Design and Construction of a Designed Ankyrin Repeat Protein (DARPin) Display Library
Marco Morselli, Marco Morselli, Thomas R. Holton, Thomas R. Holton, Matteo Pellegrini, Matteo Pellegrini, Todd O. Yeates, Todd O. Yeates, Mark A. Arbing, Mark A. Arbing
DARPin
designed ankyrin repeat protein
DNA library construction
next-generation sequencing
protein binder
protein display
Abstract
Protein display systems are powerful techniques used to identify protein molecules that bind with high affinity to target proteins of interest. The initial challenge in implementing a display system is the construction of a high-diversity naïve library. Here, we describe the methods to generate a designed ankyrin repeat protein (DARPin) display library using degenerate oligonucleotides. Specifically described is the construction of a single DARPin repeat module by overlap extension PCR, concatenation of the module by restriction enzyme digestion and ligation, and incorporation of the concatenated modules into a full-length DARPin sequence in a bacterial cloning or display vector containing the hydrophilic N- and C-terminal capping domains. Protocols for PCR amplification of DARPin sequences to estimate diversity of naïve and enriched libraries via next-generation sequencing are included, as is a simple Linux-based program for analysis of naïve and enriched sequences. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation of a single DARPin repeat by overlap extension PCR
Basic Protocol 2 : Concatenation of DARPin repeats
Basic Protocol 3 : Ligation of internal repeats into cloning/display vector containing N- and C-terminal capping repeats
Basic Protocol 4 : Estimation of library size and diversity by next-generation sequencing (NGS)
Basic Protocol 5 : NGS analysis of naïve and enriched libraries
INTRODUCTION
Isolation of antibodies and similar binding molecules, e.g., nanobodies, from immunized animals is a lengthy and intensive process (Harmansa & Affolter, 2018). Protein display techniques were developed as an alternative to simplify the process and to allow the identification of novel designed protein binders that are not naturally occurring [e.g., single-chain variable fragment (scFv) fragments, synthetic protein binders]. In vitro (mRNA and ribosome display) and in vivo (phage, bacteria, and yeast) systems are capable of displaying a broad range of molecules, from short peptides (Rice & Daugherty, 2008; Wu et al., 2016) to small globular proteins (Chao et al., 2006; McMahon et al., 2018). Cell-based systems, using bacteria, phage, or yeast, secrete and anchor the protein binder to the cell or phage surface, where it can interact with exogenous target proteins; cells displaying target-specific binders are then selected for, and enriched, using a variety of methods, including biopanning and magnetic and/or fluorescence-based cell sorting (Chao et al., 2006; see Current Protocols article: Kenrick et al., 2007). Iterative rounds of selection of increasing stringency are used to isolate protein binders with high specificity and affinity for the target protein (Chao et al., 2006). Binders, in whatever form, have a wide range of applications, including as molecular tools used in basic research as well as therapeutic molecules for the treatment of disease.
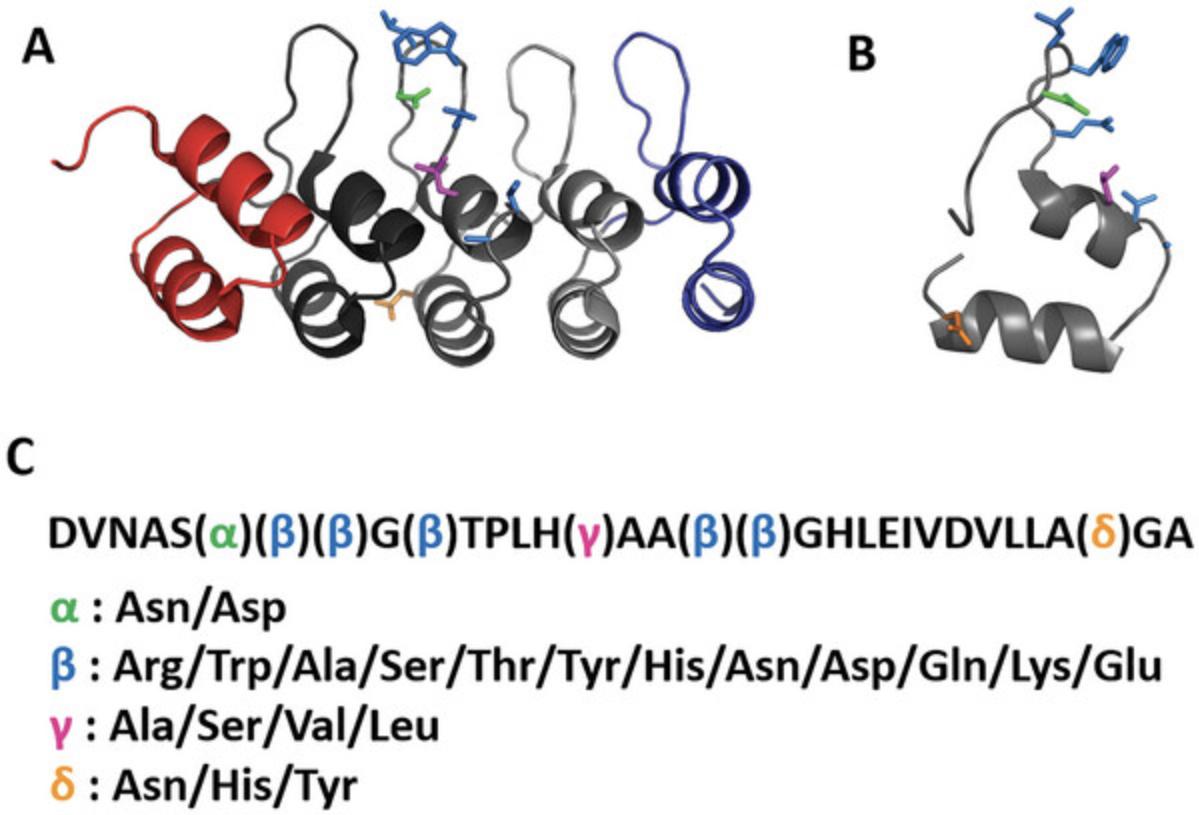
Modern recombinant molecular biology techniques have simplified the creation of highly diverse libraries that reproduce naturally occurring binder repertoires (see Current Protocols article: Schladetsch & Wiemer, 2021) that expand on protein folds used for cellular protein-protein interactions. An example of the latter is ankyrin repeat proteins, which are naturally occurring proteins composed of repeating 33 amino acid modules, each consisting of a β-turn followed by two anti-parallel α-helices (Fig. 1). The average ankyrin repeat protein has 4 to 6 modules, although proteins with up to 33 modules exist (Kumar & Balbach, 2021). The repeats generate a concave binding surface, with 6 to 8 amino acid residues of a module contributing to substrate binding (Kumar & Balbach, 2021). Binz et al. recognized the utility of the ankyrin repeat protein scaffold and developed a designed ankyrin repeat protein (DARPin) system, which has two or three repeat modules sandwiched between hydrophilic N- and C-terminal caps. Using ribosome display, they confirmed that DARPins that bind target proteins with high affinity could be selected from their synthetic DARPin library (Binz et al., 2004; Binz et al., 2003). Subsequent publications have illustrated the usefulness of DARPins as molecular and diagnostic tools (Boersma, 2018; Plückthun, 2015), imaging scaffolds (Castells-Graells et al., 2023; Liu et al., 2019), and potential therapeutic agents (Shilova & Deyev, 2019).
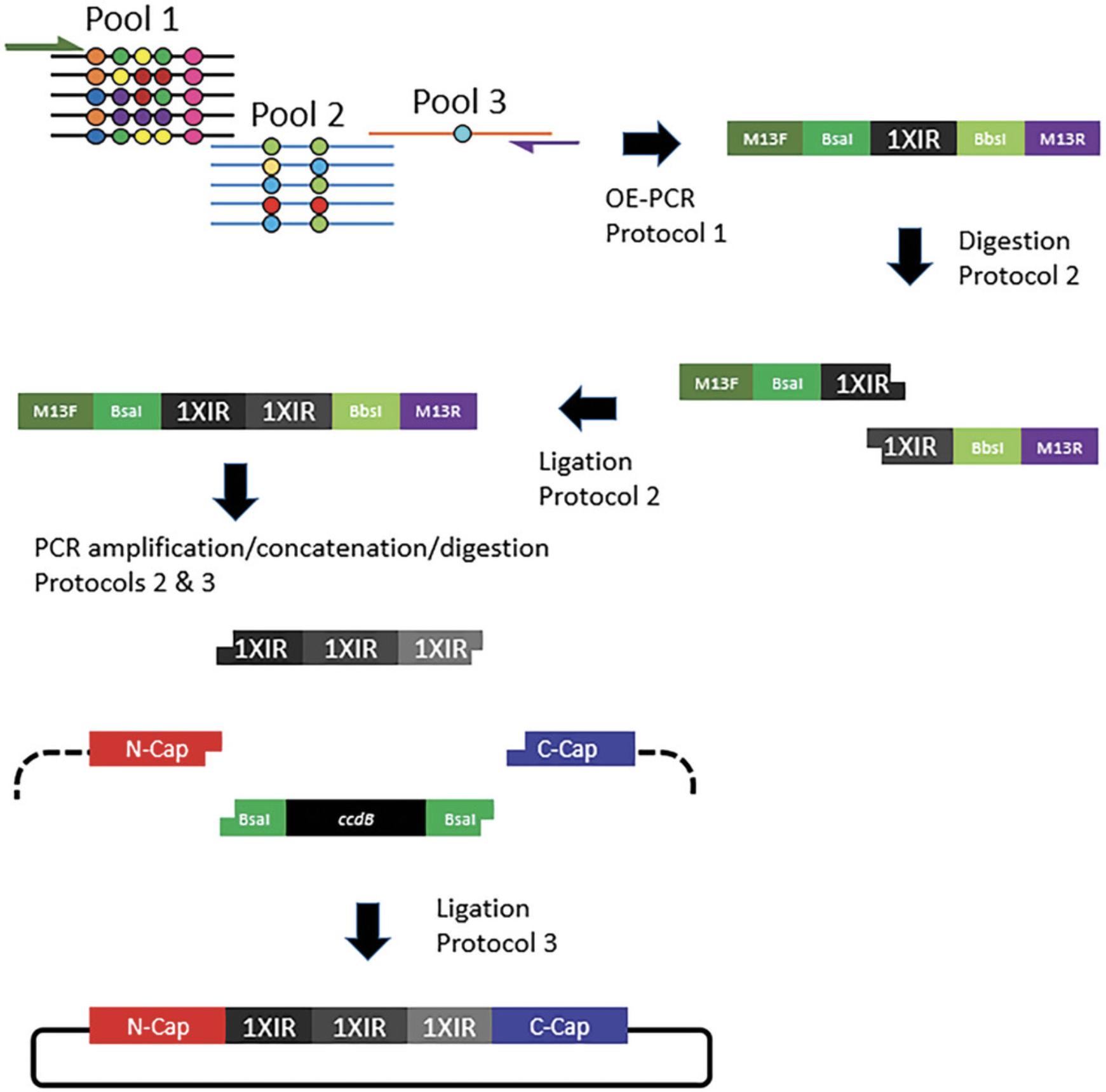
We have designed a strategy that uses basic molecular biology procedures to generate a highly diverse naïve DARPin library (≥108 unique sequences) using oligonucleotides incorporating degenerate nucleotide bases. The procedure is straightforward and inexpensive and incorporates a specific subset of amino acids into variable positions while avoiding stop codons, chemically reactive amino acids, and overrepresentation of any particular amino acid. Oligonucleotide design and considerations for amino acid diversity at variable positions are discussed. Basic Protocol 1 describes the use of overlap extension PCR (OE-PCR) and oligonucleotide pools to generate a short DNA fragment comprising a single DARPin repeat. Basic Protocol 2 details the concatenation of these repeat fragments by restriction enzyme digestion with type IIs restriction enzymes followed by ligation. Basic Protocol 3 describes the PCR amplification of the concatenated DARPin repeats and their ligation into cloning or display vectors containing the sequence-invariant N- and C-terminal caps to create a diverse library of DNA sequences encoding a complete DARPin molecule. Basic Protocol 4 describes sample preparation of PCR amplicons from naïve and enriched libraries for next-generation sequencing (NGS). Basic Protocol 5 details a method to estimate library size and diversity using the NGS data generated by Basic Protocol 4.
