Identifying Protein-Protein Interactions by Proximity Biotinylation with AirID and splitAirID
Grace A. Schaack, Grace A. Schaack, Owen M. Sullivan, Owen M. Sullivan, Andrew Mehle, Andrew Mehle
Abstract
Proteins frequently function in high-order complexes. Defining protein-protein interactions is essential to acquiring a full understanding of their activity and regulation. Proximity biotinylation has emerged as a highly specific approach to capture transient and stable interactions in living cells or organisms. Proximity biotinylation exploits promiscuous biotinylating enzymes fused to a bait protein, resulting in the biotinylation of adjacent endogenous proteins. Biotinylated interactors are purified under very strict conditions and identified by mass spectrometry to obtain a high-confidence list of candidate binding partners. AirID is a recently described biotin ligase specifically engineered for proximity labeling. This protocol details proximity biotinylation by AirID, using protein complexes that form during a type I interferon response as an example. It covers the construction and validation of AirID fusion proteins and the enrichment and identification of biotinylated interactors. We describe a variation on the protocol using splitAirID. In this case, AirID is split into two inactive fragments and ligase activity is only restored upon dimerization of the bait proteins. This permits selective detection of proteins that interact with homo- or heterodimeric forms of the bait. The protocol considers design strategies, optimization, and the properties of different biotin ligases to identify optimal conditions for each experimental question. We also discuss common pitfalls and how to troubleshoot them. These approaches allow proximity biotinylation to be a powerful tool for defining protein interactomes. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Construction and functional validation of AirID fusion proteins
Alternate Protocol : Construction and functional validation of splitAirID fusion proteins
Support Protocol : Western blot for biotinylated proteins
Basic Protocol 2 : Biotinylation, enrichment, and identification of protein interactors
INTRODUCTION
The function of a protein is determined not only by its intrinsic properties, but also by the other proteins with which it interacts. These protein-protein interactions (PPIs) can be dynamic and confer multiple, context-dependent functions to each protein. Proximity biotinylation is a technique that enables comprehensive definition of physical PPI networks (Bosch, Chen, & Perrimon, 2021). Unlike other methods that only detect stable PPIs, proximity labeling also captures transient and noncovalent interactions. The key tool in the proximity biotinylation experiment is a promiscuous biotinylating enzyme fused to the bait protein of interest. Any protein coming within approximately 10 nm of the fusion protein has the potential to be biotinylated, and proteins that physically interact with the bait are thereby permanently labeled (Kim et al., 2014). Expressing the fusion protein in a relevant cell line under conditions appropriate to the biological process under study can define the complete protein interactome of the bait in its native context. Capitalizing on the high-affinity biotin-streptavidin interaction, biotinylated proteins are rigorously purified from whole-cell extracts and identified by mass spectrometry. Quantitative mass spectrometry approaches afford even greater confidence in constructing a putative interaction network from which specific interactions may be further investigated. Multiple biotin ligases are available for use in proximity biotinylation experiments, each with their own advantages and disadvantages. Here we present protocols employing AirID, a recently developed biotin ligase with a favorable specificity and toxicity profile compared to its counterparts (Kido et al., 2020).
Standard proximity biotinylation identifies prey that interacts with a single bait protein. However, if the protein of interest functions in a context-dependent manner as part of a protein complex, it may be more useful to employ a split biotin ligase system for labeling. The split format separates the biotin ligase into two discrete polypeptides. Neither fragment has biotin ligase activity in isolation, and the fragments have low affinity for each other, preventing spontaneous complementation; rather, function is restored when the two complementary fragments are actively brought together. The reconstituted biotin ligase then labels proteins much like the full-length enzyme. This enables specific interrogation of bait dimers. The biotin ligase fragments are fused to each bait, and only upon a physical interaction of the bait proteins are the two complementary biotin ligase fragments brought into sufficient proximity to reconstitute a functional biotin ligase that labels interactors specific to this dimeric complex. Biotin ligase fragments may be fused to the same bait to study homodimers, or to different proteins to study heterodimers. We have developed a split version of AirID (splitAirID). In a splitAirID experiment, the N-terminal fragment of the AirID enzyme (AirN), is fused to one bait, and the complementary C-terminal fragment of the AirID enzyme (AirC) is fused to another bait protein. When the two baits interact with one another, AirN and AirC come together to reconstitute biotin ligase activity (Fig. 1).
