Fixation, Immunohistochemistry and in situ Hybridization of Human Lung Organoids
Morris Baumgardt, Maren Hülsemann,, Katharina Hellwig, Alina Langenhagen, Anne Voss, Simon Dökel, Achim D. Gruber, Stefan Hippenstiel, Andreas C. Hocke, Katja Hönzke
Disclaimer
Informed written consent was obtained from all volunteers and the study was approved by the Charité Ethics Committee (project 451, EA2/079/13).
Abstract
This protocol describes the fixation of infected human alveolar-like organoids, as well as the sample preparation for immunohistochemistry and immunohistology of infected tissue from alveolar-like organoids.
Before start
Steps
Fixation
Remove the organoid medium from the organoid containing well. Organoids should be collected from minimum two wells of a 24-well plate, to obtain a sufficient amount of organoids for staining.
Add 1mL cold base medium and collect Cultrex with organoids in a tube, flush well with 1mL base medium.
Place tubes with organoids at 4°C for 0h 5m 0s (to dissolve Cultrex).
Centrifuge for 300x g,4°C.
Remove supernatant, add 4% Formaldehyde solution (FA) and pipette up and down.
Store over night at 4°C in the BSL3 lab.
Export vessel from the BSL3 with organoid and FA, centrifuge for 300x g,4°C remove FA and add fresh 4% FA.
Store over night at 4°C.
Centrifuge for 300x g and remove FA.
Resolve pellet in PBS and store at 4°C or proceed with embedding.
Centrifuge for 300x g and remove PBS.
Resolve pellet in 100µL tissue Tek embedding medium (Histogel), pipette a drop on parafilm, place on ice and upon solidification place in the embedding cassette (act fast).
Close cassette, store in PBS, proceed with paraffin embedding and sectioning (sections: 2 µm).
Paraffin embedding and sectioning
Put cassette into the automatic embedding machine and start the following program:
| A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|
| Step | Solution | Concentration (%) | Duration (min:sec) | Temperature (°C) | D/V | Mix |
| 1 | Formalin | 4 | 0:00 | -- | ON | OFF |
| 2 | Water | 0:30 | 40 | ON | OFF | |
| 3 | Alcohol | 70 | 1:00 | 40 | ON | Slow |
| 4 | Alcohol | 80 | 1:00 | 40 | ON | Slow |
| 5 | Alcohol | 96 | 1:00 | 40 | ON | Slow |
| 6 | Alcohol | 96 | 1:00 | 40 | ON | Slow |
| 7 | Alcohol | 100 | 1:00 | 40 | ON | Slow |
| 8 | Alcohol | 100 | 1:00 | 40 | ON | Slow |
| 9 | Xylene | 1:00 | 40 | ON | Slow | |
| 10 | Xylene | 1:00 | 40 | ON | Slow | |
| 11 | Paraffin | 1:00 | 60 | ON | OFF | |
| 12 | Paraffin | 1:00 | 60 | ON | OFF | |
| 13 | Paraffin | 1:00 | 60 | ON | OFF | |
| 14 | Paraffin | 1:00 | 60 | ON | OFF |
Tissue-Tek VIP embedding program
In the following, after embedding, the samples are sectioned and mounted on adhesive slides. Sections of 2 µm each are prepared. Sections 1,5,9,13 are stained HE (following table) to see in which stages organoids are present, the rest is kept as blank section.
| A | B | C |
|---|---|---|
| Step | Solution | Duration (min:sec) |
| 1 | Xylene | 2x 2:00 |
| 2 | Xylene | 1x 3:00 |
| 3 | 96% ethanol | 0:30 |
| 4 | 80% ethanol | 0:30 |
| 5 | 70% ethanol | 0:30 |
| 6 | Water | 1:00 |
| 7 | Hematoxylin | 8:00 |
| 8 | Water | 0:05 |
| 9 | 70% ethanol | 0:10 |
| 10 | 80% ethanol | 0:30 |
| 11 | 96% ethanol | 0:45 |
| 12 | 100% ethanol | 1:00 |
| 13 | 100% ethanol | 1:00 |
| 14 | Xylene | 4x 1:00 |
HE staining steps
Immunohistology staining
Dewaxing over night at 60°C in Roticlear.
