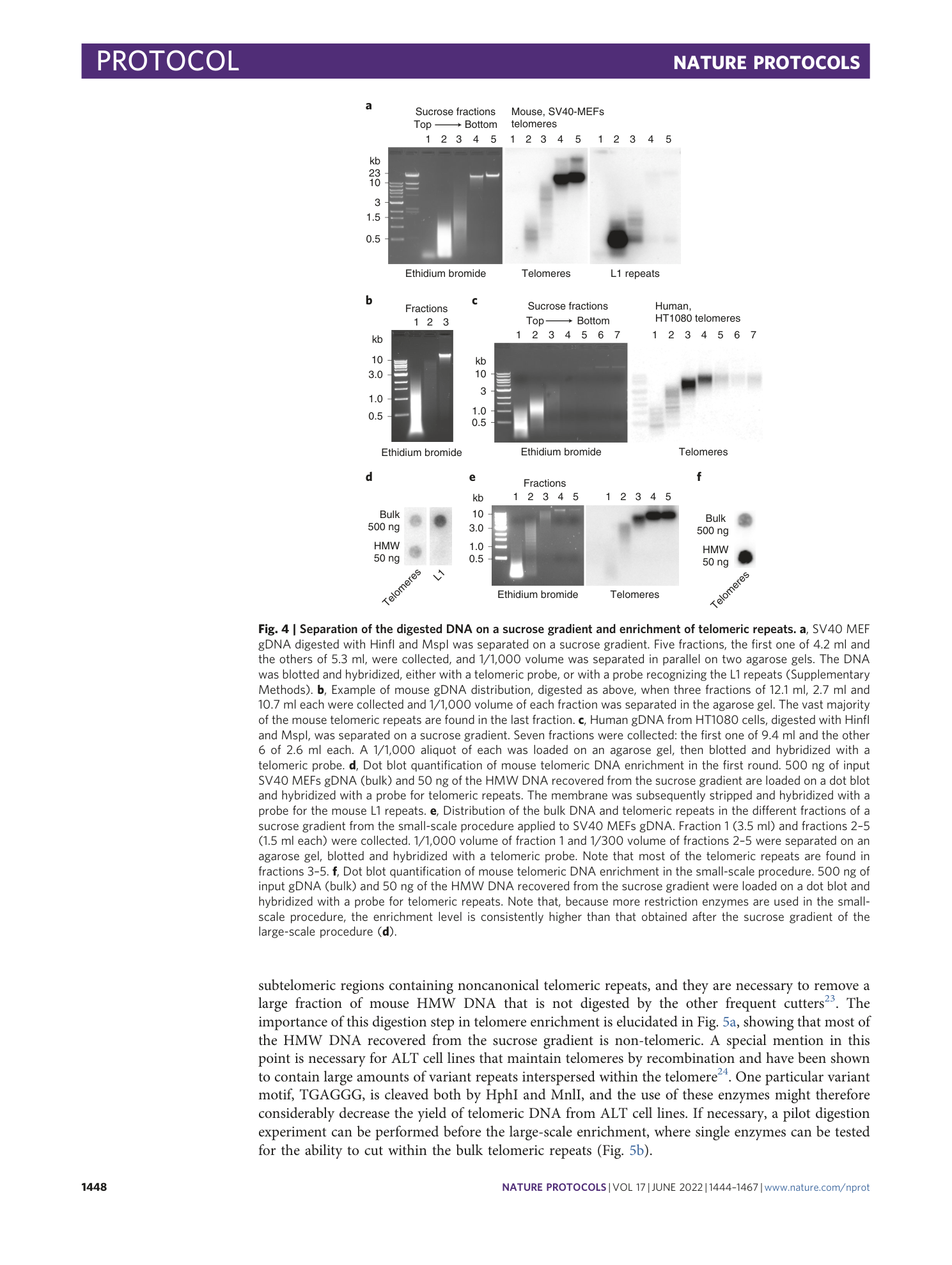

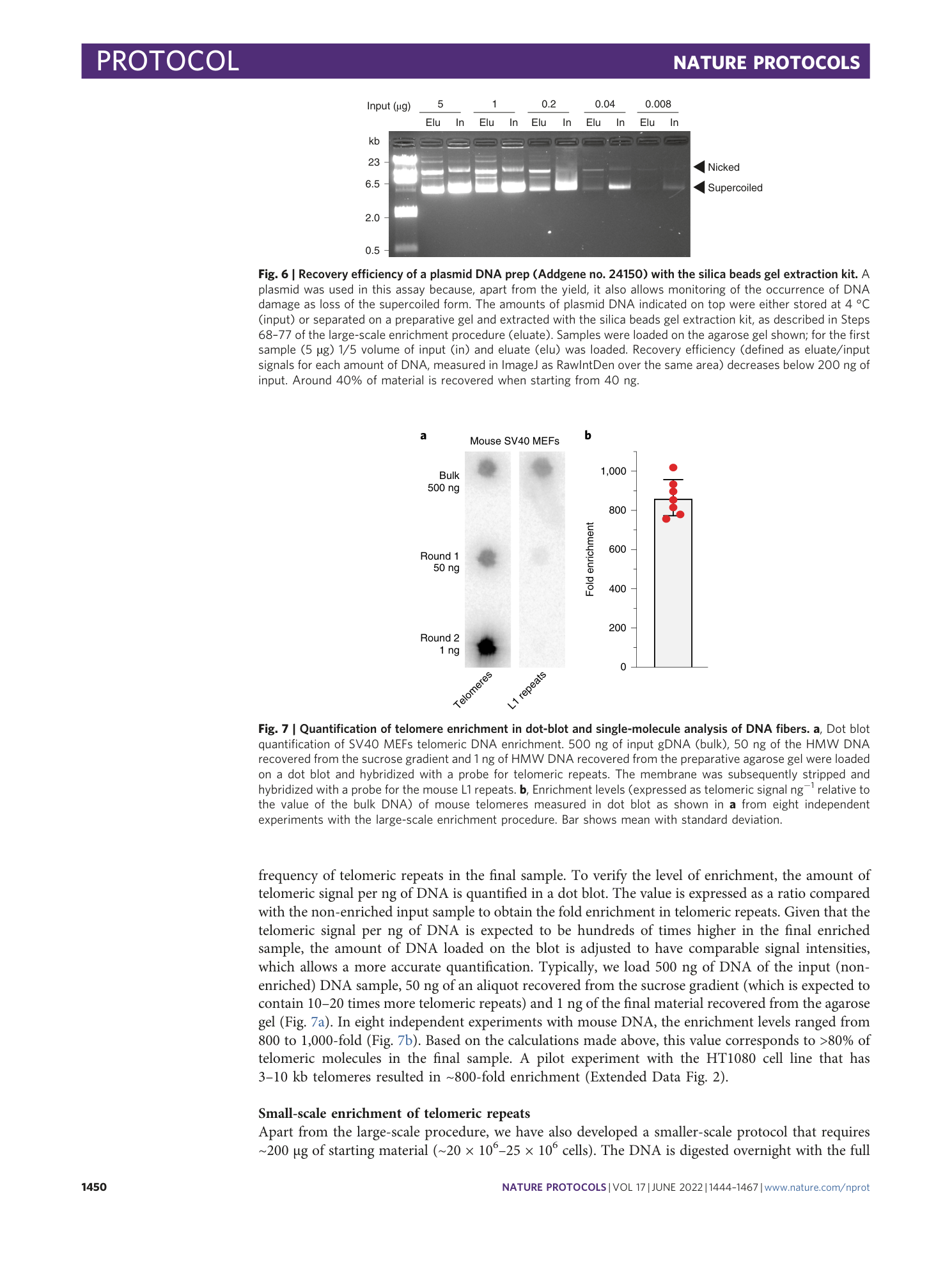

















Extended
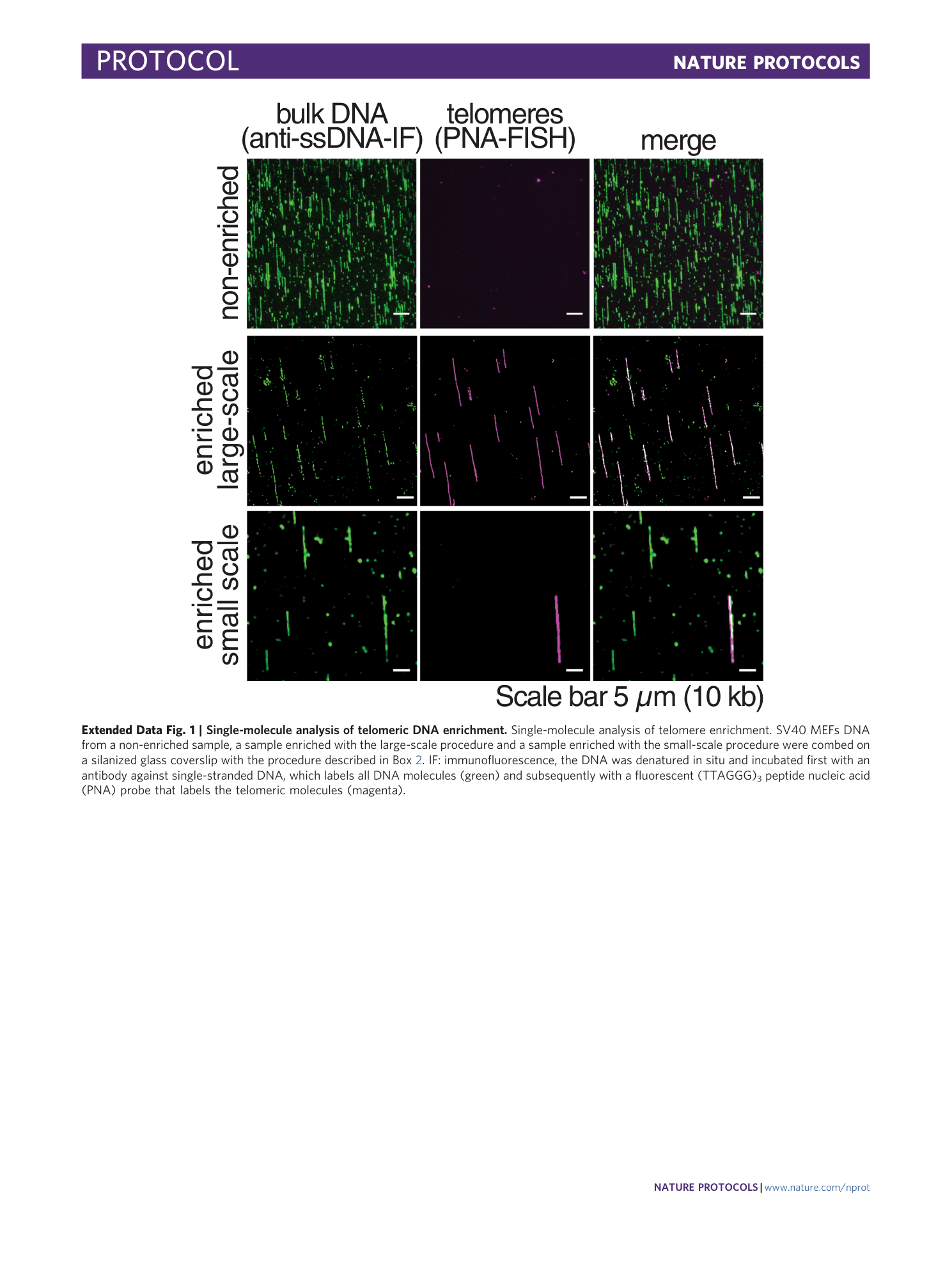
Extended Data Fig. 1 Single-molecule analysis of telomeric DNA enrichment.
Single-molecule analysis of telomere enrichment. SV40 MEFs DNA from a non-enriched sample, a sample enriched with the large-scale procedure and a sample enriched with the small-scale procedure were combed on a silanized glass coverslip with the procedure described in Box 2 . IF: immunofluorescence, the DNA was denatured in situ and incubated first with an antibody against single-stranded DNA, which labels all DNA molecules (green) and subsequently with a fluorescent (TTAGGG) 3 peptide nucleic acid (PNA) probe that labels the telomeric molecules (magenta).