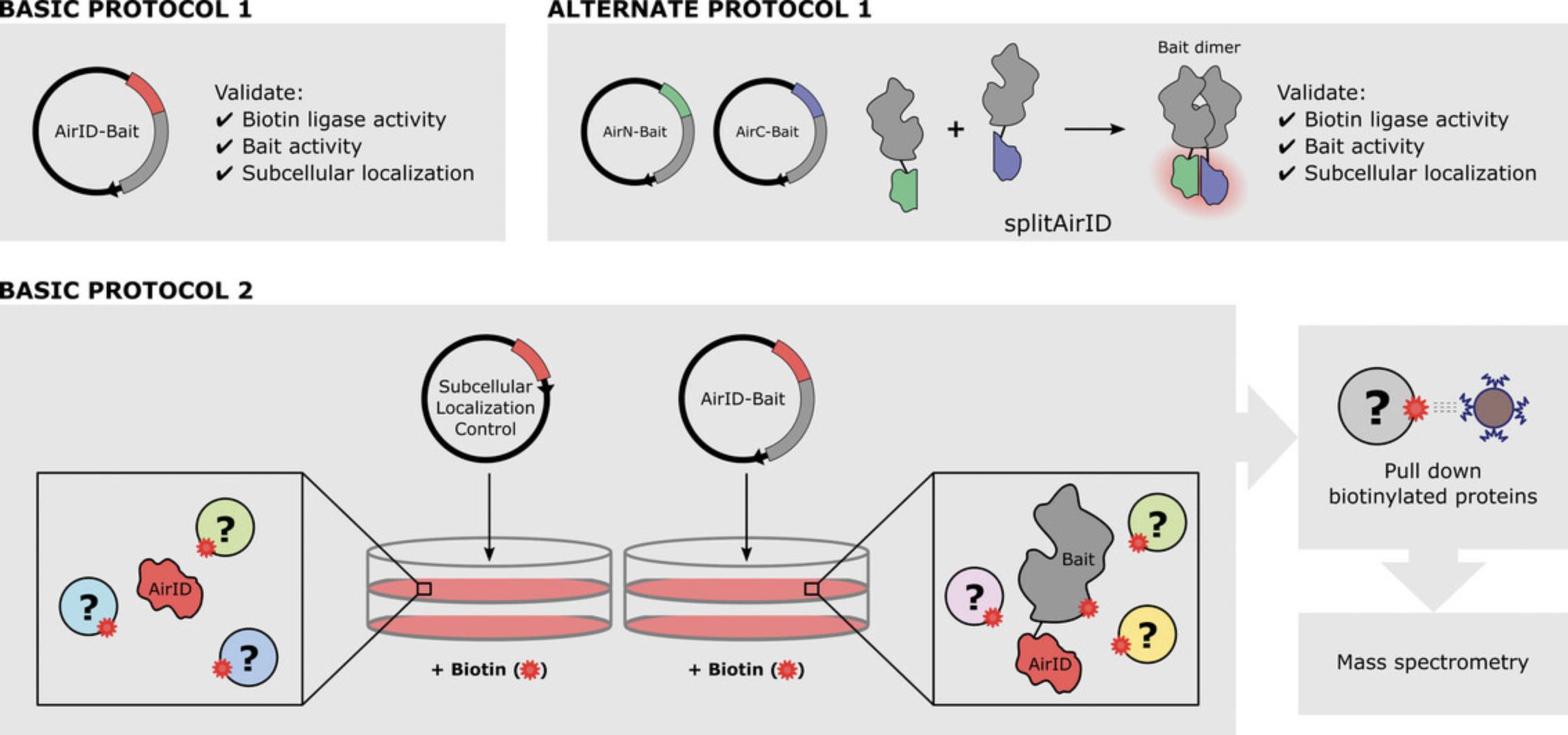
The protocols presented here cover the necessary planning and experimental steps for a proximity biotinylation experiment from the construction of AirID and/or splitAirID fusion proteins to the submission of biotinylated proteins for identification and quantification by mass spectrometry (Fig. 1). Basic Protocol 1 details the generation and validation of AirID fusion proteins. Alternate Protocol provides information on further considerations and the controls needed for splitAirID. Support Protocol outlines western blotting for biotinylated proteins, a technique that is used repeatedly in the Basic Protocols. Basic Protocol 2 describes the preparation of samples for mass spectrometry, from the expression of AirID (or splitAirID) fusion proteins to the isolation of biotinylated proteins from cell lysates. Finally, we consider common pitfalls and offer suggestions on troubleshooting.