STRATEGIC PLANNING
DARPin display was developed using in vitro display technologies (Binz et al., 2004), although expression of DARPins on the surface of bacteria (Zadravec et al., 2015), yeast (Mohan et al., 2019), and phage (Steiner et al., 2008) has been described. Initial planning should include testing whether the desired system of choice can efficiently display DARPins and/or whether modifications to the system are required for efficient display, e.g., modification of tether length or anchoring strategy. We have used a DARPin that binds maltose-binding protein (MBP) to test DARPin surface exposure, as MBP is highly soluble, easily expressed and purified, and amenable to labeling; this particular anti-MBP DARPin has not been published, but the MBP-DARPin crystal structure has been deposited into the Protein Data Bank (PDBid 5FIN). As the ligation of the variable region of the library into the display vector is a resource- and time-consuming endeavor, verification that the system of choice can display DARPins is absolutely critical.
The second major consideration is deciding on the identity of the amino acids at each variable position and choosing the degenerate codons to encode the desired residues. In the described protocols, we have used a pool of ∼80 oligonucleotides (see Supporting Information, Table 1) and seven different degenerate codons (Fig. 2, Table 1) to generate a highly diverse library with a relatively balanced distribution of amino acids at each position (Fig. 1, Table 1). Less specific degenerate codons (e.g., such as NNB and NNK, which encodes all 20 amino acids and one stop codon) could be substituted and reduce the number of required oligonucleotides at the expense of including a stop codon and incorporating potentially undesirable amino acids (e.g., cysteine, proline). Conversely, using more degenerate codons that encode fewer amino acids can provide a more desirable distribution of a specific subset of amino acids at the expense of requiring many more oligonucleotide primers.
| Position | Degenerate codon: encoded amino acids |
|---|---|
| Alpha | RAC: Asparagine (N); Aspartic acid (D) |
| Beta | WGG: Arginine (R), Tryptophan (W); DCA: Alanine (A), Serine (S), Threonine (T); NAC: Tyrosine (Y), Histidine (H), Asparagine (N), Aspartic acid (D); VAA: Glutamine (Q), Lysine (K), Glutamic acid (E) |
| Gamma | KYA: Leucine (L), Serine (S), Valine (V), Alanine (A) |
| Delta | HAC: Asparagine (N), Histidine (H), Tyrosine (Y) |
NOTE : Experiments involving PCR require extremely careful technique to prevent contamination.
Basic Protocol 1: GENERATION OF A SINGLE DARPin REPEAT BY OVERLAP EXTENSION PCR
A pool of overlapping oligonucleotides containing degenerate codons (Table 1) is used to generate a 99-bp DNA segment that encodes a 33-amino-acid DARPin repeat module. The 99-bp DARPin sequence is flanked by type IIs restriction enzyme sites, allowing the repeat to be concatenated by restriction enzyme digestion and ligation. A single repeat or concatenated repeats can be PCR-amplified using oligonucleotides (see Supporting Information, Table 1) that are complementary to the 5′ and 3′ flanking sequences, which contain the type IIs restriction enzyme sites. In addition, M13 forward and reverse primer sequences (Table 2) are appended to the “external” forward and reverse primers, respectively, allowing for Sanger sequencing of PCR products or, alternatively, another means to PCR-amplify the repeats as necessary to generate additional material for downstream manipulations (e.g., concatenation).
| Oligo name | Oligo sequence | Purpose |
|---|---|---|
| M13.DARPin.For. | 5′-GTAAAACGACGGCCAGGAAGACCTGACGTTAACGCTTC | External primer for OE-PCR |
| M13.DARPin.Rev. | 5′-CAGGAAACAGCTATGACAAGCTTCTAAGGTCTCACGTCAGC | External primer for OE-PCR |
| M13F | 5′-GTAAAACGACGGCCAG | Amplification of DARPin repeat(s); sequencing |
| M13R | 5′-CAGGAAACAGCTATGAC | Amplification of DARPin repeat(s); sequencing |
| BsaI.Vec.For. | 5′-AAAAAAGGTCTCAAAACACCGTTTGATCTGGCCATTG | Amplification of cassette-containing display vector |
| BsaI.Vec.Rev. | 5′-TATATAAGGTCTCTCATCCTGACCTGCGCTTGCTG | Amplification of cassette-containing display vector |
| LibSeqFor.1 | 5′-GCATTCAGGATGATGAAGTTCGTATTCTGATGG | NGS of library |
| LibSeqRev.1 | 5′-GCATCAGATCAAACGGTGTTTTACCAAATTTATC | NGS of library |
| LibSeqFor.2 | 5′-ATGCGTCAGGATGATGAAGTTCGTATTCTGATG | NGS of library |
| LibSeqRev.2 | 5′-ATGCGATCAAACGGTGTTTTACCAAATTTATCCTG | NGS of library |
| LibSeqFor.3 | 5′-AGTCGGTCAGGATGATGAAGTTCGTATTCTG | NGS of library |
| LibSeqRev.3 | 5′-TGCAGCCAGATCAAACGGTGTTTTACCAAATTTATCC | NGS of library |
| DARP.Amplicon.F | 5′-GATGAAGTTCGTATTCTGATGGCAAATGG | NGS of enriched DARPin libraries |
| DARP.Amplicon.R. | 5′-CGGTGTTTTACCAAATTTATCCTGGGC | NGS of enriched DARPin libraries |
Here, three oligonucleotide pools, corresponding to the 5′, middle, and 3′ ends of the DARPin repeat, in conjunction with the external 5′ and 3′ primers are used to assemble a fully synthetic double-stranded nucleic acid fragment that encodes a single DARPin internal repeat module, referred to as 1XIR, with eight variable positions specifying, via degenerate codons (Table 1), 2 to 12 amino acids (Fig. 1). The repeat is flanked by restriction enzyme recognition sites and unique sequences allowing sequencing and/or PCR amplification of the DNA fragment (Fig. 2). The protocol as described uses ∼80 oligonucleotides (see Supporting Information, Table 1), although the choice of degenerate codons can dramatically increase, or decrease, the number of required oligonucleotides. If executed properly, a limited number of PCR reactions are required with the oligonucleotide pools to generate the DARPin repeat, and thus, picomole-scale oligonucleotide synthesis in 96-well plates should provide ample starting material. We suggest testing multiple PCR reaction conditions to determine optimal parameters to generate the initial DARPin repeat. Specifically, we test multiple high-fidelity DNA polymerases, two or more annealing temperatures, and three oligonucleotide template concentrations.
Materials
- PCR components (see Table 3)
- Degenerate oligonucleotides (IDT, delivered wet in H2O at 5 μM concentration; sequences in Supporting Information, Table 1)
- High-fidelity DNA polymerases [e.g., KOD HotStart DNA Polymerase (Millipore Sigma, 71086), PrimeSTAR GXL DNA Polymerase (Takara, R050B), and Phusion High-Fidelity DNA Polymerase (New England Biolabs, M0530S)]
- Nucleic acid gel stain (GelRed® Nucleic Acid Gel Stain, Biotium, 41003)
- Spin column kit (e.g., DNA Clean & Concentrator-25, Zymo Research, D4033) or gel extraction kit (e.g., Wizard® SV Gel and PCR Clean-Up System, Promega, A9281)
- Elution buffer (10 mM Tris HCl, pH 8, or sterile nuclease-free water)
| Template concentration | |||
|---|---|---|---|
| Component | 0.2 μM | 0.04 μM | 0.01 μM |
| 10× Buffer for KOD HotStart DNA Polymerase | 5 | 5 | 5 |
| dNTPs (2 mM each; New England Biolabs, N0447S) | 5 | 5 | 5 |
| 25 mM MgSO4 | 3 | 3 | 3 |
| M13.DARPin primers (25 µM; see Table 2) | 1/1 | 1/1 | 1/1 |
| Template (see Basic Protocol 1, step 4) | 6 | 1.2 | 0.3 |
| 1 U/µl KOD HotStart DNA Polymerase (Millipore Sigma, 71086) | 1 | 1 | 1 |
| H2O | to 50 μl | to 50 μl | to 50 μl |
- a
The reaction volume is split in half, and 25-µl reactions are run at two different annealing temperatures. For KOD HotStart DNA Polymerase, we use annealing temperatures of 60°C and 65°C, annealing and extension steps of 10 s, and 10 to 15 PCR cycles.