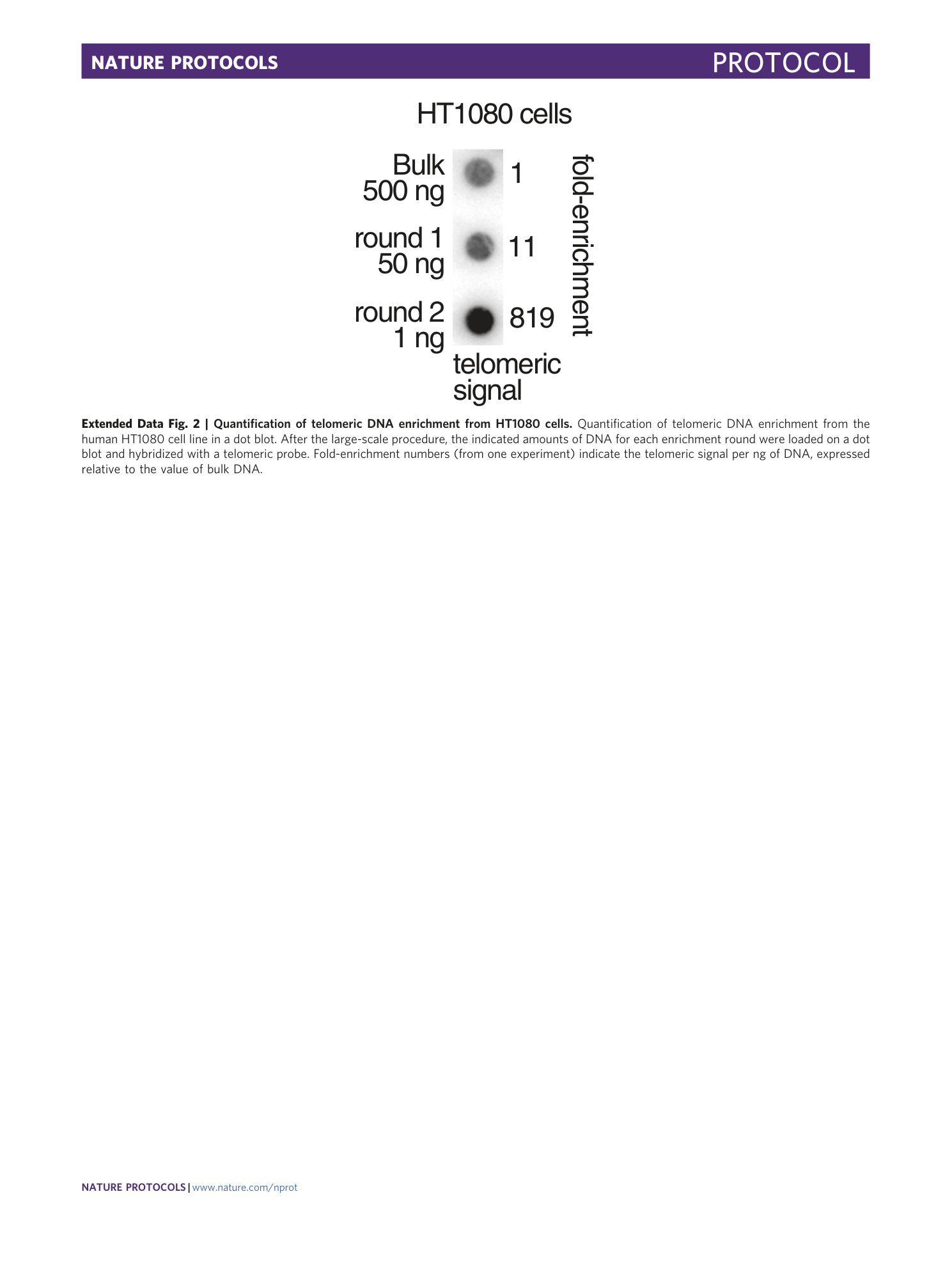
Extended Data Fig. 2 Quantification of telomeric DNA enrichment from HT1080 cells.
Quantification of telomeric DNA enrichment from the human HT1080 cell line in a dot blot. After the large-scale procedure, the indicated amounts of DNA for each enrichment round were loaded on a dot blot and hybridized with a telomeric probe. Fold-enrichment numbers (from one experiment) indicate the telomeric signal per ng of DNA, expressed relative to the value of bulk DNA.
Extended Data Fig. 3 No DNA damage induced during fractionation in sucrose gradient and DNA concentration.
Control experiment showing that separation in sucrose gradient and concentration in Amicon ultra centrifugal filters does not induce detectable DNA damage. Equal amounts of a plasmid prep (Addgene no. 71116) were either stored at 4 °C (input) or separated in a sucrose gradient, collected and concentrated in Amicon ultra centrifugal filters. As a positive control, the same plasmid DNA was incubated with 0.3 µg ml −1 ethidium bromide in TE 1× and irradiated for 10 min with 254 nm UV in a stratalinker. Equal amounts of the three samples were loaded in the agarose gel shown. Note that, in the positive control, treated with UV, all supercoiled form of the plasmid is lost, while no loss of the supercoiled form is detected after separation in sucrose gradient and sample concentration.
Supplementary information
Supplementary Information
Supplementary Methods
Supplementary Video 1
Video showing the psoralen cross-linking procedure
Supplementary Video 2
Video showing the manual preparation of sucrose gradients
Supplementary Video 3
Video showing sample loading on the sucrose gradient
Supplementary Video 4
Video showing the collection of sucrose fractions