NOTE : These protocols employ HEK 293T cells but are easily adapted to suit other cell lines as needed.
Basic Protocol 1: CONSTRUCTION AND FUNCTIONAL VALIDATION OF AirID FUSION PROTEINS
To capture biologically relevant interactions, the AirID fusion protein must mimic the native bait protein. This protocol describes the generation of fusion constructs with your protein of interest (the bait) and the AirID enzyme. Fusion proteins must be validated with respect to biotin ligase function as well as the function of the bait protein (Fig. 1). Perturbations in localization and/or functionality of the bait due to fusion with the AirID enzyme may increase artifactual results, decreasing the relevance of identified PPIs. The protocol requires basic molecular cloning, DNA transfection, western blotting, and immunofluorescence staining techniques along with any additional techniques specific to assaying for the function of your protein of interest.
Materials
-
Expression plasmid for bait protein of interest
-
AirID gene (Addgene plasmid #182210; also see Supplemental Files in Supporting Information)
-
DNA plasmid purification kit of choice (e.g., PureYield Plasmid Miniprep System, Promega, cat. no. A1222; ZymoPURETM II Plasmid Purification Kit, midiprep, Zymo Research, cat. no. D4201)
-
HEK 293T cells (or other appropriate cell line)
-
1× Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) (or other appropriate cell culture medium)
-
jetPRIME transfection reagent and buffer (PolyPlus, cat. no. 101000015)
-
100 mM biotin (see recipe)
-
1× Dulbecco's phosphate-buffered saline (DPBS) (Corning, cat. no. 21-031-CV)
-
Co-immunoprecipitation (IP) lysis buffer (see recipe) or other lysis buffer
-
Protease inhibitor cocktail (Millipore, cat. no. P2714-1BTL)
-
Acetone, high grade, suitable for immunofluorescence staining (Electron Microscopy Sciences, cat. no. 10000)
-
Methyl alcohol, high grade, suitable for immunofluorescence staining (Electron Microscopy Sciences, cat. no. 18510)
-
1%-3% (w/v) bovine serum albumin (BSA) in DPBS
-
Fluorophore-conjugated streptavidin, Alexa Fluor 488 conjugate (Invitrogen, cat. no. S11223)
-
Fluorophore-conjugated antibodies to detect bait and AirID fusion proteins (e.g., human anti-V5 epitope tag Alexa Fluor 594, Novus, cat. no. NBP2-81035AF594)
-
Mounting medium containing DAPI stain (Vectashield, H-1200)
-
6-well tissue culture plates
-
Microplate sealing tape
-
Platform rocker
-
Microcentrifuge tubes
-
Microcentrifuge
-
Glass coverslips coated with fibronectin over a PDL layer (Neuvitro, cat. no. GG-12-1.5-Fibronectin)
-
Glass microscope slides
-
Needle
-
Forceps
-
Nail polish
Generate plasmid encoding AirID fused to bait protein
The AirID amino acid and gene sequences are available from Kido et al. (Kido et al., 2020). Codon choice and other transcript features may be altered to ensure optimal expression in your system. See Supplemental Files in Supporting Information for the DNA sequence encoding AirID codon-optimized for Homo sapiens.
