Immunofluorescence-Mediated Detection of Respiratory Virus Infections in Human Airway Epithelial Cultures
Talita B. Gagliardi, Talita B. Gagliardi, Ethan Iverson, Ethan Iverson, Emma J. DeGrace, Emma J. DeGrace, Brad R. Rosenberg, Brad R. Rosenberg, Margaret A. Scull, Margaret A. Scull
Abstract
A diverse collection of viral pathogens target airway epithelial cells for infection, with effects ranging from mild upper respiratory tract symptoms to death of the infected individual. Among these pathogens are recently discovered and/or emergent viruses that sometimes fail to infect commonly used, immortalized cell lines and for which infection phenotypes in the respiratory tract remain unknown. Human airway epithelial cultures have been developed over the past several decades and have proven to be a useful model system in culturing hard-to-grow viruses and assaying various features of infection in a physiologically relevant setting. This article includes methods for the generation of well-differentiated human airway epithelial cell cultures at air-liquid interface that recapitulate the mucosal epithelium of the trachea/bronchus in vivo. We further detail inoculation of these cultures with respiratory viruses—specifically rhinovirus, influenza virus, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—and provide a protocol for the detection of double-stranded RNA or viral antigen–positive cells by immunofluorescence microscopy. These techniques, together with a post-imaging analysis, can be applied to characterize the efficiency of infection and kinetics of spread within the airway epithelium. Furthermore, these methods can be utilized in conjunction with antibodies against cellular targets to determine cell tropism and colocalization with specific host factors during infection. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation of human airway epithelial cultures at air-liquid interface (HAE-ALI)
Basic Protocol 2 : Viral inoculation of HAE-ALI
Basic Protocol 3 : Immunofluorescence (IF)-based detection of infected cells in HAE-ALI
INTRODUCTION
Viruses are obligate intracellular parasites, and virus-host interactions depend on both the particular virus and the cellular landscape in which infection occurs. For important human respiratory viruses such as influenza A virus (IAV), rhinovirus (RV) C, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), among many others, the airway epithelium is the primary site of replication (Hou et al., 2020; Long, Mistry, Haslam, & Barclay, 2019; Royston & Tapparel, 2016). Human airway epithelial cultures (HAE) are a well-characterized, in vitro model system derived from primary airway basal cells that are differentiated on permeable membrane supports to yield a pseudostratified epithelium at air-liquid interface (ALI). This system recapitulates many aspects of in vivo physiology (Bukowy-Bieryłło, 2021) and has been widely applied to the study of epithelial cell biology and respiratory viral infections (Rijsbergen, van Dijk, Engel, de Vries, & de Swart, 2021). Specifically, HAE cultures offer researchers the opportunity to identify or confirm virus-host interactions in a primary human culture system and also investigate certain aspects of infection that are not feasible to achieve in monolayer culture of homogenous, undifferentiated cells submerged in aqueous medium. This includes determination of cell tropism and the efficiency of infection and spread within the mucosal microenvironment, which can be assessed using immunostaining and fluorescence microscopy.
The generation and subsequent inoculation and analysis of infection in normal human airway cultures begins with the expansion of primary airway basal cells that have been isolated from deceased donors. During the expansion phase, methods are optimized to maximize cell numbers and simultaneously retain differentiation capacity. Cells are then transferred onto Transwells and subsequently cultured at air-liquid interface, where culture medium is removed from the apical chamber, allowing for the polarization and differentiation of multiple cell types over the course of several weeks. In fully differentiated cultures, secretory cells and ciliated cells are found that produce and transport mucus secretions, respectively, across the culture surface. At this mature stage, HAE cultures can be utilized for a variety of viral infection studies. Given the typical route of respiratory virus transmission via direct (i.e., finger-mediated) inoculation of epithelial surfaces or inhalation of aerosolized particles, inoculation is typically performed by application of the virus to the apical (lumenal) culture surface (Figs. 1 and 2; Rijsbergen et al., 2021). The specific virus under investigation, as well as the experimental question being addressed, influences whether mucus secretions are removed prior to viral challenge, as well as the titer and volume of viral suspension applied and duration of inoculation. At various time points post-inoculation, apical washes and basolateral medium can be collected for subsequent detection of cell-free virus and/or soluble mediators of the host response. The HAE cultures themselves can also be lysed for protein analysis, dissociated to obtain a single-cell suspension for flow cytometry or single-cell RNA-seq applications, or fixed and stained for viral and/or cellular proteins.
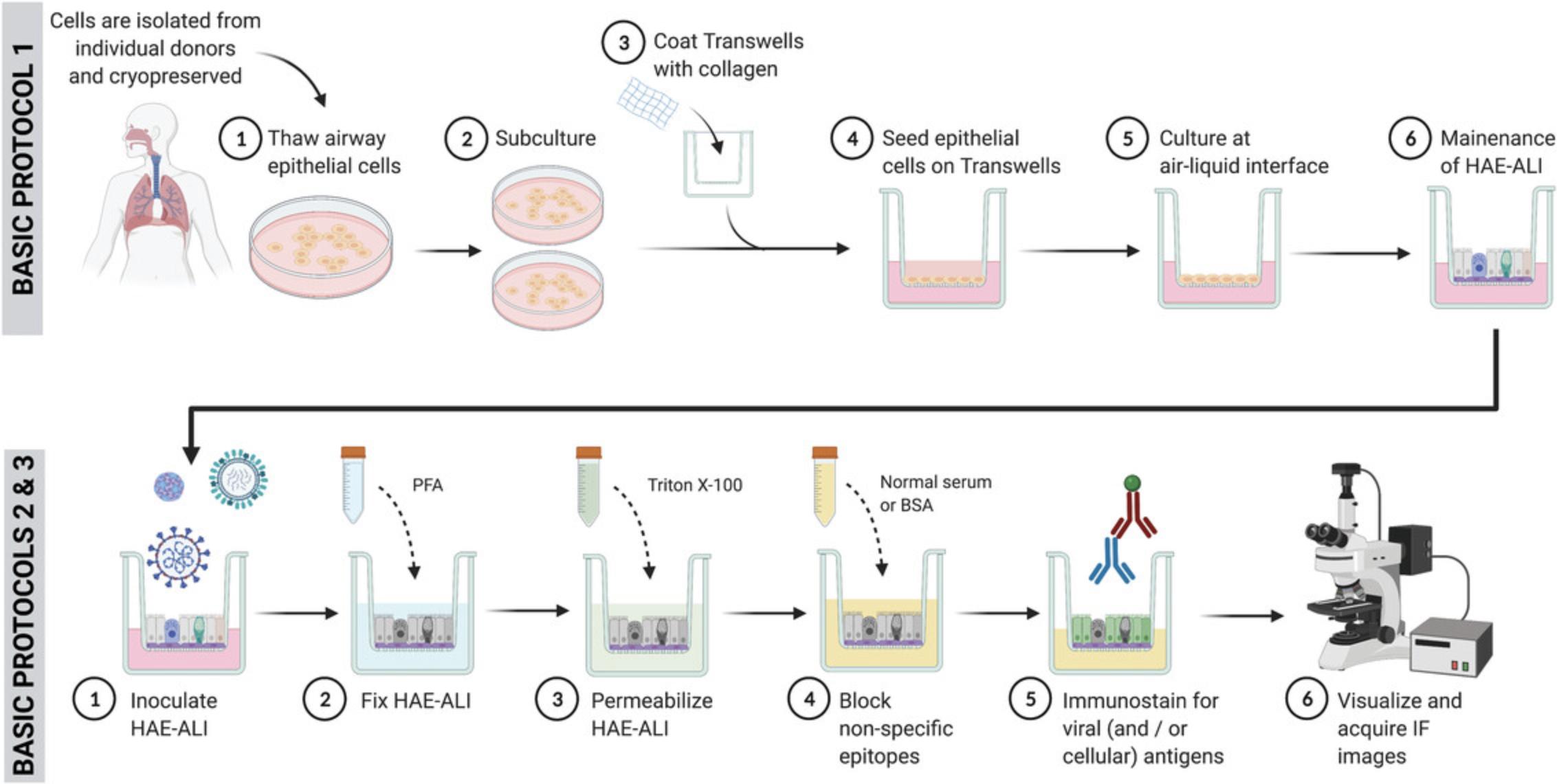
In this article, we describe protocols for the generation of HAE-ALI from commercially sourced tracheal/bronchial cells, apical inoculation of these cultures with RV-C, IAV, and SARS-CoV-2, and characterization of infection using immunofluorescence microscopy (Fig. 1). Through these protocols, researchers can expect to garner data addressing the frequency of infected cells under normal or experimental conditions, such as in the presence or absence of secreted mucus, following culture at different temperatures, or for different time periods. In addition, the types of cells infected, as well as the subcellular localization of viral components and co-localization with host proteins of interest, can be observed by pairing viral antigen detection with staining for specific cellular markers. These outputs have relevance for assessing the potential of novel viruses to infect the human respiratory tract and in defining the replication cycle of those viruses known to cause disease in humans in a relevant in vitro model system.