-
Cold block (optional)
-
PCR tubes, sterile
-
Microcentrifuge
-
Low-retention microcentrifuge tubes, sterile (Fisher Scientific, 02-681-331)
-
Thermal cycler capable of temperature gradients (e.g., Bio-Rad T100, 861096)
-
Transilluminator (e.g., Gel Doc XR+ Imager, Bio-Rad, 1708195)
-
Spectrophotometer (e.g., NanoDrop ND-1000) or fluorescence-based quantification method
-
Additional reagents and equipment for agarose gel electrophoresis (see Current Protocols article: Voytas, 2000)
Reaction setup
1.Maintain reagents (e.g., enzymes) on ice or in a cold block.
2.Add all PCR components (see Table 3), except the enzyme, to a sterile PCR tube according to the manufacturer's recommendations and mix well by inverting the tube several times. Centrifuge briefly to bring the liquid to the bottom of the tube, add the enzyme, and repeat the mixing and centrifugation steps.
3.Create three primer pools in separate sterile low-retention microcentrifuge tubes using degenerate oligonucleotides:
- For Pool 1, containing equimolar amounts of the 64 oligonucleotides encompassing the 5′ end of the DARPin repeat, as the oligos are supplied at 5 μM concentration, add equivalent volumes of each (e.g., 5 μl) to a tube to create Pool 1.
Oligonucleotide sequences are listed in Table1in theSupporting Information.
-
For Pool 2, corresponding to the middle of the DARPin repeat, add equivalent volumes of the 16 oligonucleotides comprising Pool 2 to a tube to create a 5 μM stock.
-
For Pool 3, in this DARPin design, add a single oligonucleotide at 5 μM concentration to a tube.
4.Mix the three primer pools at a 1:1:1 ratio to generate an oligonucleotide master pool containing all the oligonucleotides required to generate a single DARPin repeat.
Overlap extension PCR
5.Set up multiple test reactions with three template concentrations, different high-fidelity DNA polymerases (we typically test KOD HotStart DNA Polymerase, PrimeSTAR GXL DNA Polymerase, and Phusion High-Fidelity DNA Polymerase), and two annealing temperatures. Add external primers incorporating M13 sequences (M13.DARPin.For. and M13.DARPin.Rev.; see Table 2) at 0.5 μM to the reactions. Run PCR in a thermal cycler capable of temperature gradients.
6.Evaluate the PCR reaction by electrophoresing a 5-μl sample of the PCR product on a 2% agarose gel, staining with nucleic acid gel stain, and imaging with a transilluminator.
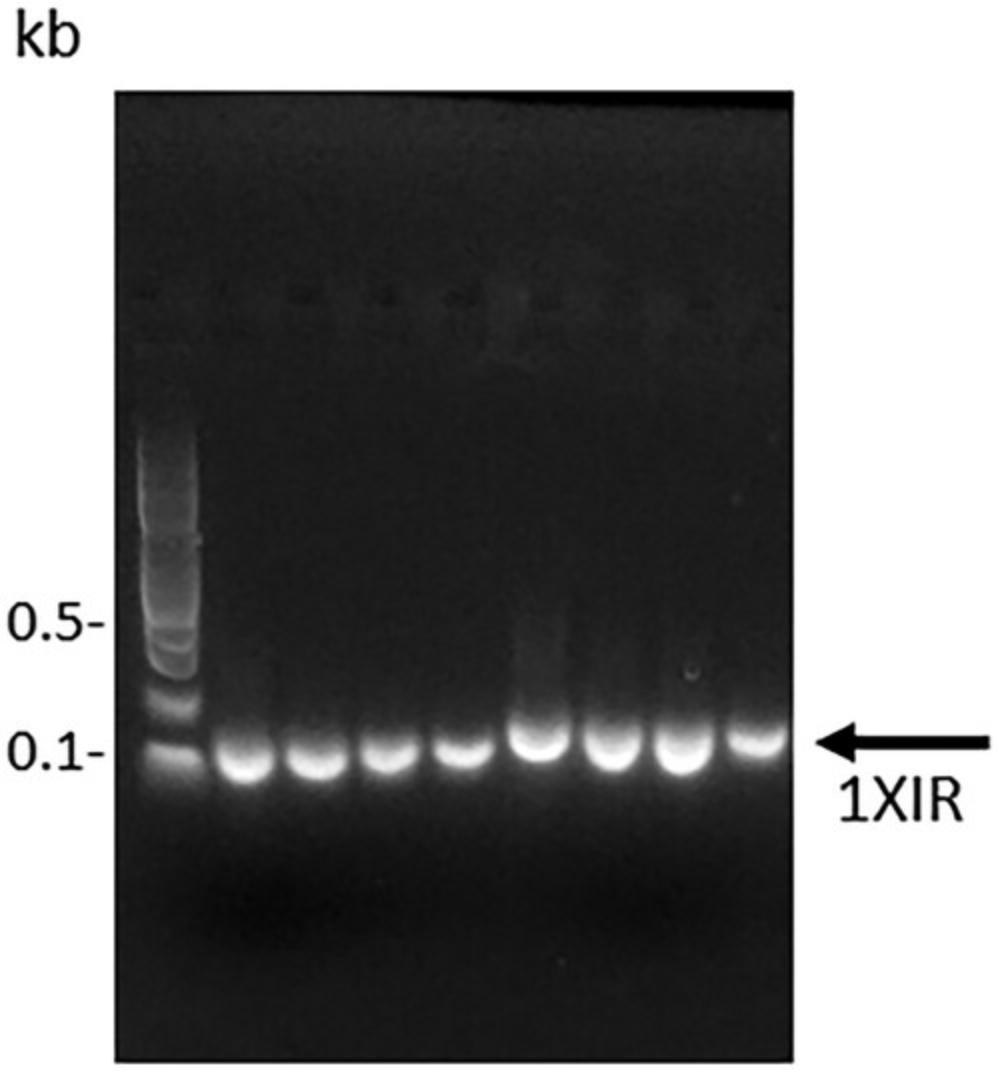
7.Scale up the PCR using conditions that generate a correctly sized PCR fragment. Purify the PCR product using a spin column kit, or if additional bands are present, purify the product by gel extraction using a gel extraction kit following the manufacturer's protocol. In either case, maximize yield by using two elutions per column: that is, after the first elution in elution buffer, add an additional volume of elution buffer to the column and incubate at room temperature for several minutes prior to centrifuging.
8.Quantify the purified DNA fragment using a spectrophotometer or, preferably, a fluorescence-based quantification method. Verify the fragment quality using a 2% agarose gel and then store the DNA fragments frozen if not proceeding directly to restriction enzyme digestion (see Basic Protocol 2).
Basic Protocol 2: CONCATENATION OF DARPin REPEATS
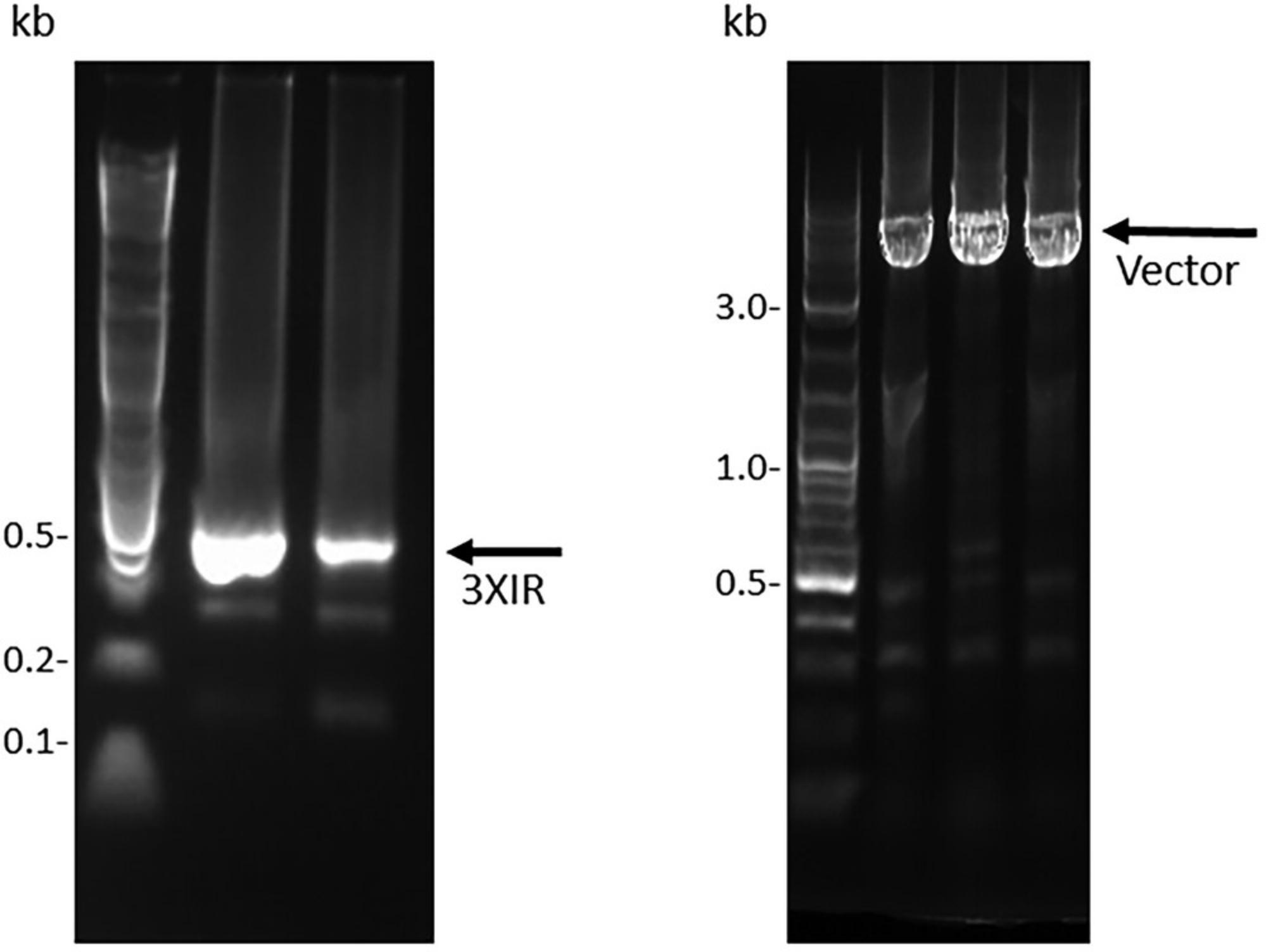
This protocol describes the concatenation of DARPin repeat modules to generate DNA fragments containing two or more consecutive in-frame repeat modules that can be inserted between the hydrophilic N- and C-terminal caps to generate a DNA fragment that encodes a complete DARPin molecule. The purified DNA fragment generated by OE-PCR (Basic Protocol 1) is the starting material for this protocol. The starting material can be amplified using the M13.DARPin.For./M13.DARPin.Rev. primers to generate additional material, which may be required because purification of the ligation products from agarose gels is typically inefficient. Two different type IIs enzymes (BbsI and BsaI) will be used to generate compatible 5′ and 3′ overhangs, and ligation of the two differentially digested fragments will generate a DNA fragment containing two consecutive DARPin modules (Figs. 2 and 4). The two-repeat (“2XIR”) module is then PCR-amplified to generate additional material that can be digested with a single type IIs enzyme (e.g., BsaI) and then ligated to a single “1XIR” module that has been digested with the compatible type IIs enzyme (e.g., BbsI) to generate a “3XIR” module containing three consecutive repeats (Fig. 5). Although most DARPin libraries employ two or three internal DARPin repeat modules, the digestion and ligation of DNA fragments could be repeated to generate any number of internal repeat modules. Large amounts of DNA fragments (∼10 μg) should be subjected to restriction enzyme digestion, as significant amounts of material are lost during the gel extraction and purification steps.