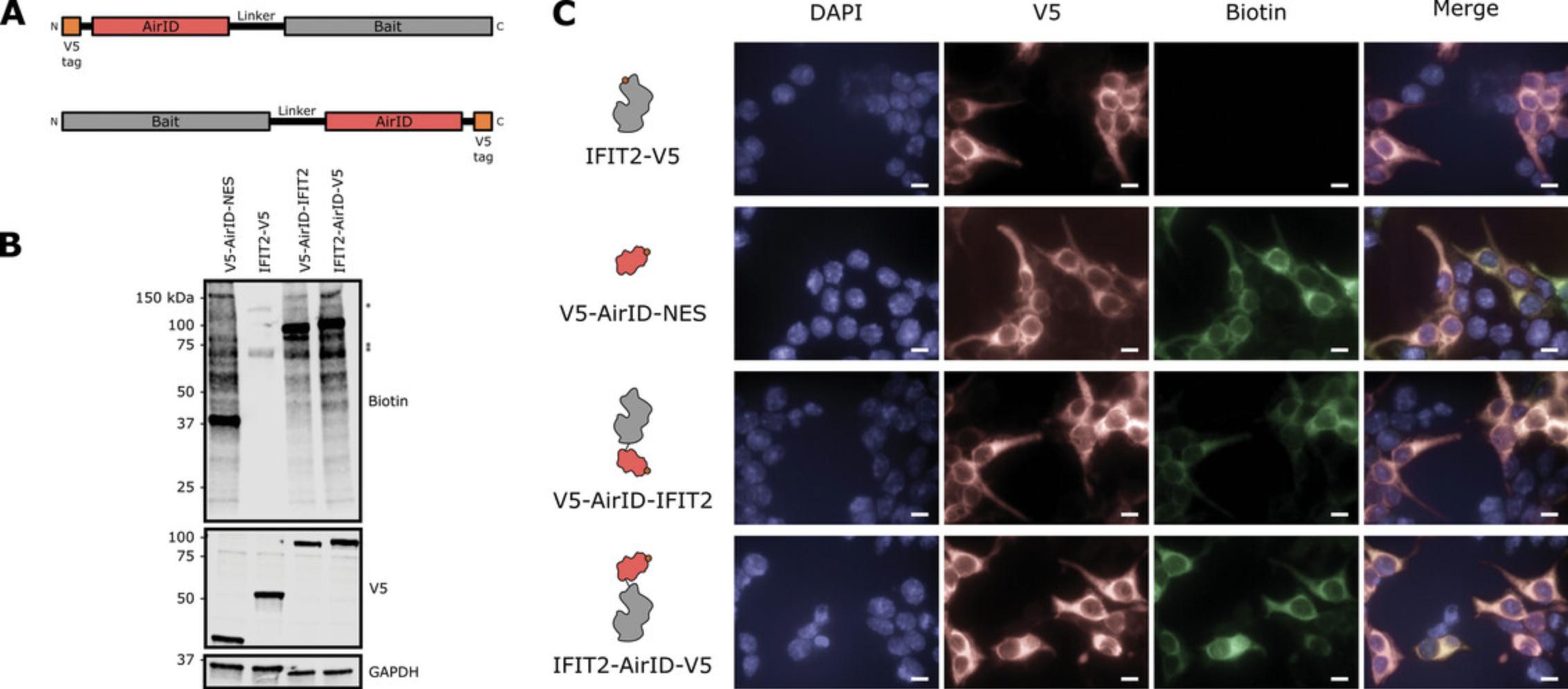
1.Use a molecular cloning technique of your choice to fuse AirID to the bait protein in an appropriate expression vector. For example, in our work, we chose to generate N-terminal and C-terminal fusions of AirID onto our bait protein and included V5 epitope tags for ease of protein detection (Fig. 2A). An overview of molecular cloning strategies is available at https://www.addgene.org/mol-bio-reference/cloning/.