CAUTION : These protocols involve work with unfixed primary human cells and infectious respiratory viruses known to cause disease in humans. Rhinovirus is a Biosafety Level 2 (BSL-2) pathogen. Severe acute respiratory syndrome coronavirus 2 is a Biosafety Level 3 (BSL-3) pathogen. Influenza A virus biosafety level depends on the particular strain, with some classified as BSL-2 and others as BSL-3. Follow all appropriate guidelines and regulations for the use and handling of human-derived materials and pathogenic microorganisms. See Current Protocols article Burnett, Lunn, & Coico (2009) for more information.
Basic Protocol 1: GENERATION OF HUMAN AIRWAY EPITHELIAL CULTURES AT AIR-LIQUID INTERFACE (HAE-ALI)
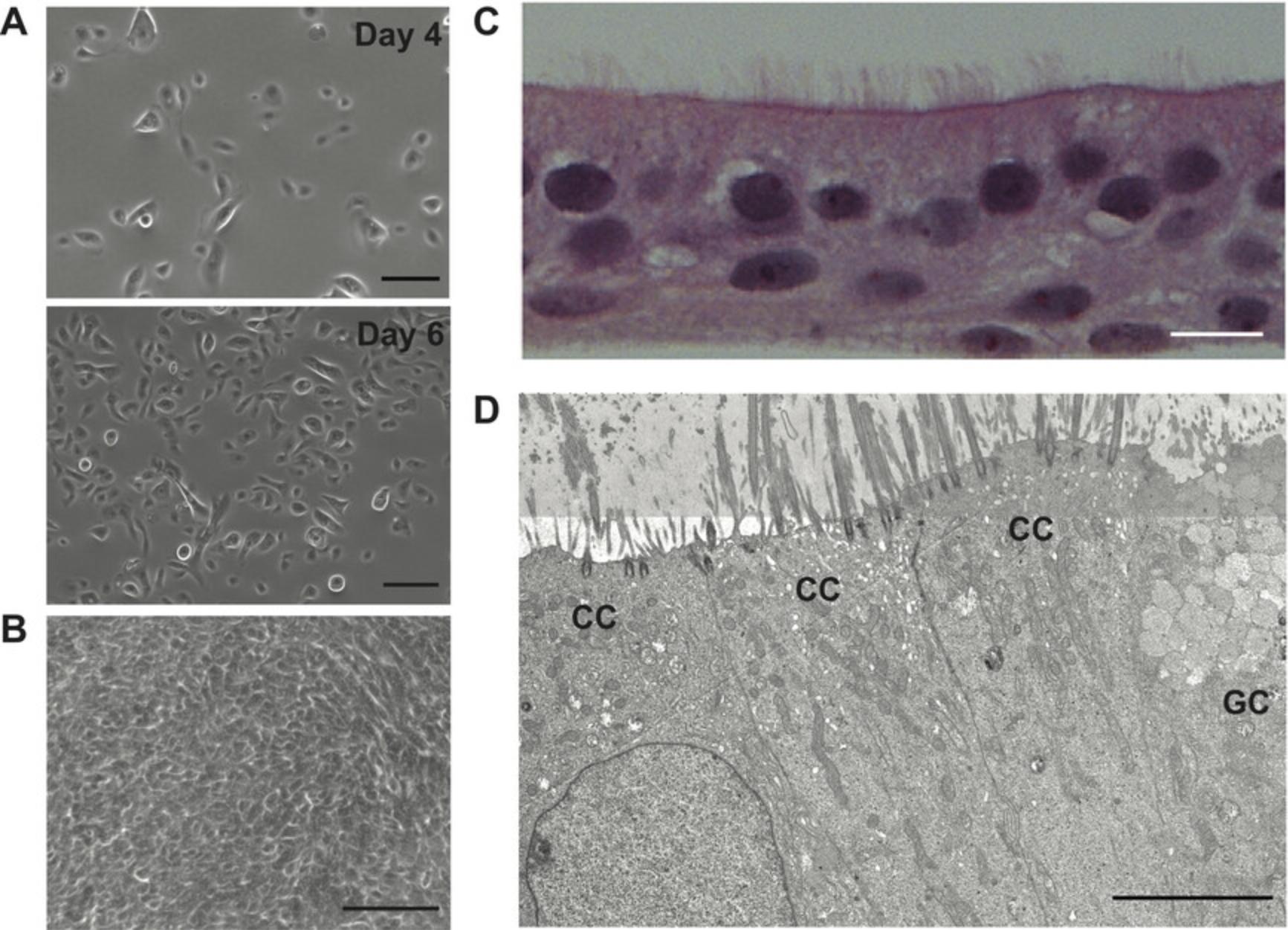
This protocol is used to obtain a well-differentiated human bronchial epithelium in vitro. In brief, undifferentiated cells are first expanded in T-175 flasks and then seeded onto permeable membrane supports to follow differentiation. Cultivating HAE under ALI conditions and with specific cell culture medium will result in a multilayered epithelium with similar morphology and physiology as observed in vivo , which includes beating cilia, production and transport of mucus, and expression of antiviral and pro-inflammatory cytokines.
Materials
-
PneumacultTM-EX Complete medium (see recipe)
-
Normal human bronchial/tracheal epithelial cells (Lonza; cat. no. CC2540S or equivalent)
-
Phosphate-buffered saline (PBS) without Ca2+ or Mg2+ (Gibco; cat. no. P5119)
-
0.025% trypsin/EDTA (see recipe)
-
Trypsin inhibitor solution (TIS; see recipe)
-
0.4% trypan blue solution (Gibco; cat. no. 15250061)
-
Collagen Type I–rat tail working solution (see recipe)
-
PneumacultTM-ALI Maintenance medium (see recipe)
-
Certified class II biological safety cabinet
-
Water baths at 37°C and 56°C
-
175-cm2 (T-175) cell culture flasks (Corning; cat. no. 132705 or equivalent)
-
Laboratory marking pens
-
Humidified incubator at 37°C with 5% CO2
-
Timer
-
Refrigerator or cold room at 4°C
-
Freezer at −20°C
-
Light microscope with 4×, 10×, 20×, and 40× objective lenses
-
Pipet-Aid
-
Sterile serological pipettes: 5 ml (Corning; cat. no. 07-200-9 or equivalent), 10 ml (Corning; cat. no. 07-200-12 or equivalent), 25 ml (Corning; cat. no. 07-200-15 or equivalent)
-
2D-Chip disposable hemocytometer (Bulldog Bio; cat. no. DHC-N002 or equivalent)
-
Centrifuge
-
Sterile polypropylene conical tubes: 15-ml (Fisherbrand; cat. no. 07-200-886 or equivalent) and 50-ml (Fisherbrand; cat. no. 05-539-13 or equivalent)
-
Micropipettes
-
Sterile 6.5-mm Transwells with 0.4-µm-pore polyester membrane inserts (Corning; cat. no. 3470)
-
24-well plates
-
Additional reagents and equipment for counting cells with trypan blue exclusion and hemocytometer (see Current Protocols article: Strober, 2001).
Phase 1: Thaw normal human bronchial/tracheal epithelial cells
1.Working in a certified class II biological safety cabinet, add 25 ml of PneumacultTM-EX Complete medium to a T-175 flask.
2.Warm the medium by incubating the flask in the 37°C incubator for 30 min.
3.Remove the epithelial cells from cryostorage and thaw rapidly in a water bath at 37°C.
4.Add one vial of cells (if using Lonza cat. no. CC2540S; or < 2 × 106 cells if thawing another cryopreserved stock) to the T-175 flask and disperse the cells by rocking the flask.
5.Label the flask with the following information: donor number/date/passage number.
6.Incubate the culture overnight at 37°C with 5% CO2.
7.The following day, discard the medium from the flask and replace it with 25 ml of fresh, warm PneumacultTM-EX Complete medium.
8.Incubate the culture overnight at 37°C with 5% CO2.
9.Every 2 days, change the medium as described above (steps 7-8).
10.Subculture the cells when the monolayer achieves 80% confluence (Fig. 2A, see “day 6” photo).
Phase 2: Sub-culture normal human bronchial/tracheal epithelial cells
Sub-cultivation is done to amplify the number of undifferentiated human airway epithelial cells, which will enable the researcher to either seed a larger number of Transwells or cryopreserve a portion of the cells for use in future experiments with cells from the same donor.
11.Discard the medium from the T-175 flask.