Additional Materials (also see Basic Protocol 1)
- Restriction enzyme digestion components (see Table 4)
- T4 DNA Ligase (New England Biolabs, M0202S)
| Digestion | ||
|---|---|---|
| Component | BsaI-HF | BbsI-HF |
| 10× CutSmart Buffer | 10 µl | 10 µl |
| BsaI-HF (BsaI-HFv2, New England Biolabs, R3733S) | 2 µl | — |
| BbsI-HF (BbsI-HF, New England Biolabs, R3539S) | — | 2 µl |
| DNA (purified single DARPin repeat; see Basic Protocol 1) | 10 µg | 10 µg |
| H2O | to 100 µl | to 100 µl |
- Razor blade
- 16°C temperature-controlled water bath (optional)
Reaction setup
1.Maintain reagents (e.g., enzymes) on ice or in a cold block.
2.Add reagents, except the enzyme, to a sterile PCR tube according to the manufacturer's recommendations and mix well by inverting the tube several times. Centrifuge briefly to bring the liquid to the bottom of the tube, add the enzyme, and repeat the mixing and centrifugation steps.
Restriction enzyme digestion
3.Set up two reactions with restriction enzyme digestion components (see Table 4), one with BbsI and the other with BsaI, to generate DNA fragments with compatible 5′ and 3′ ends. Using a thermal cycler, incubate the reactions for 1 to 6 hr at 37°C and then heat-inactivate the enzymes by incubation at 80°C for 20 min.
4.Electrophorese the entire reaction volume on a 2% agarose gel, excise the fragments with a razor blade, and subsequently column-purify using the Promega Wizard® SV Gel and PCR Clean-Up System. To maximize yield, use two elutions of 50 µl per column.
5.Quantify the purified DNA fragments using a spectrophotometer and verify that the restriction enzyme digest and subsequent purification step generated a single band using a 2% agarose gel.
Ligation of the DARPin repeats
6.Mix the BsaI- and BbsI-digested fragments at a 1:1 equimolar ratio and ligate using T4 DNA Ligase following the manufacturer's protocol. Ligate at 16°C overnight using a thermal cycler or temperature-controlled water bath.
7.Electrophorese the entire ligation mixture on a 2% agarose gel and purify the ∼200-bp band corresponding to a successfully ligated 2× DARPin repeat (2XIR) by gel extraction.
8.PCR-amplify the 2XIR fragment to generate additional material for restriction enzyme digestion.
9.Gel-extract the PCR-amplified 2XIR fragment, digest with a single type IIs enzyme (either BsaI or BbsI), and ligate to the purified single DARPin repeat that has been restriction-digested with the enzyme to generate a compatible overhang, as described in steps 6 to 7, to generate a 3XIR repeat.
10.As for the 2XIR repeat, purify the 3XIR fragment by gel extraction and PCR-amplify to generate sufficient material for ligation into a plasmid vector. Quantify the purified DNA fragment using a spectrophotometer, verify the fragment quality using a 2% agarose gel (Fig. 5), and store the fragments frozen at −20°C if not proceeding directly to Basic Protocol 3.
Basic Protocol 3: LIGATION OF INTERNAL REPEATS INTO CLONING/DISPLAY VECTOR CONTAINING N- AND C-TERMINAL CAPPING REPEATS
This protocol describes the large-scale ligation of the DARPin variable regions (Basic Protocol 2) into a vector containing the N- and C-terminal capping repeats. The vector will vary according to the chosen experimental system (bacteria, phage, yeast), although the strategy is essentially the same for the in vivo selection systems: a cassette containing the N- and C- terminal caps and a “stuffer” sequence containing the ccdB toxin gene, for negative selection of the parental template, is cloned in-frame with the 5′ and 3′ elements required for display (e.g., signal sequences, tethers, epitope tags for cell surface detection). A variety of display systems have been described for bacteria (Rice & Daugherty, 2008; Wendel et al., 2016), yeast (McMahon et al., 2018; Mohan et al., 2019), and phage (Pardon et al., 2014). As these systems have typically been used for display of nanobody or scFv fragments, optimization of the elements flanking the DARPin library may be necessary. Alternatively, the DARPin library can be assembled in a cloning vector for use in in vitro systems or for transfer into other systems that have issues with transformation efficiency. The sequence of the cassette that we use is available in the Supporting Information. Note that use of the ccdB negative selection element requires that plasmids containing the cassette be propagated in specific Escherichia coli strains (e.g., DB3.1). Alternatively, the cassette can be generated with a random DNA sequence inserted as the stuffer fragment.
Large-scale DNA ligation and subsequent electroporation are critical steps in generating large libraries and require optimization of ligation reactions and electroporation parameters. We have followed detailed ligation and electroporation protocols for the generation of bacterial peptide display (Getz et al., 2012) and scFv phage display (see Current Protocols article: Schladetsch & Wiemer, 2021) libraries to optimize parameters for our library, and we refer the reader to these excellent references.
Additional Materials (also see Basic Protocols 1 and 2)
- PCR reagents (see Table 5)
- Plasmid miniprep kit (e.g., Zymo ZR Plasmid Miniprep – Classic, D4015; optional)
- Cloning/display vector, containing DARPin N- and C-caps and ccdB gene
- DpnI restriction enzyme (New England Biolabs, R0176S; optional)
- Nuclease-free water, sterile
- Electrocompetent cells (Lucigen MC1061 F- Electrocompetent Cells, 60514-2)
| Component | Volume (µl) |
|---|---|
| 10× Buffer for KOD Hot Start DNA Polymerase | 50 |
| dNTPs (2 mM each; New England Biolabs, N0447S) | 50 |
| 25 mM MgSO4 | 30 |
| Primers (25 μM; see Table 2) | 7.5/7.5 |
| Template (3XIR; see Basic Protocol 2) | 15 |
| 1 U/µl KOD HotStart DNA Polymerase (Millipore Sigma, 71086) | 10 |
| H2O | 330 |
| Total reaction volume | 500 |
- a
The template concentration is 10 ng/µl. The reaction volume is divided into five portions of 100 µl each for PCR cycling with an annealing temperature of 62.5°C, extension time of 4 min, and 30 PCR cycles.
-
Membrane filters (Millipore “V” Series filters, Millipore-Sigma, VSWP02500)
-
Petri dish
-
Electroporation cuvettes (Gene Pulser/MicroPulser Electroporation Cuvettes: 0.1 cm gap, Bio-Rad, 1652089; 0.2 cm gap, Bio-Rad, 1652086)
-
Electroporator (e.g., Gene Pulser Xcell Electroporation System, Bio-Rad, 1652660)
-
Additional reagents and equipment for whole-plasmid sequencing and for large-scale electroporation, estimation of electroporation efficiency, and preparation of freezer stocks and library maxipreps (Getz et al., 2012; see Current Protocols article: Schladetsch & Wiemer, 2021)
Reaction setup
1.Maintain reagents (e.g., enzymes) on ice or in a cold block.
2.Add PCR reagents (see Table 5), except the enzyme, to a sterile PCR tube according to the manufacturer's recommendations and mix well by inverting the tube several times. Centrifuge briefly to bring the liquid to the bottom of the tube, add the enzyme, and repeat the mixing and centrifugation steps.
Vector preparation
3.Prepare the display vector in large quantities using a plasmid miniprep kit or by PCR amplification using the parameters in the footnote to Table 5, primers BsaI.Vec.For. and BsaI.Vec.Rev. (see Table 2), the cloning/display vector as a template, and KOD HotStart DNA Polymerase. If the vector is PCR-amplified, treat the PCR product with DpnI restriction enzyme and column-purify. Electrophorese the purified vector on a 1% agarose gel to confirm that only a single band is present (Fig. 5) and verify the sequence of the purified PCR fragment by whole-plasmid sequencing before proceeding to the next step.