Put the slide with organoids three times for 0h 15m 0s in Roticlear at 60°C (use fresh Roticlear for every step).
0h 15m 0s 100% Ethanol.
0h 1m 0s 100% Ethanol.
0h 10m 0s 96% Ethanol.
0h 5m 0s 80% Ethanol.
0h 5m 0s 70% Ethanol.
0h 5m 0s 50% Ethanol.
Shake 3x 0h 5m 0s in 0.01Molarity (M) PBS (100 U/min).
Antigen-Retrival for 0h 30m 0s in a steam bath: Tris-EDTA buffer.
Let cool down for approx. 0h 30m 0s.
Shake 3x 0h 5m 0s in 0.01Molarity (M) PBS (100 U/min).
0h 15m 0s permeabilisation in 1% Triton (in 0.01Molarity (M) PBS) (on shaker 100 U/min).
Shake 3x 0h 5m 0s in 0.01Molarity (M) PBS (100 U/min).
Blocking for 0h 30m 0s in 5% appropriate serum (in dilution medium), ca. 70µL per tissue slice.
Flush carefully with 0.01Molarity (M) PBS (Pasteur pipette).
Dilute primary antibody in dilution medium and incubate over night at 4°C (wet chamber).
Flush carefully with 0.01Molarity (M) PBS (Pasteur pipette).
Shake 3x 0h 5m 0s in 0.01Molarity (M) PBS (100 U/min).
Dilute secondary antibody in dilution medium and incubate over night at 4°C (wet chamber).
Flush carefully with 0.01Molarity (M) PBS (Pasteur pipette).
Shake 3x 0h 5m 0s in 0.01Molarity (M) PBS (100 U/min).
Incubate 0h 5m 0s with DAPI.
Shake 3x 0h 5m 0s in 0.01Molarity (M) PBS (100 U/min).
Embedding with Mowiol® and coverslip, let air dry for 0h 10m 0s and seal edges with nail polish.
Immunofluorescence is analyzed by spectral confocal microscopy using a LSM 780 [Carl-Zeiss, Jena, Germany]. Based on a spectral image lambda stack, linear unmixing of tissue autofluorescence and overlapping spectra of fluorochromes is performed using ZEN 2012 software (Carl-Zeiss, Jena, Germany). To reveal lung and cell morphology, images are combined with Differential Interference Contrast (DIC). All image sets are acquired using optimal configuration regarding resolution and signal to noise ratio. Images are processed using ZEN 2012.