12.Wash once with 10 ml of PBS (without Ca2+ or Mg2+) at room temperature.
13.Add 10 ml of warm 0.025% trypsin/EDTA.
14.Incubate the flask for 5 min at 37°C.
15.Visualize the cells using a light microscope. If the cells are detached, proceed with step 16; otherwise return the flask to the 37°C incubator and monitor every 1-2 min.
16.Add 10 ml of the trypsin inhibitor solution (TIS).
17.Gently homogenize the cell suspension using a serological pipette, then transfer the cell suspension to a 50-ml conical tube.
18.Centrifuge for 5 min at 350 × g , room temperature.
19.Discard the supernatant by pipetting.
20.Dislodge the pellet by flicking the tube and gently resuspend the cell pellet in 10 ml of warm PneumacultTM-EX Complete medium.
21.Count the number of viable cells using a hemocytometer or a cell counter.
22.Seed 1.75 × 106 cells (10,000 cells/cm2) into a T-175 flask and bring the final volume in the flask up to 20 ml with warm PneumacultTM-EX Complete medium.
23.Label the flask with the following information: donor number/date of sub-cultivation/passage number.
24.Incubate the flask at 37°C with 5% CO2.
25.Every 2 days, change the medium as described above (Phase 1, steps 7-8).
26.When the monolayer achieves 80% confluence, sub-culture one more time (Phase 2) or start the process of cell differentiation (step 33, below).
Phase 3: Coat Transwells with collagen
27.Place the 6.5-mm Transwells into 24-well plates.
28.Add 100 μl Collagen Type I–rat tail working solution to the apical chamber (on the Transwell membrane).
29.Incubate the plates for 45 min in the 37°C incubator.
30.Gently aspirate the collagen from the apical chamber using a micropipette.
31.Wash the membrane once with 150 μl of PBS followed by immediate removal.
32.Store the plates at 4°C until use.
Phase 4: Pre-differentiation/expansion phase—Seed undifferentiated epithelial cells on Transwells
33.Follow steps 11-21 from Phase 2 (sub-culturing), then resuspend the cells at 3.3 × 105 cells/ml.
34.Transfer 100 μl of cell suspension (33,000 cells) to a collagen-coated Transwell membrane.
35.Add 500 μl of warm PneumacultTM-EX Complete medium to the basolateral chamber.
36.Label the plates with the following information: donor number/date/passage number.
37.Incubate the plate at 37°C with 5% CO2.
38.The following day, aspirate and discard the medium from the apical Transwell chamber.
39.Wash the monolayer by adding 100 μl of PBS at room temperature to the apical Transwell chamber and then removing it by aspiration.
40.Add 200 μl warm PneumacultTM-EX Complete medium to the apical chamber.
41.Discard the basolateral medium and replace it with 500 μl fresh, warm PneumacultTM-EX Complete medium.
42.Incubate the cultures at 37°C with 5% CO2.
43.Change the medium in both the apical and basolateral chambers every 2 days, as described above (steps 38, 40-42).
44.Visualize the cultures daily under a light microscope to monitor expansion and growth on the Transwell.
45.When the monolayer is 100% confluent, proceed to Phase 5 (differentiation).
Phase 5: Differentiation phase—Culture human airway epithelial cells at air-liquid interface (HAE-ALI)
46.When the monolayer achieves 100% confluency (Fig. 2B), discard the medium from both the apical and basolateral chambers.
47.Add 500 μl warm PneumaCult-ALI Maintenance medium to the basolateral chamber. The apical chamber will remain dry (air-lift).
48.Mark the plate with the date of air-lift.
49.Incubate the plates at 37°C with 5% CO2.
50.Every 2 days, discard the medium from the basolateral chamber and replace it with 500 μl warm PneumaCult-ALI Maintenance medium. Return cultures to the 37°C incubator with 5% CO2.
51.Visualize the cultures under a microscope frequently. You will be able to visualize cilia beating when the process of differentiation is complete (Video 1).
Basic Protocol 2: VIRAL INOCULATION OF HAE-ALI
This protocol is used to infect differentiated HAE-ALI with viruses at the apical (lumenal) surface. Prior to inoculation, the apical surface of differentiated HAE-ALI is washed to remove mucus secretions and facilitate virus access to underlying epithelial cells. Viral inoculum is then added to the culture surface at the desired dose and returned to the CO2 incubator for viral adsorption. The inocula can be kept on, or removed from the culture by washing the apical surface with PBS.
Materials
-
Fully differentiated HAE-ALI cultures (see Basic Protocol 1)
-
Phosphate-buffered saline (PBS) without Ca2+ or Mg2+ (Gibco; cat. no. P5119)
-
PneumacultTM-ALI Maintenance medium (see recipe)
-
Stock of infectious virus (RV, IAV, SARS-CoV-2, or other) with known titer
-
Viruses typically require ultra-cold (–80°C) storage to preserve infectivity. Ensure access to proper storage facilities, if required.
-
Certified class II biological safety cabinet
-
Humidified incubator at 37°C with 5% CO2
-
Water bath at 37°C
-
Barrier pipette tips: 20 μl (Thermo Fisher Scientific; cat. no. 2149P or equivalent), 200 μl (Thermo Fisher Scientific; cat. no. 2069 or equivalent), 1000 μl (Thermo Fisher Scientific; cat. no. 2079E or equivalent)
-
Micropipettes
Phase 1: Prepare HAE-ALI and viral inoculum
1.Select desired temperature conditions for your experiment.
2.Optional : Immediately prior to inoculation, remove excessive mucus from the apical surface of HAE-ALI by washing twice with room temperature PBS (without Ca2+ or Mg2+): add 100 μl PBS to the apical chamber, incubate for 15 min at 37°C, and discard the supernatant.
3.Remove the basolateral medium and replace with 500 μl of warm Pneumacult-ALI Maintenance medium.
4.Dilute virus and prepare controls in ≤50 μl final volume.
Phase 2: Inoculate HAE-ALI
5.Add the inoculum (up to 50 μl; prepared in Phase 1, step 4) to the apical surface of the appropriate cultures and return the cultures to the 5% CO2 incubator at the desired temperature.
6.After viral adsorption, aspirate the inoculum and wash the apical surface three times using 100 μl room temperature PBS (no incubation is needed). Alternatively, if the inoculum volume is low (≤10 μl), it may be left on the culture for the duration of the experiment.
7.Return cultures to the 5% CO2 incubator at the desired temperature until experimental time points are reached.
Basic Protocol 3: IMMUNOFLUORESCENCE (IF)-BASED DETECTION OF INFECTED CELLS IN HAE-ALI
This protocol is used to detect double-stranded RNA (dsRNA), a marker of single-stranded RNA viral replication, as well as viral proteins in HAE-ALI, by indirect immunostaining. Cultures previously inoculated with virus as described in Basic Protocol 2 are fixed at various experiment time points of interest, and infection is assessed through immunostaining, followed by visualization and image acquisition using a fluorescence or confocal microscope. Additional notes are provided to facilitate adaptation of this protocol for direct immunostaining or staining of multiple targets (e.g., both viral and cellular antigens).
Materials
- Fully differentiated HAE-ALI cultures inoculated with virus, and controls (see Basic Protocol 2)
- Phosphate-buffered saline (PBS) without Ca2+ or Mg2+ (Gibco; cat. no. P5119)
- 4% Paraformaldehyde (PFA; see recipe)
- Quenching solution (see recipe)
- Permeabilization solution (see recipe)
- Blocking solution (see recipe)
- Antibodies (see recipe and Table 1)
- PBS (Gibco; cat. no. P5119) with 1% bovine serum albumin (BSA; Sigma-Aldrich; cat. no. 9048-46-8)
| Target antigen | Primary antibody | Secondary antibody | Type of assay |
|---|---|---|---|
| dsRNA | Mouse IgG2a anti-dsRNA (J2) monoclonal antibody (SCICONS; cat. no. 10010200; 1:1000) | Goat anti-mouse IgG2a secondary antibody AlexaFluor 555-conjugated (Thermo Fisher Scientific; cat. no. A-21137; 1:200) | Indirect |
| Influenza A virus nucleoprotein (IAV-NP) | Anti-Influenza A Antibody, nucleoprotein, clones A1, A3 blend (Millipore; cat. no. MAB8251; 1:100) | Donkey anti-mouse IgG H&L AlexaFluor-488 conjugated (Thermo Fisher Scientific; cat. no. A-21202; 1:500) | Indirect |
| SARS-CoV-2 nucleocapsid protein (SARS2-N) | Mouse IgG2b monoclonal anti-SARS-CoV-2 antibody, nucleocapsid protein, clone 1C7 (Bioss Antibodies; cat. no. BSM-41411M; 1:250) | Donkey anti-mouse IgG H&L AlexaFluor-488 conjugated (Thermo Fisher Scientific; cat. no. A-21202; 1:500) | Indirect |
| Motile cilia | Mouse IgG2b anti-acetylated alpha-tubulin (6-11B-1) monoclonal antibody AlexaFluor 647-conjugated (Santa Cruz Biotechnology; cat. no. sc-23950; 1:50) | N/A | Direct |
-
Nuclei stain: 1 μg/ml bisBenzimide H 33342 trihydrochloride (Hoechst 33342; Sigma-Aldrich; cat. no. 875756-97-1) or 10 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich; cat. no. 28718-90-3) diluted in PBS
-
VectaShield antifade mounting medium (Vector Laboratories; cat. no. H1000-10)
-
Nail polish without formaldehyde (to seal the slide).