Restriction enzyme digestion and analysis
4.Restriction-digest the 3XIR DARPin library (purified, concatenated 3XIR DARPin repeats generated in Basic Protocol 2) with BsaI and BbsI (see Table 6) while the vector (see step 3) is digested with BsaI alone, as the cassette is designed to generate compatible ends using a single enzyme (see Fig. 2). Simultaneously dephosphorylate the vector with rSAP to reduce background. Incubate the reactions in a thermal cycler for 8 hr at 37°C and then heat-inactivate the enzymes by incubation at 80°C for 20 min.
| Component | 3XIR repeats | Vector |
|---|---|---|
| 10× CutSmart Buffer | 10 µl | 10 µl |
| BsaI-HF (BsaI-HFv2, New England Biolabs, R3733S) | 1.5 µl | — |
| BbsI-HF (BbsI-HF, New England Biolabs, R3539S) | 1.5 µl | 5 µl |
| rSAP (Recombinant Shrimp Alkaline Phosphatase, New England Biolabs, M0371S) | — | 5 µl |
| DNA (purified, concatenated 3XIR DARPin repeats generated in Basic Protocol 2 or display/cloning vector) | 10 µg | 15 µg |
| H2O | to 100 µl | to 150 µl |
5.Column-purify the 3XIR repeats. Gel-extract the miniprepped and digested vector to eliminate partially digested vector and the stuffer sequence.
6.Quantify the purified DNA fragments using a spectrophotometer and verify the fragment quality using a 1% agarose gel for the large DNA fragments and 2% agarose gel for small fragments (Fig. 5).
Test and large-scale ligations
7.After performing test ligations to determine optimal ligation ratios and minimize background (which have been described and will not be covered here; Getz et al., 2012; see Current Protocols article: Schladetsch & Wiemer, 2021), set up large-scale ligations as in Table 7 (which describes a 3:1 insert-to-vector ratio). Ligate at 16°C overnight using a thermal cycler or temperature-controlled water bath.
| Component | 3XIR repeats |
|---|---|
| 10× Ligase Buffer | 50 µl |
| Vector | 6.5 μg |
| Insert | 1 μg |
| T4 DNA Ligase (New England Biolabs, M0202S) | 25 µl |
| H2O | to 500 µl |
8.Column-purify the ligated library using the Zymo DNA Clean & Concentrator-25 using two elutions of 30 μl sterile nuclease-free water to maximize DNA yield. Further desalt the samples by drop dialysis for 2 hr using membrane filters floated on sterile water in a petri dish according to the manufacturer's instructions. Pipet the samples off the filters and then wash the filters with 25 μl water, add this water to the dialyzed sample, and quantify the DNA yield/recovery with a spectrophotometer.
9.Perform large-scale electroporation, estimation of electroporation efficiency, and preparation of freezer stocks and library maxipreps as previously described in detail (Getz et al., 2012; see Current Protocols article: Schladetsch & Wiemer, 2021) using electrocompetent cells, electroporation cuvettes, and an electroporator.
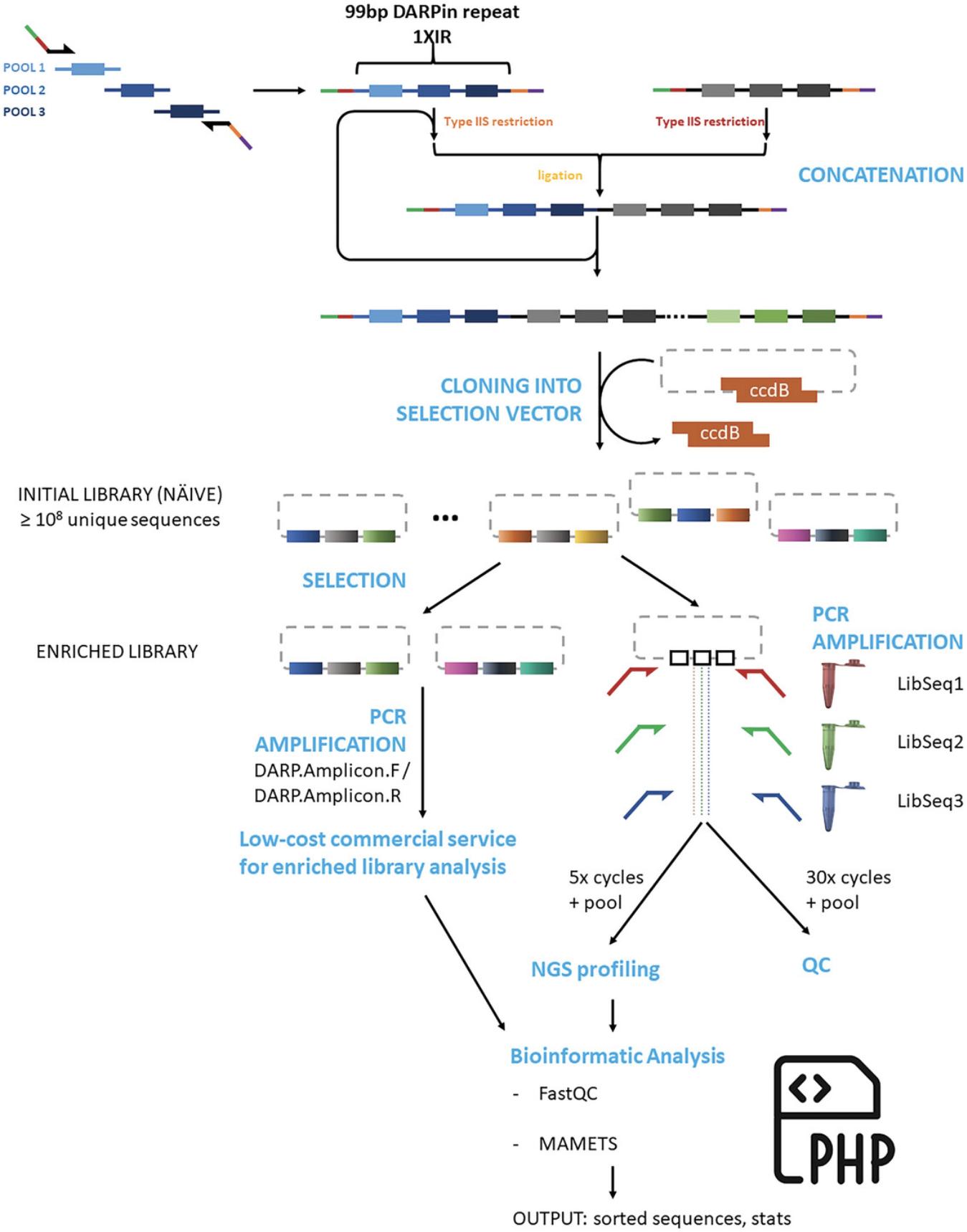
Basic Protocol 4: ESTIMATION OF LIBRARY SIZE AND DIVERSITY BY NEXT-GENERATION SEQUENCING (NGS)
An estimate of the library size can be calculated from the number of colony-forming units obtained after electroporation of the library into bacterial cells (Getz et al., 2012) (Basic Protocol 3). However, these measurements provide no information on library diversity, the correctness of the final assembled library, and the presence or absence of non-productive DNA species (e.g., cloning artifacts, frameshifts) The following protocol uses PCR amplification of the DARPin repeats from the plasmid maxiprep in Basic Protocol 3 to generate an NGS template. To avoid introduction of bias and other artifacts (see Current Protocols article: Podnar et al., 2014), the number of PCR cycles is limited to five, although an identical reaction of 25 to 30 PCR cycles is run in parallel to ensure that a DNA band of the correct size, as visualized by agarose gel electrophoresis, is generated by the PCR reaction.
The protocol will generate high-quality DNA fragments suitable for NGS analysis that will provide enough data to assess the library diversity. We suggest two potential instruments for NGS analysis, MiSeq and NovaSeq (both Illumina instruments), which are capable of the read lengths necessary for the full coverage of DARPin variable regions (∼300 bp); in most instances, researchers will access these instruments through core facilities or commercial entities, so instrument choice may differ.
Additional Materials (also see Basic Protocol 1)
- PCR reagents (see Table 8)
- SPRISelect beads (Beckman-Coulter, B23317)
- 80% (v/v) ethanol (from 200-proof stock; make fresh)
- 10 mM Tris·HCl, pH 8 (Thermo Scientific, J22638.AP)
- High-Sensitivity D1000 Assay (D1000 High Sensitivity ScreenTape, Agilent Technologies, 5067-5584, and D1000 High Sensitivity Reagents, Agilent Technologies, 5067-5585)
- NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, E7645S; alternatively, NEBNext Ultra II DNA Library Prep Kit for Illumina with Sample Purification Beads, New England Biolabs, E7103S, can be used and SPRISelect beads omitted)
- NEBNext® Multiplex Oligos for Illumina (Unique Dual Index UMI Adaptors DNA Set 1, New England Biolabs, E7395S; alternatively, use New England Biolabs, E7874S, E7876S, or E7878S)
- Qubit HS dsDNA Assay or equivalent
- Qubit 1× dsDNA HS Assay Kit (Thermo Fisher Scientific, Q33230) or equivalent (qPCR-based library quantification methods can be used, e.g., KAPA Library Quantification Kits - Complete kit (Universal), Roche Sequencing, KK4824-07960140001, or NEBNext Library Quant Kit for Illumina, New England Biolabs, E7630S)
- PhiX sequencing control (Illumina, FC-110-3001)
- DARP.Amplicon.F/DARP.Amplicon.R primer pair (Table 2)
| Component | Volume | Cycling parametersa | ||
|---|---|---|---|---|
| 10× Buffer for KOD Hot Start DNA Polymerase | 10 µl | Lid temperature | 105°C | |
| dNTPs (2 mM each; New England Biolabs, N0447S) | 10 µl | Polymerase activation | 95°C | 2 min |
| 25 mM MgSO4 | 10 µl | Denaturation | 95°C | 20 s |
| Forward and reverse primers (10 µM stock; see Table 2) | 3/3 µl | Annealing | 63°C | 10 s |
| Plasmid template (maxiprep DARPin library generated in Basic Protocol 3) | 100 ng per reaction | Extension | 70°C | 10 s |
| PCR-grade H2O | X µl | Final extension | 70°C | 20 s |
| 1 U/µl KOD HotStart DNA Polymerase (Millipore Sigma, 71086) | 2 µl | Hold | 4°C | |
| Final volume | 100 µl | |||
- a
Perform five cycles of amplification for library generation and 25 to 30 cycles for agarose gel electrophoresis.