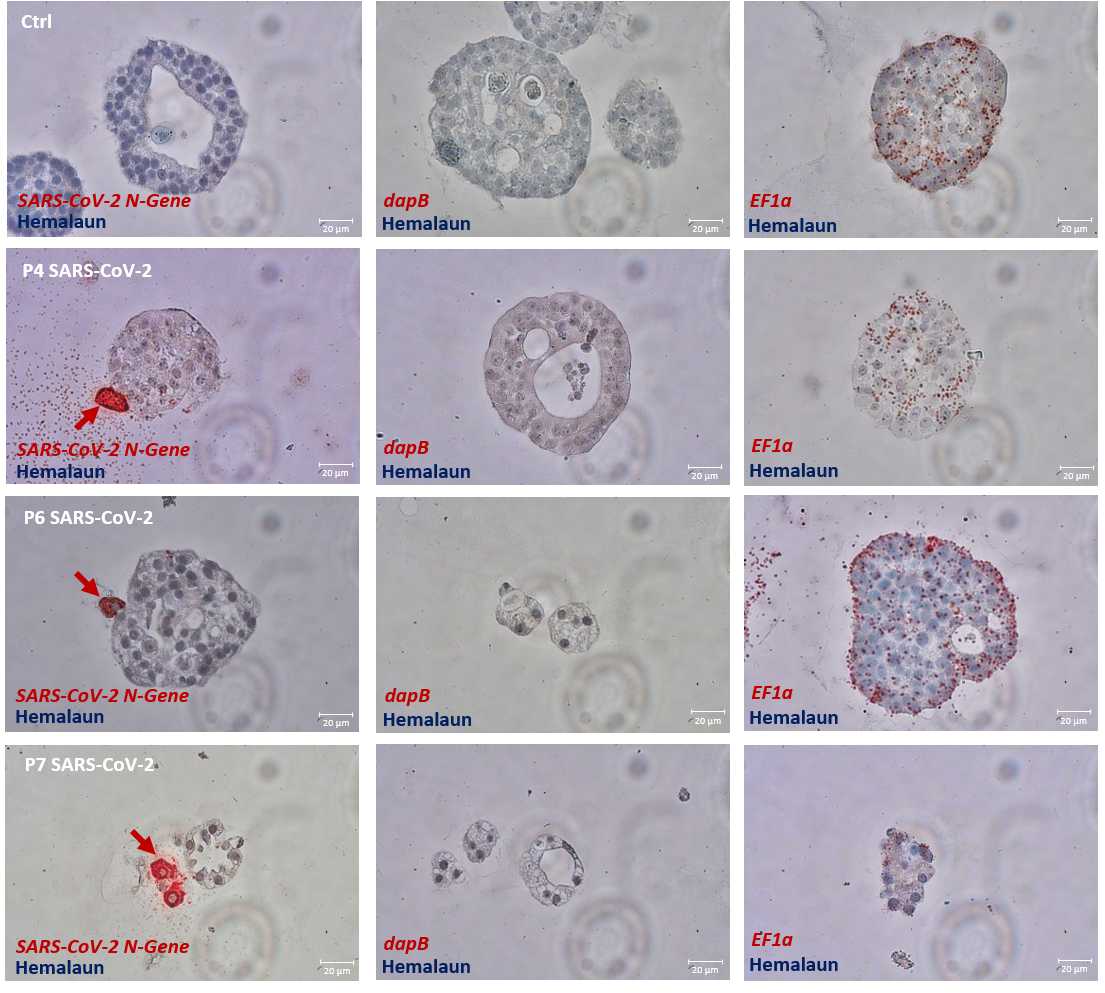
Viral in situ hybridization
We used a maximum of 12 slides at a time for the procedure. The size of the FFPE section should not exceed 2/3 of the slide and at least 5 mm away from each slide boarder.
Baking:
Bake the slides for 1h 0m 0s at 60°C in a dry oven.
Direct after baking remove paraffin with a tissue as much as you can, but do not touch the tissue.
Deparaffinization:
Pour 200mL of xylene into a staining dish.
Transfer the baked slides to the staining dish and incubate the slides at room temperature for 0h 5m 0s. Agitate frequently by moving the rack up and down.
Repeat the xylene washing with fresh xylene for 2 times.
Remove the slides from the xylene and wash the slides twice with 200mL of 100% ethanol for 0h 5m 0swith frequent agitation.
Remove the slides from the ethanol and place them face up on a paper towel to air dry for 0h 5m 0s at room temperature.
Hydrophobic barrier:
Before using the hydrophobic barrier pen dap it on a paper towel several times to ensure proper flow of the hydrophobic solution.
To create a hydrophobic barrier draw a rectangle around the organoid section, repeat it 2-4 times to ensure a solid seal.
Let the barrier dry for ~0h 25m 0s (until it is really dry).
Heat Pretreatment:
Heat the 1x Pretreatment Solution to 90°C in a porcelain staining jar (do not boil the Solution).
Incubate the slides for 0h 10m 0s at 90°C in the 1x Pretreatment Solution (time depends on tissue sample).
Remove the slide from the jar and submerge it directly into a clear staining dish containing 200mL of full desalted autoclaved water (VE water), at this point the slides should not dry out.