-
Immersion oil (Carl Zeiss; cat. no. 444960-0000-000 or equivalent)
-
Ensure the type of oil used is compatible with your imaging system.
-
Certified class II biological safety cabinet
-
Barrier pipet tips: 20 μl (Thermo Fisher Scientific; cat. no. 2149P or equivalent), 200 μl (Thermo Fisher Scientific; cat. no. 2069 or equivalent), 1000 μl (Thermo Fisher Scientific; cat. no. 2079E or equivalent)
-
Micropipettes
-
Timer
-
Refrigerator or cold room at 4°C
-
Freezer at −20°C
-
Rocker or belly dancer
-
Parafilm
-
35-mm dish with 14-mm glass-bottomed insert (Mattek; cat. no. P35G-1.5-14-C)
-
Fluorescence microscope (Zeiss Axio Observer 3) with 5×, 10×, 20×, 40×, and 63× objective lens, camera, and AIM-Zen 2007 software
-
(Optional) Confocal microscope with 10×, 20×, 40×, and 63× objective lens
-
0.1875-in. Humboldt H-9663 plated brass cork borer with handle (“#2 T-bar”)
-
Forceps
-
1 mm-thick precleaned microscope glass slides (Fisherbrand; cat. no. 22-265-446 or equivalent)
-
24 × 50 mm, thickness #1.5 (0.16-0.19 mm) rectangular coverslips (Fisherbrand; cat. no. 12550-100 or equivalent)
Phase 1: Fix HAE-ALI
The fixation and permeabilization of cell membranes are critical steps, as they can influence antibody access to target antigens and binding to epitopes. Because of this, the specific protocol selected depends on the subcellular location of the antigen (plasma membrane, cytoplasm, nucleus, etc.). This protocol uses paraformaldehyde during fixation, which enables detection of dsRNA (e.g., during RV infection), IAV-NP, and SARS2-N (Fig. 3A and 3B). This protocol is also compatible with many other viral and cellular proteins, including acetylated α-tubulin, a marker for motile cilia that can be used to identify mature ciliated cells (Fig. 3C and 3D). If needed, an alternative fixative reagent can be used; however, this will likely impact the time and temperature at which fixation is performed, as well as other downstream steps. In addition, alternative fixation reagents (e.g., methanol:acetone) may impact the integrity of cellular structures (Fig. 3C).
![Details are in the caption following the image Immunofluorescence-based detection of viral and cellular antigens in differentiated HAE-ALI. (A) Differentiated HAE-ALI infected with rhinovirus-C15 (10<sup>10</sup> RNA copies) and probed for dsRNA at 12 hr post-infection (20× magnification; scale bar = 100 µm). Cells were fixed with 4% paraformaldehyde (15 min incubation), permeabilized with 0.2% Triton X-100, and blocked with 10% normal goat serum followed by a dsRNA indirect immunostaining assay [1:1000 mouse IgG2a anti-dsRNA antibody (J2), diluted in PBS with 1% BSA and 0.2% Tween-20; 1:200 goat anti-mouse IgG2a secondary antibody AlexaFluor 555-conjugated, diluted in PBS with 1% BSA] and Hoechst 33342 nuclei stain. (B) Differentiated HAE-ALI infected with influenza A virus (A/Puerto Rico/8/34; 5 × 10<sup>4</sup> plaque forming units) or SARS-CoV-2 (USA-WA1/2020; 1 × 10<sup>6</sup> plaque forming units) and probed for viral antigen at 48 and 72 hr post-infection, respectively. *Indicates the starting point of a “comet” of antigen-positive cells (10× magnification; scale bars = 100 µm). Cells were fixed with 4% paraformaldehyde for 15 min (IAV) or ≥24 hr (SARS-CoV-2), permeabilized with 2.5% Triton X-100, and blocked with 3% BSA followed by an indirect immunostaining assay [1:100 anti-IAV NP clones A1, A3 antibody blend or 1:500 anti-SARS N clone 1C7 antibody (kindly provided by Thomas Moran) diluted in PBS with 1% BSA; 1:500 donkey anti-mouse IgG H&L AlexaFluor 488-conjugated secondary antibody diluted in PBS with 1% BSA] and Hoechst 33342 nuclei staining. (C) Differentiated HAE-ALI were fixed with methanol:acetone (20 min incubation at −20°C) or 4% PFA (15 min incubation at room temperature), permeabilized with 0.2% Triton X-100, and blocked with 10% normal goat serum followed by an acetylated alpha-tubulin direct immunostaining assay [1:50 mouse IgG2b anti-acetylated alpha-tubulin (6-11B-1) monoclonal antibody AlexaFluor 647-conjugated diluted in PBS with 1% BSA and 0.2% Tween-20] and Hoechst 33342 nuclei staining (63× magnification; scale bar = 10 µm). (D) Z-stack (XY, XZ, and 3D views) of differentiated HAE-ALI 12 hr post-infection with 10<sup>10</sup> RNA copies of rhinovirus-C15 immunostained sequentially for dsRNA (indirect assay), acetylated alpha-tubulin (direct assay), and nuclei (63× magnification; scale bar = 10 µm). Images in (A) and (B) were taken using a Zeiss Axio Observer 3 inverted fluorescence microscope equipped with 10× and 20× LWD objectives, Zeiss Axiocam 503 monochrome camera, and AIM-Zen 2007 software, while images in (C) and (D) were obtained after analysis of z-series optical sections (at 1 µm intervals) acquired using LSM710 Zeiss Laser Scanning confocal microscope equipped with a 63×/1.4NA Oil/0.190 nm WD objective, argon laser, and AIM-Zen 2009 software. Z-sections were analyzed using Fiji–ImageJ v.2.1.0/1.53c software (Schindelin et al., 2012).](https://static.yanyin.tech/literature_test/cpz1453-fig-0003-m.jpg)
1.Discard the medium from the basolateral chamber of the culture from Basic Protocol 2.
2.Wash once with PBS: add 100 and 500 μl PBS (no Ca2+ or Mg2+) to the apical and basolateral chambers, respectively.
3.Discard the PBS and, to fix the culture, add 100 μl and 500 μl 4% (v/v) PFA to the apical and basolateral chambers, respectively.
4.Incubate the culture for 15 min at room temperature.
5.Discard the PFA from both sides of the chamber and wash the culture with PBS: add 100 and 500 μl PBS to the apical and basolateral chambers, respectively. Incubate for 3 min at room temperature with constant agitation by placing the cultures on a rocker or belly dancer.
6.Discard the PBS and repeat step 5 twice, for a total of three washes.
7.Add 100 and 500 μl quenching solution to the apical and basolateral chambers, respectively, then incubate the plate for 10 min at room temperature.
8.Discard the quenching solution from both sides of the chamber and wash three times with PBS as in step 5.
9.Discard the PBS and add 200 and 1000 μl PBS to the apical and basolateral chambers, respectively.
10.Seal the plate with parafilm and store at 4°C until use.
Phase 2: Permeabilize cell membranes
11.Discard the PBS from both sides of the chamber and add 100 and 500 μl permeabilization solution to the apical and basolateral chambers, respectively, then incubate for 15 min at room temperature.
12.Discard the permeabilization solution from both sides of the chamber and wash the culture with PBS: add 100 μl and 500 μl PBS to the apical and basolateral chambers, respectively. Incubate for 3 min at room temperature with constant agitation by placing the cultures on a rocker or belly dancer.
13.Discard the PBS and repeat step 12 twice, for a total of three washes.
14.Discard the PBS and add 100 and 500 μl PBS to the apical and basolateral chambers, respectively.
15.Seal the plate with parafilm and store at 4°C until use.
Phase 3: Block nonspecific epitopes
16.Discard the PBS from both sides of the culture and add 100 and 500 μl blocking solution (10% normal serum diluted in PBS with 0.2% Triton X-100) to the apical and basolateral chambers, respectively.
17.Incubate the plate for 15-60 min at room temperature.
18.Discard the blocking solution from the apical chamber only.