-
Magnetic stand for PCR tubes (e.g., PCR Strip MagStand, Zymo Research, 3DP-1002)
-
Agilent TapeStation (4150, G2992AA, or 4200, G2991BA; alternatively, use any sensitive DNA analysis instrument, e.g., Agilent BioAnalyzer, Perkin Elmer LabChip GX, or similar)
-
Qubit fluorometer (Thermo Fisher Scientific, Q33238, or earlier versions) or real-time machine (if opting for qPCR-based quantification method)
-
NovaSeq6000 SP flowcell
-
Additional reagents and equipment for colony PCR (Bonnet et al., 2013; see Current Protocols article: Woodman et al., 2016) and NGS
Sample preparation for naïve library analysis
1.Prepare PCR reactions in sterile PCR tubes and perform PCR according to the PCR reagents and protocol in Table 8, using three sets of primers (Table 2: LibSeq1-3) in separate PCR reactions to prepare samples for NGS sequencing. Subject identical PCR reactions to 5 (100 μl volume) and 30 cycles (25 to 50 μl volume) of PCR amplification, with the 30-cycle PCR reaction subsequently analyzed by agarose gel electrophoresis to ensure that the reagents are functioning properly and that the desired product is amplified.
2.Assuming bands of the correct size, approximately 340 to 350 bp, are obtained (as determined by analysis of the 30-cycle PCR reactions), purify the 5-cycle reactions from low-molecular-weight DNA fragments (<200 bp).
3.To begin concentration, mix the PCR product with 0.8 volumes of SPRISelect beads (e.g., 80 μl SPRISelect for 100 μl PCR reaction). Incubate at room temperature for 5 to 10 min.
4.Transfer the tube to a magnetic stand for PCR tubes and incubate for 5 to 10 min or until the solution is clear.
5.While the tube is still on the magnet, carefully remove the supernatant (containing fragments <200 bp), add 200 μl freshly prepared 80% ethanol solution, and incubate for 30 s.
6.Keep the tube on the magnetic stand and discard the ethanol-containing supernatant.
7.Repeat steps 5 and 6 an additional time for a total of two washes with 80% ethanol. Completely remove any traces of ethanol and allow the samples to air-dry for 5 min or until the bead pellet is matte (not shiny).
8.Remove the tube from the magnetic stand and resuspend the bead pellet with 55 μl of 10 mM Tris·HCl, pH 8.Incubate for 5 min at room temperature.
9.Transfer the tube into the magnetic stand and incubate for ≥1 min or until the solution is clear.
10.Measure the size distribution and concentration of the purified, PCR-amplified DARPin libraries using 2 μl for the High-Sensitivity D1000 Assay on an Agilent TapeStation.
11.Transfer 50 μl into a new tube for NGS library preparation following the manufacturer's instructions for the NEBNext Ultra II DNA Library Prep Kit for Illumina using NEBNext® Multiplex Oligos for Illumina during the ligation step.
12.Use the starting amount of DNA measured in step 10 to calculate the approximate number of PCR cycles for the final amplification step (according to the manufacturer's recommendations).
13.Purify the final library with SPRISelect beads similarly to steps 3 to 9, except that the ethanol-washed beads are resuspended in 20 μl of 10 mM Tris·HCl, pH 8.
14.Subject the final libraries to quality control using the High-Sensitivity D1000 Assay according to the manufacturer's instructions and quantify with the Qubit 1× dsDNA HS Assay Kit or equivalent and Qubit fluorometer or real-time machine. Then, dilute final libraries according to Illumina's recommendations and sequence on a NovaSeq6000 SP flowcell as 2 × 250 bases after spiking in 1% to 5% PhiX sequencing control in order to increase the complexity of the sequenced libraries.
Sample preparation for enriched library analysis
15.For analysis of enriched libraries after multiple selection rounds, use the same PCR reaction parameters as for the naïve library but PCR-amplify the DARPin-encoding region from individual colonies or a cell suspension by colony PCR (Bonnet et al., 2013; see Current Protocols article: Woodman et al., 2016) using the DARP.Amplicon.F/DARP.Amplicon.R primer pair (Table 2) to generate a ∼365-bp PCR product.
16.Visualize, gel-extract, and quantify the PCR amplicon as in Basic Protocol 1.
17.Perform NGS to analyze enriched populations.
Basic Protocol 5: NGS ANALYSIS OF NAÏVE AND ENRICHED LIBRARIES
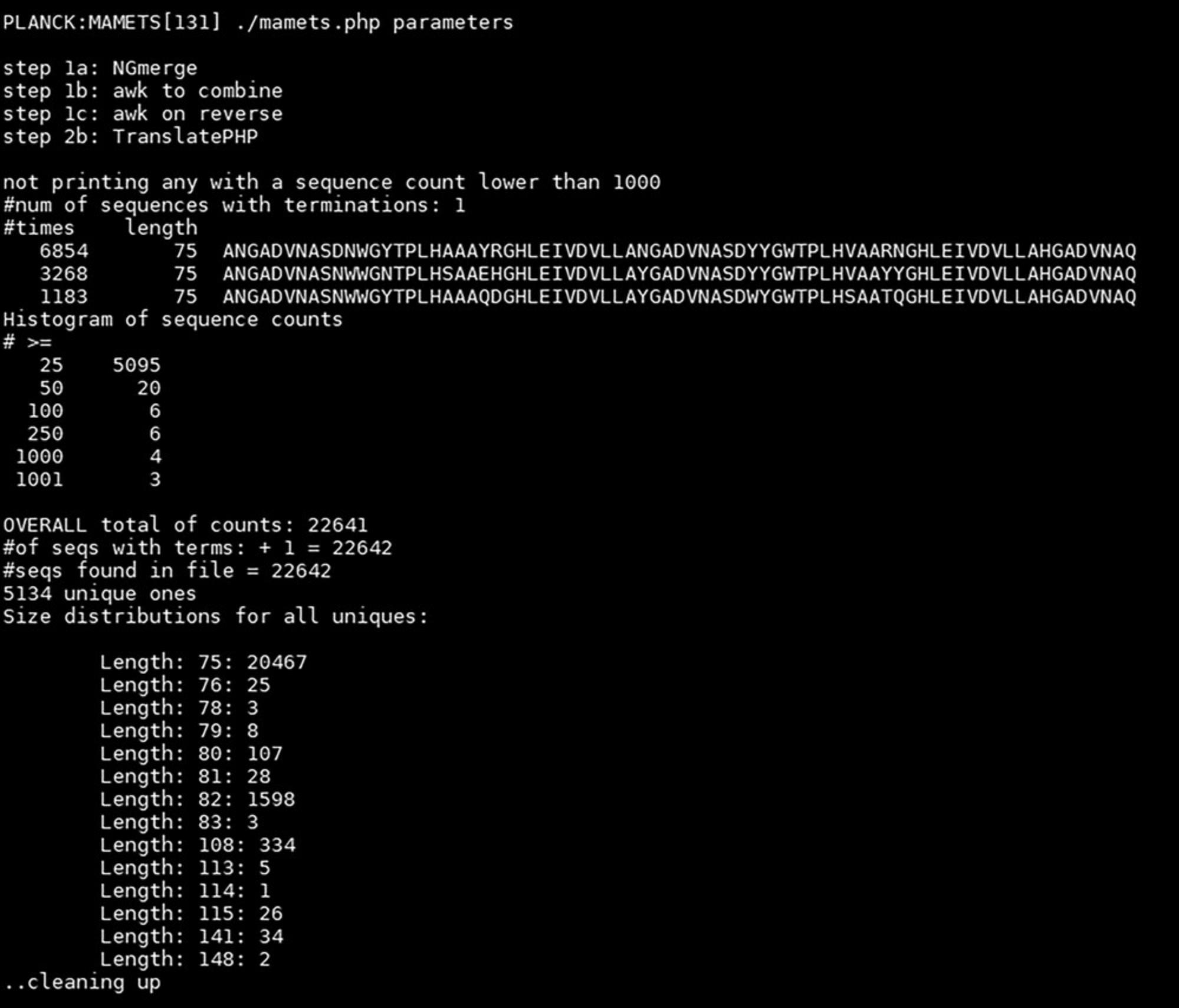
Analysis of paired-end read data from NGS of DARPin PCR amplicons is complicated by the high sequence identity of internal DARPin repeats (75 of the 99 bp of a repeat are conserved, as they represent invariant amino acids), and programs that create merged sequences often discard sequences or incorrectly merge sequences (e.g., a DARPin sequence that contains three repeats is incorrectly assembled into a merged sequence containing two internal repeats with the sequence of one repeat discarded). Experimentally, this problem can be solved by using a single-read sequencing technology or by altering the library design such that internal repeats are generated with distinct primer pools that use different synonymous codons for invariant positions. Fortunately, NGmerge (Gaspar, 2018), an open-source program, is a capable of merging repetitive DARPin sequences.
Bioinformatic analysis of NGS sequence data requires a significant amount of expertise for complicated datasets. In contrast, basic DARPin library analysis to determine the number of unique sequences in naïve libraries, e.g., library diversity, and abundance of specific sequences in enriched libraries, e.g., enriched DNA sequences of the target-specific binders, is relatively simple. Data quality can be assessed with free tools (e.g., FastQC), the paired-end reads assembled into a single read with NGmerge, and merged DNA sequences translated and sorted using basic php scripts. We have written a simple program (MAMETS) that uses demultiplexed files as input and that merges paired-end reads, removes the adapter sequences used by sequencing instruments, reverse-complements reverse sequences, translates DNA sequences into one-letter amino acid code, sorts sequences by abundance, and writes out basic statistics on sequence abundance and characteristics. Parameters are easily modifiable for analysis of other library types (e.g., nanobodies).