Wash for 0h 1m 0s with frequent agitation.
Repeat the wash step 46.3 and 46.4 one more time with 200mL of fresh VE water.
Transfer the slides to a clear staining dish containing 0.01Molarity (M) PBS.
Protease digestion and fixation:
Dilute the Protease (all in ViewRNA Tissue Kit) 1:100 in pre warmed 1X PBS (40°C) and briefly vortex to mix.
Remove each slide out of the dish and flick it to remove excess PBS. Do not let the slides dry out.
Place each slide face up on an object carrier box (OCB; pre tempered to 40°C / pre wetted with VE water) and immediately add 200µL of the working protease solution onto the tissue section. Make sure that the tissue section is covered with working protease solution.
Incubate the OCB in an isotemp oven (Thermo Fisher) at 40°C for 0h 20m 0s (Time depends on organoid sample).
After incubation decant the working protease solution from the slides and wash the slides two times with PBS in a staining dish for 0h 1m 0s.
Remove each slide out of the dish and flick it to remove excess PBS. Do not let the slides dry out.
Place each slide on a paper towel under a fume hood and incubate with 4% PFA for 0h 5m 0s at room temperature.
After incubation decant the PFA from the slides and wash the slides two times with PBS in a staining dish for 0h 1m 0s.
Hybridization:
Dilute the viewRNA probe set 1:40 in pre warmed probe set diluent and briefly vortex to mix. For positive control we use a EF1α probe set (also possible is ACTB or GAPDH ) and for negative control we use a DapB probe set.
Remove each slide out of the dish and flick it to remove excess PBS. Do not let the slides dry out.
Place each slide face up on an OCB and immediately add 200µL of the diluted probe set solution onto the tissue section. Make sure that the tissue section is covered with solution.
Incubate the OCB in an isotemp oven at 40°C for 2h 0m 0s.
Prepare 2L wash buffer with VE water, 18mL wash comp1 and 5mL wash comp2.
After incubation decant the probe set solution and wash the slides in a staining dish with wash buffer for 0h 2m 0s. Repeat washing for 2 times more with fresh washing buffer.
Optionally the slides can be stored at this point in 200mL storage buffer (60mL comp2 and 140mL VE water) for 24h 0m 0s at RT.
After storage the slides must be washed again 3 times with wash buffer.
Pre amplifier hybridization:
Pre warm the pre amplifier mix and swirl it to mix the solution.
Remove each slide out of the dish and flick it to remove excess liquid. Do not let the slides dry out.
Place each slide face up on an OCB and immediately add200µL of the pre amplifier mix onto the tissue section. Make sure that the tissue section is covered with solution.
Incubate the OCB in an isotemp oven at 40°C for 0h 25m 0s.
After incubation decant the pre amplifier mix and wash the slides in a staining dish with wash buffer for 0h 2m 0s. Repeat washing for 2 times more with fresh washing buffer.
Amplifier hybridization:
Pre warm the amplifier mix and swirl it to mix the solution.
Remove each slide out of the dish and flick it to remove excess liquid. Do not let the slides dry out.
Place each slide face up on an OCB and immediately add 200µL of the amplifier mix onto the tissue section. Make sure that the tissue section is covered with solution.
Incubate the OCB in an isotemp oven at 40°C for 0h 15m 0s
After incubation decant the amplifier mix and wash the slides in a staining dish with wash buffer for 0h 2m 0s. Repeat washing for 2 times more with fresh washing buffer.
Label Probe 1-AP hybridization:
After incubation decant the amplifier mix and wash the slides in a staining dish with wash buffer for 0h 2m 0s.
Repeat washing for 2 times more with fresh washing buffer.
Remove each slide out of the dish and flick it to remove excess liquid. Do not let the slides dry out.
Place each slide face up on an OCB and immediately add 200µL of the label probe 1-AP solution onto the tissue section. Make sure that the tissue section is covered with solution.
Incubate the OCB in an isotemp oven at 40°C for 0h 15m 0s.