19.Wash with PBS: add 100 μl PBS to the apical chamber; incubate for 3 min at room temperature and with constant agitation. Discard the PBS and repeat for a total of 3 washes.
20.Discard the PBS and add 100 μl PBS to the apical chamber. Store the plate at 4°C until use.
Phase 4: Immunostaining of viral (and cellular) antigens
This protocol can be done as a single procedure using fluorophore-conjugated primary antibodies (direct IF assay), or as a two-stage process in which the sample is first incubated with primary antibody followed by fluorophore-conjugated secondary antibodies (indirect IF staining). In addition, as a multiplex IF assay, several targets can be probed in the same HAE-ALI culture through sequential rounds of immunostaining (Fig. 3D). In all workflows, nuclei are stained immediately prior to mounting on slides (optional) and visualization.
21.Discard the PBS from the apical chamber.
22.Add 100 μl primary antibody diluted in PBS with 1% BSA to the apical chamber.
23.Incubate the plate overnight at 4°C, protected from the light.
24.Discard the primary antibody solution from the apical chamber.
25.Wash with PBS: add 100 μl PBS to the apical chamber; incubate for 3 min at room temperature and with constant agitation. Discard the PBS and repeat this step for a total of three washes.
26.Discard the PBS and add 100 μl secondary antibody diluted in PBS with 1% BSA to the apical chamber.
27.Incubate for 2 hr at room temperature, protected from the light.
28.Discard the secondary antibody solution.
29.Wash once with PBS: add 100 μl PBS to the apical chamber; incubate for 3 min at room temperature and with constant agitation.
30.Discard the PBS and add 100 μl of nuclei staining solution to the apical chamber.
31.Incubate for 5 min at room temperature, protected from the light.
32.Discard the nuclei staining solution.
33.Wash with PBS: add 100 μl PBS to the apical chamber; incubate for 3 min at room temperature and with constant agitation. Discard the PBS and repeat this step for a total of two washes.
34.Discard the PBS and remove the Transwell membrane using the #2 T-bar.
35.Place the Transwell membrane on a glass microscope slide with the epithelium facing up.
36.Add one drop of antifade mounting medium on top of the membrane.
37.Cover the membrane with the coverslip and seal with nail polish.
38.Store the slide at 4°C and protect from the light until use.
Phase 5: Visualization and acquisition of IF images
It is essential that your IF assay uses dyes that can be detected by the fluorescence microscope available. This information is obtained by checking microscope laser, filter, and detector configurations. In this protocol, we describe steps to acquire images using the Zeiss Axio Observer 3 inverted fluorescence microscope equipped with AIM-Zen 2007 software. This protocol can be adapted for use with other microscope models.
39.Turn on the microscope and open the AIM-Zen 2007 software.
40.Place the positive control slide in the slide holder with the coverslip facing the objective.
41.Select the “observation mode”, turn on the transmitted light, and, using a 10× objective lens, search for the epithelium.
42.Correct the focus by gently moving the focus knob up and down.
43.Turn off the transmitted light and turn on the reflected light with the proper filter for your fluorophore.
44.Search for detectable fluorescence in the positive control and correct the focus.
45.If needed, switch to higher observation magnification (e.g., 40×, 63× objective lens) and correct the focus.
46.Switch to “camera mode” and begin to optimize the image using AIM-Zen 2007 software.
47.Keep the “auto-exposure” mode selected.
48.Place your sample on the viewer by moving the XY knob and correcting the focus.
49.Under “image acquisition settings”, set gain for 1× and binning mode for 1 × 1.
50.Acquire the image and take note of the exposure time.
51.Turn off the fluorescence light on the sample temporarily.
52.On the histogram, set the minimum and maximum signal intensity values, trying to remove background and eliminate saturation. Take note of both values.
53.Switch the slide to the negative control.
54.Locate the sample on the slide, as above, then turn on the fluorescence light and correct the focus if needed.
55.Keep the same exposure time, gain, and binning mode previously used for the positive control.
56.Acquire the image and turn off the fluorescence light on the sample temporarily.
57.On the histogram, check if the previous values set at step 52 fit the negative control and correct it if not.
58.If you change the minimum and/or maximum values in step 57, update these values on the image taken from the positive control.
59.Finally, if the photo quality is still poor (e.g., with saturated signal or high background), acquire a new image starting again with the positive control and adjusting the exposure time.
60.For multiplex assays, repeat this procedure (steps 44-59) for the next target using the proper filter for each fluorophore. Leave nuclei detection for last.
61.After setting the parameters for all fluorophores/nuclei, acquire the final images from your experiment by choosing a spot of interest on your sample and taking pictures of each target without changing the X/Y position. You are only allowed to change filters and correct the focus (Z-drive), if needed.
62.After image acquisition, add scale bars to the photos as “μm.”
63.Export the photo as a .tiff or .czi file.
REAGENTS AND SOLUTIONS
Antibodies
Dilute primary antibodies in PBS (Gibco; cat. no. P5119) with 1% BSA (Sigma-Aldrich; cat. no. 9048-46-8) and optional 0.2% Tween 20 (Sigma-Aldrich; cat. no. 9005-64-5).
Dilute fluorophore-conjugated secondary antibodies in PBS (Sigma-Aldrich; cat. no. 9005-64-5) with 1% BSA (Sigma-Aldrich; cat. no. 9048-46-8).
Blocking solutions
- 10% normal serum from the same species as the secondary antibody in PBS (Gibco; cat. no. P5119) with 0.2% Triton X-100 (Sigma-Aldrich; cat. no. 9036-19-5).
- Normal donkey serum (Jackson ImmunoResearch Labs; cat. no. 017-000-121) and normal goat serum (Jackson; cat. no. 005-000-121) are used in Basic Protocol 3.
- 3% bovine serum albumin (BSA; Sigma-Aldrich; cat. no. 9048-46-8) in PBS (Gibco; cat. no. P5119) with 0.2% Tween 20 (Sigma-Aldrich; cat. no. 9005-64-5).
- Prepare fresh.
Collagen Type I–rat tail working solution
Dilute Collagen Type I–rat tail (cat. no. 354236; Corning) to 30 µg/ml in PBS (Gibco; cat. no. P5119). Store up to product expiration date at 4°C.
Fetal bovine serum, heat-inactivated
- Fetal bovine serum (FBS; Gibco; cat. no. 16000044)
- Incubate the serum for 1 hr in a water bath at 56°C to inactivate the serum. Let the serum cool down at room temperature. Make 50-ml aliquots and store up to product expiration date at −20°C.
Paraformaldehyde, 4%
8% paraformaldehyde (Electron Microscopy Sciences; cat. no. 157-8-100) diluted 1:1 (v/v) with PBS (Gibco; cat. no. P5119).
Permeabilization reagent
0.2% Triton X-100 (Sigma-Aldrich; cat. no. 9036-19-5) in PBS (Gibco; cat. no. P5119). Store up to several months at 4°C.
PneumacultTM-ALI Complete Base medium (final volume of 500 ml)
- 450 ml PneumacultTM-ALI medium (Stemcell Technologies; cat. no. 05001)
- 50 ml PneumacultTM-ALI 10× Supplement (Stemcell Technologies; included with cat. no. 05001)
- Store up to product expiration date at 4°C protected from light
PneumacultTM-ALI Maintenance medium (final volume of 100 ml)
- 98.3 ml PneumaCultTM-ALI Complete Base medium (see recipe)
- 1 ml PneumaCultTM-ALI Maintenance Supplement (Stemcell Technologies; included with cat. no. 05001)
- 500 μl hydrocortisone stock solution (96 µg/ml; Stemcell Technologies; cat. no. 07925)
- 200 μl 0.2% heparin solution (Stemcell Technologies; cat. no. 07980)
- Make 10-ml working aliquots in 15-ml sterile polypropylene conical tubes
- Store up to product expiration date at −20°C
PneumacultTM-EX Complete medium (final volume of 500 ml)
- 490 ml PneumacultTM-EX Basal medium (Stemcell Technologies; cat. no. 05008)
- 10 ml PneumacultTM-EX 50× Supplement (Stemcell Technologies; included with cat. no. 05008)
- 0.5 ml hydrocortisone stock solution (96 µg/ml; Stemcell Technologies; cat. no. 07925)
- Store up to product expiration date at 4°C protected from light
Quenching solution
25 mM ammonium chloride (NH4Cl) in PBS (Gibco; cat. no. P5119). Store at 4°C.
Trypsin/EDTA, 0.025%
0.05% trypsin/EDTA (Gibco; cat. no. 25300-062) diluted 1:1 with Hanks’ Balanced Salt Solution (HBSS; Thermo Scientific Pierce; cat. no. 88284). Store up to 2 weeks at 4°C.