Materials
- Demultiplexed NGS dataset (provided by sequencing service; see Basic Protocol 4)
- Computer with Linux and php interpreter installed
Procedure
1.Download demultiplexed NGS dataset from sequencing service/facility and perform simple quality-control checks using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Specifically, verify that the sequence length and %GC content match the anticipated values.
2.Download MAMETS (https://srv.mbi.ucla.edu/MAMETS) and extract/unzip into an accessible directory. Review the instructions for running the program in the README.txt file.
3.Edit the parameter file (labeled parameters) to indicate the location of the NGS sequence files (inputR1 and inputR2).
4.Modify the parameter file to include the 5′ and 3′ invariant nucleotide sequences that flank the variable library region for sequences in both the forward and reverse directions.
5.Set the NGmerge mismatch parameter (probability) for file merging.
6.Edit the parameter file to specify the minimum sequence length of the merged files for subsequent analysis.
7.Specify the reading frame in the parameter file for translation of merged nucleotide sequences into one-letter amino acid sequences.
8.Modify the skipN parameter in the parameter file to specify the threshold for writing out translated amino acid sequences.
9.Run the script from the MAMETS directory using “./mamets.php parameters”.
COMMENTARY
Background Information
Library generation is a time-consuming endeavor that requires considerable planning in order to generate a highly diverse library capable of producing high-affinity binders. The first consideration is which DARPin scaffold to use, as there are a number of variations (Schilling et al., 2014; Seeger et al., 2013) on the original scaffold (Binz et al., 2004); the approach described herein uses a second-generation scaffold (Seeger et al., 2013). The second consideration is the choice of amino acids at each variable position and the degenerate codons to be used to incorporate the desired amino acids. Many protein binder libraries have chosen to exclude cysteine, due to its chemical reactivity, and proline, due to conformational restraints. Excellent discussions on the choice of amino acids to include at variable positions in DARPin and nanobody libraries have been published (Chen et al., 2021; McMahon et al., 2018; Seeger et al., 2013; Valdés-Tresanco et al., 2022). Using degenerate codons to encode amino acids at variable positions has the potential to bias the amino acid composition at each position toward amino acids encoded by four or more codons (e.g., arginine, serine, leucine) at the expense of those amino acids encoded by two or fewer codons; the use of a different construction method, e.g., gene synthesis using nucleoside phosphoramidites, allows fine control of amino acid distribution but is significantly more expensive. Various programs (Jacobs et al., 2015) and web-based services (http://algo.tcnj.edu/decodoncalc/) are available to assess and evaluate the possible codon choices. The simplest choice is to use NNK or NNB degenerate codons, which encode all 20 amino acids, but at the expense of skewed amino acid frequencies and the inclusion of a stop codon (TAG). A recent synthetic nanobody library was generating using NNB codons but included an in vitro translation technique to eliminate transcripts that included stop codons (Chen et al., 2021); this procedure could be adapted to the creation of a DARPin library. Our approach uses a number of different degenerate codons at each variable position to produce a balanced distribution of amino acids with desired chemical properties and no stop codons and which requires ∼90 oligonucleotide primers.
Finally, the desired library diversity and thus size must be considered. The general consensus is that a library size of >108 unique sequences is sufficient to identify high-affinity binders to the target of interest. High diversity at the molecular level is relatively easy to achieve (1 ng of a 99-bp DNA fragment encoding a DARPin repeat is 1 × 1010 molecules) and is part of the allure of cell-free systems (e.g., ribosome and mRNA display), which can easily achieve library sizes in excess of 1012 unique sequences (He & Taussig, 2002). In vivo systems are, however, limited by transformation efficiency of the organism of choice, which typically restricts library sizes to 108 to 1010.
Critical Parameters
Beyond the initial design of the library, the most critical factors in the described experiments are minimizing bias in the library and generating sufficient quantities of input DNA for each protocol. Bias in the library can be introduced by PCR amplification during library generation and in amplicon preparation for NGS analysis through a number of mechanisms (Kebschull & Zador, 2015; Krehenwinkel et al., 2017). Use of high-fidelity DNA polymerases and minimization of the number of PCR amplification cycles, for all protocols, can significantly reduce the introduction of bias into the naïve library or PCR amplicons prepared for NGS. Large amounts of input DNA (in excess of 10 µg) are required for most of the library construction protocols, so PCR reactions should be scaled accordingly, taking into consideration that significant amounts of material are lost during manipulation (purification, restriction enzyme digestion, and ligation) of the DNA fragments.
Troubleshooting
Common problems in executing the protocols are listed in Table 9.
| Problem | Possible cause | Solution |
|---|---|---|
| Basic Protocol 1: No band of appropriate size present | Reaction parameters not optimized | Use the gradient function of the thermal cycler to simultaneously evaluate annealing temperatures and template concentrations |
| Chosen polymerase may not be optimal | Try using a different high-fidelity polymerase | |
| Basic Protocol 2: Low yield from gel extraction step | Gel extraction/purification is inefficient | Increase input DNA in ligation step |
| Basic Protocol 2: Multiple bands are present after gel extraction/purification | Contamination of desired ligation product with input DNA | Clean gel electrophoresis apparatus; electrophorese samples for longer period of time at lower voltage to ensure complete separation of fragments |
| Basic Protocol 3: Low transformation efficiency | Ratios in ligation reaction incorrect; ligation reaction not properly desalted; electrocompetent cells incorrectly prepared or stored | Optimize using small-scale reactions; desalt ligation reactions ≥2 hr; compare self-prepared cells with purchased cells |
| Basic Protocol 5: Low number of merged output sequences | Wrong dataset; sequencing failure; incorrect merging parameters | Evaluate number and average length of sequences in sequencing instrument output with FastQC; manually examine ∼10 sequences to look for error source; modify mismatch parameter for sequence merging |
Understanding Results
This series of protocols explains a relatively straightforward method to generate a diverse DARPin library and a simple program (MAMETS) to analyze library diversity. Enriched libraries obtained using the display technique of choice are analyzed using the same program to determine which binder sequences have been enriched. The MAMETS program can also be applied to other library types (e.g., nanobody or scFv) by modifying sequence-specific parameters in the parameters file.
Library construction
The library is constructed using standard procedures (PCR, restriction enzyme digestion, ligation) common in labs that routinely use molecular biology techniques. Basic Protocols 1 and 2 generate about 100- to 300-bp DNA fragments that encode single or concatenated DARPin repeat modules, and Basic Protocol 3 incorporates the library into an appropriate display vector. The results from these protocols will be DNA fragments or plasmids that are visualized by agarose gel electrophoresis (Figs. 3 and 5) to ensure that the DNA fragments are the correct size. The presence of M13 primer sequences on the 5′ and 3′ ends of the DARPin repeats and vector-specific sequencing primers, for display vectors, allows Sanger sequencing of the DARPin repeats as they are amplified, concatenated, and ligated into the display vector to estimate whether library construction is proceeding correctly. Sanger sequencing results can verify whether the fragments are in-frame and if concatenated repeats are correctly assembled (note that invariant bases will be correctly assigned, whereas variable bases will be assigned two or more nucleotide bases depending on the degenerate base).
Library analysis by next-generation sequencing
Sample preparation of the naïve library for NGS analysis requires similar molecular biology techniques (PCR, ligation) as for library construction but also requires bead-based DNA fragment purification and sensitive DNA quantification methods and instrumentation. Biochemistry labs that lack the necessary expertise and instrumentation may elect to provide the PCR amplicons generated in the first two steps of Basic Protocol 4 to a sequencing facility for sample preparation and sequencing. The result from Basic Protocol 4 is raw sequencing data; inexpensive amplicon sequencing of enriched libraries will generate 50 to 200K reads, whereas the thorough analysis of naïve libraries using a NovaSeq6000 SP flowcell, as detailed in this protocol, will produce in excess of 1.3 × 109 reads.
Basic Protocol 5 uses the raw sequencing data generated by Basic Protocol 4 as input, with the result being an easy-to-interpret collection of statistics (Fig. 6). Enriched libraries will have a relatively small number of sequences that are very abundant and that represent likely binders or false positives that bind to a component of the display system. The results for analysis of the naïve library will, optimally, show no enrichment of any particular sequence and a very large number of unique sequences, indicating high diversity.
Time Considerations
The choice and design of the library of interest should be carefully considered, as a significant amount of time will be required to generate the final library. Basic Protocol 1 can be accomplished in 1 week if multiple parameters are evaluated early (e.g., polymerase and PCR reaction conditions). Basic Protocols 2 and 3 may take an experienced researcher several months to accomplish, as restriction enzyme digestions, ligations, and subsequent amplification of DNA products can take considerable time due to the inefficiency of the gel extraction and purification steps. Subsequently, the large-scale ligation requires large amounts of products to be prepared and small-scale test ligations and transformations to be carried out and optimized prior to scaling up the reactions. Preparation of sufficient amounts of electrocompetent cells in-house can also be time consuming; this step could be expedited by purchasing commercially prepared cells (which may be prohibitively expensive for very large libraries). Preparation of amplicons for NGS analysis (Basic Protocol 4) can be completed in 1 week, although the actual sequencing may take 1 to 2 weeks depending on the sequencing facility's work schedule. The analysis of the NGS data (Basic Protocol 5) can be completed in 1 day.
Acknowledgments
This work was supported by the United States Department of Energy grant DE-FC02-02ER63421. We thank Calin Plesa, Cliff Boldridge, Guillaume Urtecho, and Andrew Hausrath for helpful discussions about DNA library construction.
Author Contributions
Marco Morselli : Investigation; methodology; writing—review and editing. Thomas R. Holton : Data curation; formal analysis; methodology; software; writing—review and editing. Matteo Pellegrini : Methodology; resources; writing—review and editing. Todd O. Yeates : Conceptualization; funding acquisition; resources; writing—review and editing. Mark A. Arbing : Conceptualization; data curation; formal analysis; investigation; methodology; project administration; validation; visualization; writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Data available on request from the authors.