1.Based on what is known about bait protein function, is it preferable to fuse AirID to the N- or the C-terminus of the bait protein? Have other fusion proteins been described in the literature to serve as models for the fusion protein design (e.g., protein of interest fused to a fluorescent protein)? 2.Epitope tag(s) can be incorporated on the bait or AirID domains and may be useful for downstream detection of your fusion protein by western blotting and immunofluorescence staining. 3.The length and flexibility of the linker between AirID and the bait protein will impact protein function and the labeling radius. 4.Ensure that sequences encoding bait, AirID, and epitope tag(s) remain in the same reading frame.
2.Generate an unfused AirID that localizes to the same subcellular compartment(s) as the bait to serve as a control for baseline AirID labeling. Create this subcellular localization control plasmid in the same vector backbone as your fusion construct. Determine the subcellular localization of the bait protein (via literature search or experimentally by immunofluorescence assays or subcellular cell fractionation), and add a corresponding subcellular localization signal to the AirID sequence (e.g., nuclear localization signal, nuclear export signal, mitochondrial targeting sequence, etc.).
3.Sequence plasmids to confirm the accuracy of the cloning procedure.
4.Prepare transfection-quality plasmids using a DNA plasmid purification kit of your choice.
Assess AirID biotin ligase function
The goal of this part of the protocol is to confirm that the AirID portion of your fusion protein retains biotin ligase enzymatic activity. To assess biotin ligase function, fusion proteins are expressed in cells for a sufficient duration to allow robust biotinylation to occur, then biotinylated proteins are detected in cell lysates by western blot. This is essentially a scaled-down version of the ultimate proximity biotinylation experiment.
5.Seed a 6-well tissue culture plate (one well per condition) with 5 × 105 HEK 293T cells in 500 µl DMEM with 10% FBS per well. Allow cells to grow to ∼80% confluency. For example, to generate the data shown in Figure 2B, 4 wells were seeded to accommodate the subcellular localization control condition (V5-AirID-NES), a negative-control condition with unfused bait protein (IFIT2-V5), and two experimental conditions consisting of AirID-fused bait proteins (V5-AirID-IFIT2 and IFIT2-AirID-V5).
6.Transfect each well with appropriate plasmids for each condition using jetPRIME per manufacturer's instructions. Include the subcellular localization control condition (AirID without bait) and a negative-control condition with unfused bait protein.
Optional : Immediately prior to transfection, remove spent medium and replace with medium supplemented with biotin to 50 μM.
7.At 24 hr post-transfection, carefully remove the media from each well and gently wash once with cold DPBS (∼500 µl per well) to remove excess biotin. Proceed directly to cell lysis or seal the plate with microplate sealing tape and store at −80°C until ready to lyse cells.
8.Lyse cells in co-IP lysis buffer. Add 500 μl co-IP lysis buffer supplemented with protease inhibitor cocktail to each well. Rock plates 20-30 min at 4°C, transfer lysates to microcentrifuge tubes, and clarify lysates by centrifugation for 15 min at 15,000 × g , 4°C. Transfer supernatants to fresh tubes.
9.Perform western blotting according to Support Protocol to detect biotinylated proteins in the lysates and to assess expression and stability of the AirID fusion proteins (Fig. 2B).
Assay for bait protein function
In addition to confirming that the AirID portion of the fusion protein retains function, it is important to confirm that the bait protein also remains functional despite fusion to the AirID enzyme. Relevant assays will be specific to your protein of interest, but if an established functional assay for the bait protein is available, use this to assess activity of the fusion protein compared to unfused bait protein.