Trypsin inhibitor solution (TIS; final volume of 10 ml)
- 1.5 ml heat-inactivated FBS (see recipe)
- 8.5 ml Hanks’ Balanced Salt Solution (HBSS; Thermo Scientific Pierce; cat. no. 88284)
- Filter-sterilize the solution using tube-top vacuum filter with 0.22-μm membrane (Corning; cat. no. 430420 or equivalent). Store up to 4 weeks at 4°C.
COMMENTARY
Background Information
Tissue culture is an important tool for the study of viral pathogens, with immortalized cell lines being the most commonly used platform because they are easy to handle, require relatively affordable supplies, and can be routinely scaled up to facilitate many biological replicates or high-throughput screens. Nonetheless, even with the plethora of immortalized cell lines available, monolayer culture of homogenous, undifferentiated cells submerged in liquid medium does not allow researchers to address questions pertaining to certain aspects of respiratory viral infection, such as cell tropism. Indeed, immortalized cell lines used for propagation and interrogation of virus-host interactions in vitro are frequently unrelated to the cell type in which the virus replicates in vivo. HeLa cells (human endocervix epithelial cells; ATCC-CCL2), for example, have been used to study various respiratory viruses, including rhinovirus (Amineva, Aminev, Gern, & Palmenberg, 2011). Furthermore, aqueous medium constitutes a very different extracellular environment when compared to the secreted mucus and underlying periciliary layers that respiratory viruses must breach to infect epithelial cells in the lung. With little to no barrier function, or directionality of flow, it is difficult to ascertain information about the dynamics of infection and spread.
As a result, many groups have contributed to the development of airway cultures with emergent properties. Beginning with tissue explants, which have a limited life-span ex vivo and suffer from frequent contamination (Bukowy-Bieryłło, 2021), techniques then evolved to enzymatically dissociate the cells, cultivate undifferentiated primary cells, and subsequently grow the cells under air-liquid interface (Gruenert, Finkbeiner, & Widdicombe, 1995; Whitcutt, Adler, & Wu, 1988). The latter technique allows the cells to develop as a polarized multilayered epithelium with physiology and morphology similar to in vivo.
HAE-ALI cultures have distinct apical and basolateral surfaces, are composed of different cell types (e.g., ciliated cells, secretory cells, basal cells, etc.), and secrete mucus which is propelled across the apical surface of the cells as a function of underlying beating cilia driving active mucociliary transport. The current HAE system has been developed over the course of several decades, and continues to evolve. Additional medium formulations, alternative culturing techniques (Bukowy-Bieryłło, 2021), and the generation of immortalized airway epithelial cells that retain their multipotent capacity (Walters et al., 2013) have all enabled further subculturing prior to differentiation, allowing researchers to increase cell yield and opening a larger window of time to manipulate the cells. Notably, additional protocols are tailored for cells from different regions of the airway (e.g., nasal, tracheal, alveolar) as well as from donors with underlying lung disease (e.g., cystic fibrosis, asthma and chronic obstructive pulmonary disorder (Baldassi, Gabold, & Merkel, 2021)). Collectively, these airway epithelial model systems allow for the determination of cell tropism within specific regions of the respiratory tract as well as the polarity of viral entry and exit, and recapitulation of viral replication and spread dynamics in mucosal microenvironments that is reflective of health or disease. Furthermore, they often support infection by viruses that fail to infect or amplify well in immortalized cell lines, such as RV-C (Hao et al., 2012) and HKU1 (Pyrc et al., 2010), enabling researchers to interrogate specific virus-host interactions of these and other respiratory viruses in primary human cells.
As is the case with all model systems, HAE cultures also have some drawbacks. At the outset, the number of cells sourced from a single donor is finite, and the financial and time investment required to establish HAE-ALI cultures is significant. Further, despite improvements in expanding undifferentiated cells prior to differentiation, clonal expansion is not currently feasible and transfection or transduction post-differentiation remains a challenge. HAE-ALI cultures also lack 3D architecture, inclusion of other cell types (e.g., immune cells, endothelial cells), and dynamic forces characteristic of the lung, such as cyclical stretch, that other in vitro models have successfully integrated (Iverson et al., 2020). Finally, the protocol provided here for viral inoculation requires the addition of fluid on the apical culture surface. While this ensures efficient infection, this delivery method differs from the natural deposition of aerosolized particles throughout the respiratory tract by inhalation. How these delivery methods impact the mucus barrier and infection kinetics is not well understood. Inoculation of HAE-ALI with aerosolized viruses is feasible, but requires additional, highly specialized equipment not available in most laboratories.
Following infection of either cell lines or model systems such as HAE-ALI cultures that exhibit emergent properties, numerous methodologies exist to characterize and quantify infection kinetics. Typical methods include quantitative PCR of viral genomes and the assay of infectious particles using plaque assays or limiting-dilution assays. These assays offer insight into viral growth kinetics over time; however, the data acquired represent an average measure of the culture as a whole, and thus, alone, cannot yield a complete picture of infection dynamics. The application of immunofluorescence microscopy following infection enables single-cell analysis and retention of spatial information within the cells as well as across the culture as a whole, and can be paired with image-analysis tools to investigate co-localization of viral and/or host proteins or correlate expression of viral and cellular markers at the single-cell level. This is especially useful in cases where few cells are permissive for infection; here, whole-culture analyses, or analyses that only assay a portion of the cells (e.g., flow cytometry), may mask or ‘dilute’ effects of the virus. Immunostaining methods in HAE allow detection of single infected cells, which can confirm infection even when few cells may be permissive, identify cell tropism when paired with cell type–specific markers, and resolve spread dynamics that can be influenced by cell tropism, air-surface liquid properties, and cilia beat function. Furthermore, immunofluorescence-mediated detection of infection in HAE-ALI can be applied to study the efficacy of inhibitors that impede entry or spread in the native microenvironment (Matrosovich, Matrosovich, Gray, Roberts, & Klenk, 2004). In sum, these methods allow for the evaluation of respiratory viral infection in a physiological setting.
Critical Parameters
HAE-ALI cultures are derived from primary cells, isolated from individual donors; thus, variation in the exact timing to differentiation, ratio of different cell types, and mucus production should be expected. Indeed, due to this variation in the frequency of each cell type between donors and even individual cultures and viral tropism for specific cell types, multiplicity of infection cannot be accurately determined in HAE-ALI. Thus, we recommend that researchers report the total amount of virus applied and the surface area of the airway culture.
To minimize non-biological variation across experiments, special attention should be paid to the cell passage number used for differentiation, keeping the passage number as low as possible and consistently using cells of the same passage number. Furthermore, the type of extracellular matrix, specific membranes used for air-liquid interface culture, and source of culture medium (e.g., Stemcell Technologies, Lonza, etc.) will impact cell growth parameters and the histological, transcriptomic, and physiological characteristics of HAE-ALI (Rayner, Makena, Prasad, & Cormet-Boyaka, 2019; Saint-Criq et al., 2020); thus, we highly recommend that users ensure availability of all materials and reagents to avoid any unintentional product substitution mid-way through the protocol.
To generate rigorous and reproducible data on viral infection in HAE-ALI, it is critical to utilize cultures from multiple donors and to recognize that the degree of culture differentiation, amount of mucus secretion and overall hydration, as well as the duration of viral inoculation and temperature at which the experiment is performed will all influence the efficiency of initial infection and spread kinetics. Some of these parameters may depend on the virus under study and specific research question being addressed. For example, rhinovirus and SARS-CoV-2 replication may be more efficient at temperatures of the upper respiratory tract (Royston & Tapparel, 2016; V'kovski et al., 2021) while influenza A virus infections are routinely carried out at higher temperature (Davis et al., 2015). In all cases, removing excess mucus improves viral particle access to the cell surface and thus increases the likelihood of viral entry. However, researchers may wish to preserve the secreted mucus barrier when asking questions related to infection efficiency and the ability of viruses to overcome this barrier.
Finally, investigation of virus-host interactions in HAE-ALI may require immunofluorescence-based detection of several target antigens in one culture. In this case, it is essential that each set of antibodies be tested for antigen specificity and compatibility with the chosen fixation and permeabilization conditions before multiplexing. Furthermore, primary antibodies from different species should be selected to minimize cross-detection of secondary antibodies. If this is not possible, we suggest that researchers opt for at least one antibody (ideally targeting the epitope with highest availability) that is pre-conjugated to the fluorophore and perform the direct IF assay last. Alternatively, if sequential indirect IF assays must be performed using primary antibodies from the same species, researchers can decrease the incubation time for the secondary antibody in the second assay from 2 hr to 20 min. Finally, it is important to select photostable fluorophores and pay attention to excitation/emission wavelengths to avoid bleed-through during imaging and obtain optimal results.