Supporting Information
| Filename | Description |
|---|---|
| cpz1960-sup-0001-SuppMat.docx14.6 KB | Sequences of oligonucleotides required to generate an internal DARPin repeat module by overlap extension PCR and DNA sequence encoding the N- and C-terminal capping repeats, in yellow and cyan, respectively. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Binz, H. K., Amstutz, P., Kohl, A., Stumpp, M. T., Briand, C., Forrer, P., Grütter, M. G., & Plückthun, A. (2004). High-affinity binders selected from designed ankyrin repeat protein libraries. Nature Biotechnology , 22(5), 575–582. https://doi.org/10.1038/nbt962
- Binz, H. K., Stumpp, M. T., Forrer, P., Amstutz, P., & Plückthun, A. (2003). Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. Journal of Molecular Biology , 332(2), 489–503. https://doi.org/10.1016/S0022-2836(03)00896-9
- Boersma, Y. L. (2018). Advances in the application of Designed Ankyrin Repeat Proteins (DARPins) as research tools and protein therapeutics. Methods in Molecular Biology , 1798, 307–327. https://doi.org/10.1007/978-1-4939-7893-9_23
- Bonnet, C., Rigaud, C., Chanteclaire, E., Blandais, C., Tassy-Freches, E., Arico, C., & Javaud, C. (2013). PCR on yeast colonies: An improved method for glyco-engineered Saccharomyces cerevisiae. BMC Research Notes , 6(1), 201. https://doi.org/10.1186/1756-0500-6-201
- Castells-Graells, R., Meador, K., Arbing, M. A., Sawaya, M. R., Gee, M., Cascio, D., Gleave, E., Debreczeni, J. É., Breed, J., Leopold, K., Patel, A., Jahagirdar, D., Lyons, B., Subramaniam, S., Phillips, C., & Yeates, T. O. (2023). Cryo-EM structure determination of small therapeutic protein targets at 3 Å-resolution using a rigid imaging scaffold. Proceedings of the National Academy of Sciences , 120(37), e2305494120. https://doi.org/10.1073/pnas.2305494120
- Chao, G., Lau, W. L., Hackel, B. J., Sazinsky, S. L., Lippow, S. M., & Wittrup, K. D. (2006). Isolating and engineering human antibodies using yeast surface display. Nature Protocols , 1(2), 755–768. https://doi.org/10.1038/nprot.2006.94
- Chen, X., Gentili, M., Hacohen, N., & Regev, A. (2021). A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing nanobodies. Nature Communications , 12(1), 1. https://doi.org/10.1038/s41467-021-25777-z
- Gaspar, J. M. (2018). NGmerge: Merging paired-end reads via novel empirically-derived models of sequencing errors. BMC Bioinformatics , 19(1), 536. https://doi.org/10.1186/s12859-018-2579-2
- Getz, J. A., Schoep, T. D., & Daugherty, P. S. (2012). Peptide discovery using bacterial display and flow cytometry. In K. D. Wittrup & G. L. Verdine (Eds.), Methods in enzymology (Vol. 503, pp. 75–97). Academic Press. https://doi.org/10.1016/B978-0-12-396962-0.00004-5
- Harmansa, S., & Affolter, M. (2018). Protein binders and their applications in developmental biology. Development , 145(2), dev148874. https://doi.org/10.1242/dev.148874
- He, M., & Taussig, M. J. (2002). Ribosome display: Cell-free protein display technology. Briefings in Functional Genomics , 1(2), 204–212. https://doi.org/10.1093/bfgp/1.2.204
- Jacobs, T. M., Yumerefendi, H., Kuhlman, B., & Leaver-Fay, A. (2015). SwiftLib: Rapid degenerate-codon-library optimization through dynamic programming. Nucleic Acids Research , 43(5), e34. https://doi.org/10.1093/nar/gku1323
- Kebschull, J. M., & Zador, A. M. (2015). Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Research , 43(21), e143. https://doi.org/10.1093/nar/gkv717
- Kenrick, S., Rice, J., & Daugherty, P. (2007). Flow cytometric sorting of bacterial surface-displayed libraries. Current Protocols in Cytometry , 42, 4.6.1–4.6.27. https://doi.org/10.1002/0471142956.cy0406s42
- Krehenwinkel, H., Wolf, M., Lim, J. Y., Rominger, A. J., Simison, W. B., & Gillespie, R. G. (2017). Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Scientific Reports , 7(1), 1. https://doi.org/10.1038/s41598-017-17333-x
- Kumar, A., & Balbach, J. (2021). Folding and stability of ankyrin repeats control biological protein function. Biomolecules , 11(6), 6. https://doi.org/10.3390/biom11060840
- Liu, Y., Huynh, D. T., & Yeates, T. O. (2019). A 3.8 Å resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nature Communications , 10(1), 1864. https://doi.org/10.1038/s41467-019-09836-0
- McMahon, C., Baier, A. S., Pascolutti, R., Wegrecki, M., Zheng, S., Ong, J. X., Erlandson, S. C., Hilger, D., Rasmussen, S. G. F., Ring, A. M., Manglik, A., & Kruse, A. C. (2018). Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nature Structural & Molecular Biology, 25(3), 289–296. https://doi.org/10.1038/s41594-018-0028-6
- Mohan, K., Ueda, G., Kim, A. R., Jude, K. M., Fallas, J. A., Guo, Y., Hafer, M., Miao, Y., Saxton, R. A., Piehler, J., Sankaran, V. G., Baker, D., & Garcia, K. C. (2019). Topological control of cytokine receptor signaling induces differential effects in hematopoiesis. Science , 364(6442), eaav7532. https://doi.org/10.1126/science.aav7532
- Pardon, E., Laeremans, T., Triest, S., Rasmussen, S. G. F., Wohlkönig, A., Ruf, A., Muyldermans, S., Hol, W. G. J., Kobilka, B. K., & Steyaert, J. (2014). A general protocol for the generation of nanobodies for structural biology. Nature Protocols , 9(3), 674–693. https://doi.org/10.1038/nprot.2014.039
- Plückthun, A. (2015). Designed Ankyrin Repeat Proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annual Review of Pharmacology and Toxicology , 55(1), 489–511. https://doi.org/10.1146/annurev-pharmtox-010611-134654
- Podnar, J., Deiderick, H., & Hunicke-Smith, S. (2014). Next-generation sequencing fragment library construction. Current Protocols in Molecular Biology , 107(1), 7.17.1–7.17.16. https://doi.org/10.1002/0471142727.mb0717s107
- Rice, J. J., & Daugherty, P. S. (2008). Directed evolution of a biterminal bacterial display scaffold enhances the display of diverse peptides. Protein Engineering, Design & Selection: PEDS, 21(7), 435–442. https://doi.org/10.1093/protein/gzn020
- Schilling, J., Schöppe, J., & Plückthun, A. (2014). From DARPins to LoopDARPins: Novel LoopDARPin design allows the selection of low picomolar binders in a single round of ribosome display. Journal of Molecular Biology , 426(3), 691–721. https://doi.org/10.1016/j.jmb.2013.10.026
- Schladetsch, M. A., & Wiemer, A. J. (2021). Generation of single-chain variable fragment (scFv) libraries for use in phage display. Current Protocols , 1(7), e182. https://doi.org/10.1002/cpz1.182
- Seeger, M. A., Zbinden, R., Flütsch, A., Gutte, P. G. M., Engeler, S., Roschitzki-Voser, H., & Grütter, M. G. (2013). Design, construction, and characterization of a second-generation DARP in library with reduced hydrophobicity. Protein Science: A Publication of the Protein Society , 22(9), 1239–1257. https://doi.org/10.1002/pro.2312
- Shilova, O. N., & Deyev, S. M. (2019). DARPins: promising scaffolds for theranostics. Acta Naturae , 11(4), 42–53. https://doi.org/10.32607/20758251-2019-11-4-42-53
- Steiner, D., Forrer, P., & Plückthun, A. (2008). Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. Journal of Molecular Biology , 382(5), 1211–1227. https://doi.org/10.1016/j.jmb.2008.07.085
- Valdés-Tresanco, M. S., Molina-Zapata, A., Pose, A. G., & Moreno, E. (2022). Structural insights into the design of synthetic nanobody libraries. Molecules , 27(7), 2198. https://doi.org/10.3390/molecules27072198
- Voytas, D. (2000). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51(1), 2.5A.1–2.5A.9. https://doi.org/10.1002/0471142727.mb0205as51
- Wendel, S., Fischer, E. C., Martínez, V., Seppälä, S., & Nørholm, M. H. H. (2016). A nanobody:GFP bacterial platform that enables functional enzyme display and easy quantification of display capacity. Microbial Cell Factories , 15, 71. https://doi.org/10.1186/s12934-016-0474-y
- Woodman, M. E., Savage, C. R., Arnold, W. K., & Stevenson, B. (2016). Direct PCR of intact bacteria (colony PCR). Current Protocols in Microbiology , 42(1), A.3D.1–A.3D.7. https://doi.org/10.1002/cpmc.14
- Wu, C.-H., Liu, I.-J., Lu, R.-M., & Wu, H.-C. (2016). Advancement and applications of peptide phage display technology in biomedical science. Journal of Biomedical Science , 23(1), 8. https://doi.org/10.1186/s12929-016-0223-x
- Zadravec, P., Štrukelj, B., & Berlec, A. (2015). Improvement of LysM-mediated surface display of Designed Ankyrin Repeat Proteins (DARPins) in recombinant and nonrecombinant strains of Lactococcus lactis and Lactobacillus species. Applied and Environmental Microbiology , 81(6), 2098–2106. https://doi.org/10.1128/AEM.03694-14
Citing Literature
Number of times cited according to CrossRef: 1
- Matthew P. Agdanowski, Roger Castells-Graells, Michael R. Sawaya, Duilio Cascio, Todd O. Yeates, Mark A. Arbing, X‐ray crystal structure of a designed rigidified imaging scaffold in the ligand‐free conformation, Acta Crystallographica Section F, 10.1107/S2053230X2400414X, 80 , 5, (107-115), (2024).