Assess subcellular localization by immunofluorescence assays
The goal of this part of the protocol is to visualize the impact of fusion to AirID on the subcellular localization of the bait protein. Cells are seeded onto glass coverslips, transfected with plasmids to express AirID fusion or control proteins, then fixed and stained to assess protein subcellular localization and biotinylation by immunofluorescence microscopy. The general tissue culture conditions and transfection protocol are very similar to the protocol above for assessing biotin ligase activity (note similarities to steps 5 and 6 above), but here cells are seeded onto glass coverslips to facilitate downstream microscopy.
10.Place a fibronectin-coated glass coverslip at the bottom of each well of a 6-well plate (one well per condition), then seed with 5 × 105 HEK 293T cells in 500 µl DMEM with 10% FBS per well. Allow cells to grow to ∼60% confluency. For example, to generate the data shown in Figure 2C, 5 wells were seeded to accommodate an empty-vector control condition to establish background signal, the subcellular localization control condition (V5-AirID-NES), a negative-control condition with unfused bait protein (IFIT2-V5), and two experimental conditions consisting of AirID-fused bait proteins (V5-AirID-IFIT2 and IFIT2-AirID-V5).
11.Transfect each well with appropriate DNA for each condition using jetPRIME per manufacturer's instructions. Include the following control conditions: empty vector control to establish background signal, unfused bait protein (bait without AirID), and the subcellular localization control (AirID without bait).
Optional : Immediately prior to transfection, remove spent medium and replace with medium supplemented with biotin to 50 μM.
12.At 24 hr post-transfection, carefully remove the medium from each well and wash gently with cold DPBS (∼500 µl per well) to remove excess media and free biotin.
13.Carefully transfer coverslips to a fresh 6-well plate.
14.Fix and permeabilize cells in a cold 1:1 solution of acetone and methanol for 15 min at −20°C.
15.Wash once briefly in cold DPBS. Add enough DPBS to each well to completely cover the coverslips. Then, perform two further washes, rocking at 4°C for ∼5 min per wash.
16.Block in 1%-3% BSA in DPBS. Rock for at least 30 min at 4°C.
17.Remove blocking solution and incubate with fluorophore-conjugated streptavidin and fluorophore-conjugated antibody against the AirID fusion protein(s), each diluted 1:1000 in fresh blocking solution. Rock overnight (∼16 hr) in the dark at 4°C.
18.Wash once briefly in cold DPBS. Add enough DPBS to each well to completely cover the coverslips. Then perform two further washes, rocking at 4°C for ∼5 min per wash.
19.Mount coverslips cell-side down on glass microscope slides with a DAPI-containing mounting medium. Seal with nail polish if desired.
20.Store slides at 4°C, protected from light, until ready to view by fluorescence microscopy (Fig. 2C).
Alternate Protocol: CONSTRUCTION AND FUNCTIONAL VALIDATION OF splitAirID FUSION PROTEINS
SplitAirID allows selective detection of proteins that interact with a higher-order bait complex. This protocol describes the generation of fusion constructs of your protein(s) of interest and the splitAirID enzyme system. As in the case of full-length AirID, these fusion proteins must be validated with respect to biotin ligase function as well as bait protein function and localization. All techniques and experimental workflow for splitAirID are the same as described in Basic Protocol 1 for full-length AirID but with the inclusion of additional considerations and controls.
Additional Materials (also see Basic Protocol 1)
- Genes for AirN and AirC polypeptides (see Supplemental Files in Supporting Information)
Generate plasmid encoding splitAirID components fused to bait protein(s)
AirN and AirC sequences, codon-optimized for Homo sapiens , can be found in Supporting Information. Codon choice and other transcript features may be altered to ensure optimal expression in your system.