Troubleshooting
A troubleshooting guide listing common problems and solutions is provided in Table 2.
| Problem | Probable cause | Solution | |
|---|---|---|---|
| Generation of HAE-ALI | Cell death at 24 hr after thawing | Cells not stored under ideal conditions; incorrect cell density; multiple freeze/thaw cycles | Thaw a new vial of undifferentiated cells |
| Absent or slow proliferation of undifferentiated cells | Low number of cells seeded into the flask; contamination with bacteria/fungi | Use a new stock of medium; continue routine maintenance and track culture proliferation; treat with penicillin (100 U per ml) and streptomycin (100 µg per ml) or amphotericin B (0.25 to 2.5 µg per ml); monitor confluence during subculture prior to seeding on Transwells | |
| Cultures are persistently leaky (fluid is observed on the apical surface) | Culture did not reach 100% confluency prior to air-lift or loss of epithelium integrity | Repeat differentiation using a new stock of collagen and freshly coated Transwells; ensure cell count is accurate; ensure cells have reached confluency prior to removing the apical medium | |
| Cultures fail to become ciliated | Cells have lost their differentiation capacity due to extensive passaging | Use cells at a lower passage | |
| Epithelium is not flat (ridges or cysts are observed) | Failure to wash the cultures routinely; aberrant cell growth | Avoid using cultures with these features for imaging purposes. If all cultures in a batch exhibit these alterations, initiate a new differentiation. | |
| Bacterial or fungal contamination | Poor aseptic technique | Dispose of any potentially contaminated reagents. Remove surviving cultures to a new 24-well plate using forceps or a sterile pipette tip threaded through the Transwell basket to transfer the cultures. Monitor daily to ensure all contaminated cultures have been removed. As a last resort, you can treat the survivors with penicillin (100 U per ml) and streptomycin (100 µg per ml) or amphotericin B (0.25 to 2.5 µg per ml). Note that the treatment of differentiated HAE-ALI with antibiotics and antimycotics may interfere with epithelium homeostasis, which includes its selective permeability. | |
| Excessive medium acidification or discoloration | Rapid growth immediately post-air-lift; culture contamination; fluid or medium left on the apical surface; cell growth within the basolateral chamber | Refresh basolateral medium a day early; check for visible contaminants and discard the culture or treat with penicillin (100 U per ml) and streptomycin (100 µg per ml) or amphotericin B (0.25 to 2.5 µg per ml); ensure medium does not unintentionally drip into the apical chamber during feeding; carefully add cells to the Transwell without spilling; check for growth on the basolateral surface periodically; and transfer cultures to a new 24-well plate as needed | |
| Cell or culture integrity loss during routine maintenance | Apical cell loss during washing | Test new donor integrity against the standard washing procedure; if cell rounding or loss continues: reduce washing time, incubate at room temperature, wash more frequently during the week for less time | |
| Viral inoculation | Cytopathic effects are observed in the mock/negative controls | Diluent is cytotoxic or poorly tolerated; too much fluid has been left on the cultures |
Dialyze the virus stock against PBS; remove inoculum or reduce the volume if leaving the inoculum on for the duration of the experiment. Diluent compatibility can also be tested ahead of time by apical inoculation in HAE-ALI followed by observation under a light microscope; measuring of cytotoxic effects by quantifying lactate dehydrogenase (LDH) release and/or transepithelial electrical resistance (TEER; Gagliardi et al., 2022; Srinivasan et al., 2015). |
| Few cells are infected | Cultures are not fully differentiated; insufficient time for infection; excessive mucus remaining on the culture | Allow cultures more time to reach maturity; consider lowering the inoculation volume and leave the inoculum on to allow maximal time for infection; increased the number of washes prior to inoculation | |
| IF assay/microscopy | No viral antigen (e.g., dsRNA, IAV-NP, SARS2-N) signal is detected | Time point selected is not optimal for detection of this viral antigen | Fix cultures at different time points to determine the optimal timeframe in which to detect the viral antigen |
| Weak signal | Low levels of antigen present; insufficient antigen access; insufficient antibody incubation time | Increase amount of virus in the inoculum; change the detergent in the permeabilization step; increase the % Triton X-100 used during permeabilization (up to 2.5%); add detergent in the primary antibody solution; increase the concentration of the primary antibody; use an indirect immunofluorescence staining approach; try a different antibody (brand or epitope) | |
| Bright spots are observed during imaging | Secondary antibody precipitation; excessive mucus present on the culture | Centrifuge the secondary antibody at high speed for 1 min prior to use; wash the cultures regularly to remove accumulated mucus | |
| High background signal | Autofluorescence; excess mucus | Use farther red-shifted secondary antibodies; increase washes prior to fixing the culture; decrease the concentration of the secondary antibody. |
Understanding the Results
HAE-ALI cultures can be obtained by following the steps listed in Basic Protocol 1. Once fully differentiated, HAE-ALI cultures can be used to address a variety of questions regarding epithelium physiology, drug discovery, delivery, and toxicology, as well as infection and host response. Notably, this protocol specifically describes the generation of cultures that mimic the large airways (bronchial epithelium); since epithelial characteristics vary from proximal to distal airways, it is important to interpret data accordingly. For example, cell tropism and infection kinetics may differ in nasal or alveolar cells. Certain viral infection phenotypes may also be specific to certain host backgrounds. For example, cadherin-related family member 3 (CDHR3) is a receptor for RV-C, and its expression at the cellular membrane is influenced by a non-synonymous single nucleotide polymorphism (SNP; rs6967330[A]) (Bønnelykke et al., 2014). Thus, host genetics can impact the rate of viral infection, and cultures from multiple donors should be used to rule out donor-specific effects.
Basic Protocol 3 describes an IF assay to detect viral proteins and dsRNA. The availability of antibodies against viral epitopes is limited, and the target availability during an infection can also vary during the replication cycle. For example, dsRNA is used as a marker for rhinovirus replication in differentiated HAE-ALI, but its detection is limited to 8 to 24 hr post-infection (Gagliardi et al., 2022). In all cases, the detection of dsRNA or viral antigen does not indicate whether or not progeny infectious particles are produced. Nonetheless, viral antigen–positive cells can often be found in a “comet-like” pattern (Fig. 3B, marked by the asterisk). This is typical after initiating infection with a low dose of virus, and is indicative of infectious progeny production and viral spread, influenced by mucociliary transport.
Multiplex assays with cell and viral markers allow evaluation of cell tropism by the acquisition of 2D images. For example, co-staining infected HAE-ALI cultures for viral antigen and alpha-acetylated tubulin is a useful method to identify ciliated cells and evaluate whether these cells are permissive for viral replication. However, this marker is only present in mature ciliated cells; thus, the presence of viral antigen in an alpha-acetylated tubulin-negative cell may indicate tropism for pre-ciliated cells or another cell lineage altogether (e.g., club, goblet etc.). If the IF assay is done at different time-points during viral replication, the 2D images acquired over time can be used to understand the dynamics of the infection. For example, the tropism of SARS-CoV-2 to epithelial ciliated cells and goblet cells in differentiated HAE-ALI was shown by multiplex assay using antibodies against SARS2-N, beta tubulin-IV, and MUC5AC (Hao et al., 2020). Still, it is important to remember that these methods do not allow for a kinetic analysis within a single culture; thus, staining replicate cultures at each time point is essential to gain an accurate picture of replication and spread dynamics.
Sometimes, 2D images can indicate a potential interaction between two or more targets, which can represent a site for viral replication or a pathway activated/inhibited by the virus. However, this interaction can only be confirmed after colocalization analysis done with 3D images. Considering 2D images taken from RV-C-infected HAE-ALI in which red and green markers were used for the detection of dsRNA (viral genome replication) and giantin (Golgi vesicles marker), respectively, the visualization of yellow color suggested Golgi as a site for RV-C replication. However, colocalization analysis of 3D images did not support this finding (Gagliardi et al., 2022).
Time Considerations
The generation of HAE-ALI cultures takes 4-6 weeks, including initial expansion of commercially sourced normal human tracheal/bronchial cells prior to differentiation. The amount of “hands-on” time required varies over the course of the protocol, with the more sensitive and labor-intensive steps being concentrated during the first 2 weeks (i.e., expansion, subculture, and initial culture on Transwells). Researchers should take note of holidays and other potential disruptions to routine lab work in planning when to thaw new cells and initiate culture differentiation.