After incubation decant the amplifier mix and wash the slides in a staining dish with wash buffer for 0h 3m 0s.
Repeat washing for 2 times more with fresh washing buffer.
Fast Red Substrate:
Remove the slides from the dish and place them face up on a paper towel, immediately add 200µL of the AP Enhancer Solution to each slide and incubate for 0h 5m 0s.
Decant the AP Enhancer Solution after incubation, place each slide face up on an OCB and add directly 200µL of fresh prepared fast red mix (for 2.5mL fast red buffer add 40µL fast red substrate1, vortex then add 40µL substrate2, vortex at least add 40µL substrate3, vortex).
Incubate the OCB at RT in the dark for 1h 0m 0s.
After incubation decant the fast red mix and wash the slides in a staining dish with PBS for 0h 1m 0s.
Repeat washing for one time more with fresh PBS.
Counter staining:
Optionally counter stain for 0h 0m 45swith Gill's hematoxylin.
Wash with water for 0h 5m 0s.
Mount the slides with fluoromount (Roth) by using a robotic coverslipper.
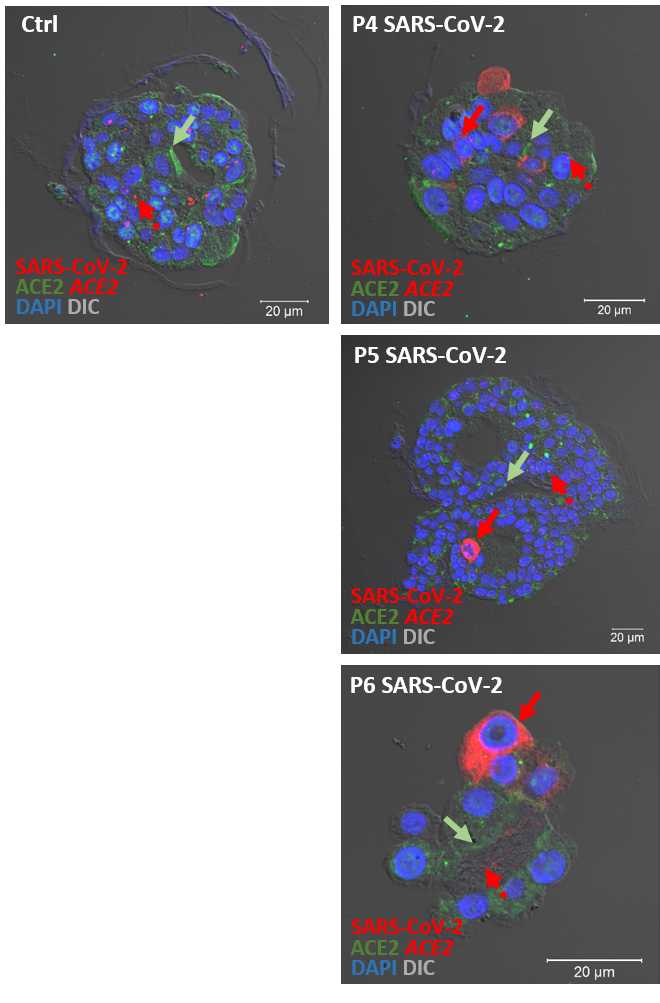
ACE2 in situ hybridization
For ACE2 in situ hybridization proceed with the "RNAscope® Multiplex Fluorescent Reagent Kit v2 Assay":
USM-323100_Multiplex_Fluorescent_v2_User_Manual_10282019.pdf
The following changes were made using the USM-323100 protocol:
Page 14, step 5: a third xylene incubation step is implemented (5 min at RT).
Page 15, target retrieval: a digital heat plate is used instead of a steamer.
Page 17: a staining cuvette is used instead of the slide holder.
After finishing the steps for "FFPE sample preparation and pretreatment", continue with chapter 4 on page 28.
Page 31: Opal is used with a dilution of 1:1500.
Exemplary results are shown at step 41 .