Subsequent viral inoculation of HAE cultures takes 1-5 hr, and the duration of the experiment will vary depending on viral infection kinetics and investigator-determined time points of interest. Inoculation with a high dose or cytopathic virus may limit the duration of the time course. Immunofluorescence-mediated detection of dsRNA or viral antigen can then be performed over 2 days as a single assay, adding 1 extra day for each additional antigen probed. Time required for imaging depends on the user's experience in manipulating the microscope and the type of image to be acquired. Acquiring images from single assays is faster than multiplex assays, and 2D images are acquired faster than 3D images. Researchers should plan carefully to ensure imaging is done soon after staining to avoid loss of signal, and that all samples are imaged together within the shortest time frame possible to ensure consistent imaging conditions and allow for appropriate comparisons between samples.
Acknowledgments
Support for the development of these protocols was provided by the National Institute of Allergy and Infectious Diseases (R21 AI163816, to M.A.S; R21 AI149180, to M.A.S and B.R.R; R01 AI151029 and U01 AI150748, to B.R.R.) and the National Heart, Lung, and Blood Institute (R01 HL151840, to M.A.S).
The authors thank Drs. James Gern, Yuri A. Bochkov, and Ann Palmenberg from University of Wisconsin–Madison (UW, Madison) for providing the rhinovirus-C15 plasmid, Dr. Adolfo Garcia-Sastre (Icahn School of Medicine at Mount Sinai) for providing the reverse genetics system for A/Puerto Rico/8/34, and Michael Schotsaert (Icahn School of Medicine at Mount Sinai) for the SARS-CoV-2 virus stock (SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281, originally deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH), as well as Thomas Moran (Center for Therapeutic Antibody Discovery at the Icahn School of Medicine at Mount Sinai) for kindly providing anti-SARS-CoV nucleocapsid antibody. The authors are also grateful to the directors and teams at the Imaging Core and the Laboratory for Biological Ultrastructure at the University of Maryland, College Park, for their assistance.
Author Contributions
Talita B. Gagliardi : conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing original draft, writing review and editing; Ethan Iverson : data curation, formal analysis, investigation, methodology, validation, visualization, writing review and editing; Emma J. DeGrace : data curation, formal analysis, investigation, writing review and editing; Brad R. Rosenberg : conceptualization, funding acquisition, project administration, supervision, validation, writing review and editing; Margaret A. Scull : conceptualization, funding acquisition, methodology, project administration, resources, supervision, validation, writing original draft, writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Literature Cited
- Amineva, S. P., Aminev, A. G., Gern, J. E., & Palmenberg, A. C. (2011). Comparison of rhinovirus A infection in human primary epithelial and HeLa cells. The Journal of General Virology , 92, 2549–2557. doi: 10.1099/vir.0.031302-0.
- Baldassi, D., Gabold, B., & Merkel, O. (2021). Air-liquid interface cultures of the healthy and diseased human respiratory tract: Promises, challenges and future directions. Advanced Nanobiomed Research , 1, 2000111. doi: 10.1002/anbr.202000111.
- Bønnelykke, K., Sleiman, P., Nielsen, K., Kreiner-Møller, E., Mercader, J. M., Belgrave, D., … Bisgaard, H. (2014). A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nature Genetics , 46, 51–55. doi: 10.1038/ng.2830.
- Bukowy-Bieryłło, Z. (2021). Long-term differentiating primary human airway epithelial cell cultures: How far are we? Cell Communication and Signaling , 19, 63. doi: 10.1186/s12964-021-00740-z.
- Burnett, L. C., Lunn, G., & Coico, R. (2009). Biosafety: Guidelines for working with pathogenic and infectious microorganisms. Current Protocols in Microbiology , 13(1), 111–1114. doi: 10.1002/9780471729259.mc01a01s13.
- Davis, A. S., Chertow, D. S., Moyer, J. E., Suzich, J., Sandouk, A., Dorward, D. W., … Taubenberger, J. K. (2015). Validation of normal human bronchial epithelial cells as a model for influenza A infections in human distal trachea. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society , 63, 312–328. doi: 10.1369/0022155415570968.
- Gagliardi, T. B., Goldstein, M. E., Song, D., Gray, K. M., Jung, J. W., Ignacio, M. A., … Scull, M. A. (2022). Rhinovirus C replication is associated with the endoplasmic reticulum and triggers cytopathic effects in an in vitro model of human airway epithelium. PLoS Pathogens , 18, e1010159. doi: 10.1371/journal.ppat.1010159.
- Gruenert, D. C., Finkbeiner, W. E., & Widdicombe, J. H. (1995). Culture and transformation of human airway epithelial cells. The American Journal of Physiology , 268, L347–60.
- Hao, S., Ning, K., Kuz, C. A., Vorhies, K., Yan, Z., & Qiu, J. (2020). Long-term modeling of SARSCoV-2 infection of cultured polarized human airway epithelium. mBio , 11, e02852–20. doi: 10.1128/mBio.02852-20.
- Hao, W., Bernard, K., Patel, N., Ulbrandt, N., Feng, H., Svabek, C., … Zhu, Q. (2012). Infection and propagation of human rhinovirus C in human airway epithelial cells. Journal of Virology , 86, 13524–13532. doi: 10.1128/JVI.02094-12.
- Hou, Y. J., Okuda, K., Edwards, C. E., Martinez, D. R., Asakura, T., Dinnon, K. H. 3rd, … Baric, R. S. (2020). SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell , 182, 429–446.e14. doi: 10.1016/j.cell.2020.05.042.
- Iverson, E., Kaler, L., Agostino, E. L., Song, D., Duncan, G. A., & Scull, M. A. (2020). Leveraging 3D model systems to understand viral interactions with the respiratory mucosa. Viruses , 12, 1425. doi: 10.3390/v12121425.
- Long, J. S., Mistry, B., Haslam, S. M., & Barclay, W. S. (2019). Host and viral determinants of influenza A virus species specificity. Nature Reviews. Microbiology , 17, 67–81. doi: 10.1038/s41579-018-0115-z.
- Matrosovich, M. N., Matrosovich, T. Y., Gray, T., Roberts, N. A., & Klenk, H.-D. (2004). Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. Journal of Virology , 78, 12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004.
- Pyrc, K., Sims, A. C., Dijkman, R., Jebbink, M., Long, C., Deming, D., … Pickles, R. (2010). Culturing the unculturable: Human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. Journal of Virology , 84, 11255–11263. doi: 10.1128/JVI.00947-10.
- Rayner, R. E., Makena, P., Prasad, G. L., & Cormet-Boyaka, E. (2019). Optimization of Normal Human Bronchial Epithelial (NHBE) cell 3D cultures for in vitro lung model studies. Scientific Reports , 9, 500. doi: 10.1038/s41598-018-36735-z.
- Rijsbergen, L. C., van Dijk, L. L. A., Engel, M. F. M., de Vries, R. D., & de Swart, R. L. (2021). Modelling of respiratory virus infections in human airway epithelial cells–a systematic review. Frontiers in Immunology , 12, 683002. doi: 10.3389/fimmu.2021.683002.
- Royston, L., & Tapparel, C. (2016). Rhinoviruses and respiratory enteroviruses: Not as simple as ABC. Viruses , 8, 16. doi: 10.3390/v8010016.
- Saint-Criq, V., Delpiano, L., Casement, J., Onuora, J. C., Lin, J., & Gray, M. A. (2020). Choice of differentiation media significantly impacts cell lineage and response to CFTR modulators in fully differentiated primary cultures of cystic fibrosis human airway epithelial cells. Cells , 9, 2137. doi: 10.3390/cells9092137.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., … Cardona, A. (2012). Fiji: An open-source platform for biological image analysis. Nature Methods , 9, 676–682. doi: 10.1038/nmeth.2019.
- Srinivasan, B., Kolli, A. R., Esch, M. B., Abaci, H. E., Shuler, M. L., & Hickman, J. J. (2015). TEER measurement techniques for in vitro barrier model systems. Journal of Laboratory Automation , 20, 107–126. doi: 10.1177/2211068214561025.
- V'kovski, P., Gultom, M., Kelly, J. N., Steiner, S., Russeil, J., Mangeat, B., … Dijkman, R. (2021). Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biology , 19, e3001158. doi: 10.1371/journal.pbio.3001158.
- de Vries, M., Mohamed, A. S., Prescott, R. A., Valero-Jimenez, A. M., Desvignes, L., O'Connor, R., … Dittmann, M. (2021). A comparative analysis of SARSCoV-2 antivirals characterizes 3CL inhibitor PF-00835231 as a potential new treatment for COVID-19. Journal of Virology , 95(10), e01819–20. doi: 10.1128/JVI.01819-20.
- Walters, M. S., Gomi, K., Ashbridge, B., Moore, M. A. S., Arbelaez, V., Heldrich, J., … Crystal, R. G. (2013). Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respiratory Research , 14, 135. doi: 10.1186/1465-9921-14-135.
- Whitcutt, M. J., Adler, K. B., & Wu, R. (1988). A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cellular & Developmental Biology, 24, 420–428.