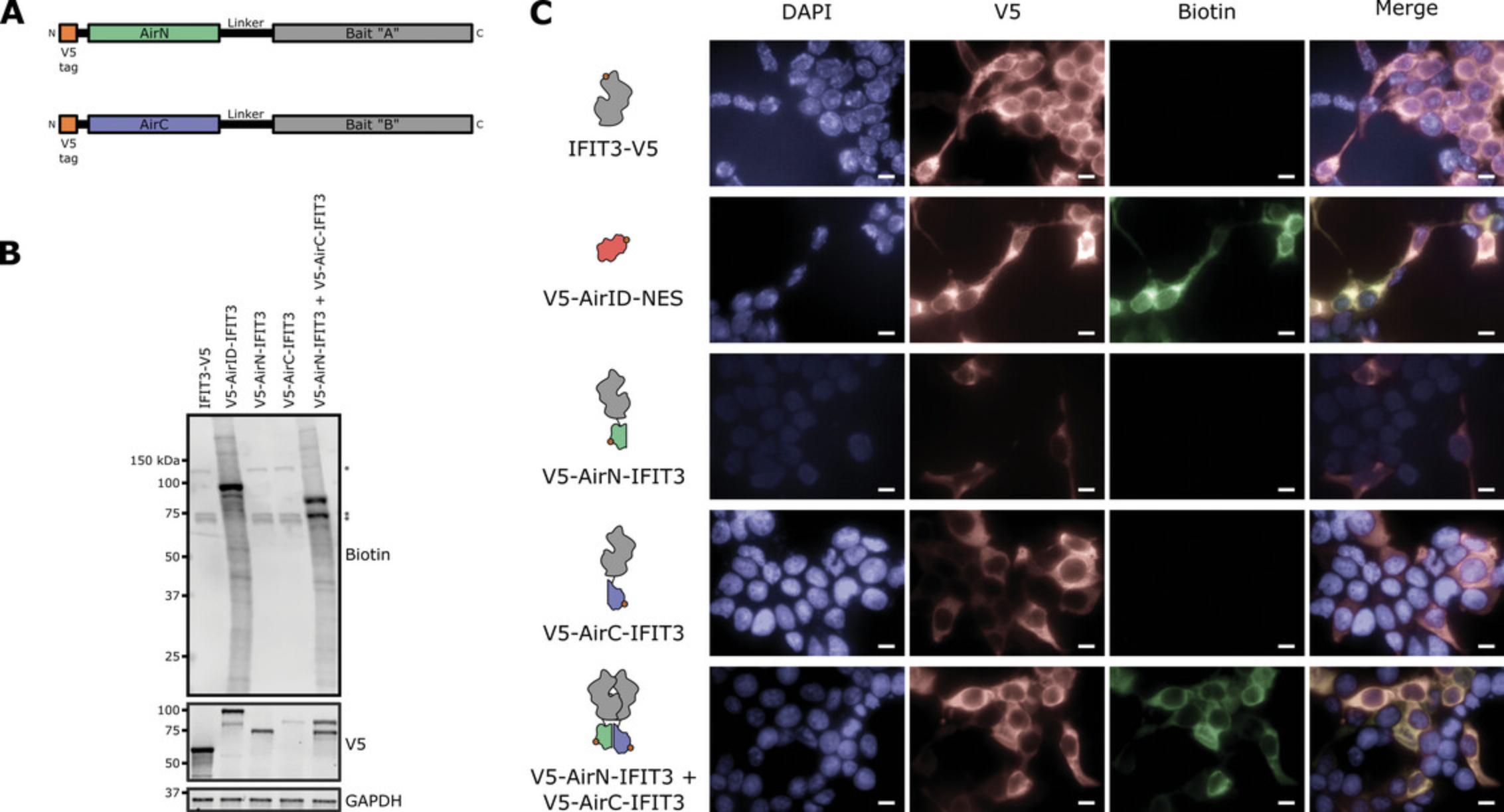
1.Construct plasmids encoding the bait protein(s) of interest fused with AirN and AirC as described above in Basic Protocol 1 for full-length AirID (Fig. 3A).
1.Consider the spatial orientation of your interacting bait proteins. Is it preferable to append AirN/AirC to the N- or C-terminus of each bait? Will AirN and AirC be able to interact when the bait proteins dimerize? Is it preferable to fuse the smaller AirN (∼11 kDa) or the larger AirC (∼27 kDa) to a particular bait? It may be necessary to test multiple combinations of N- and C-terminal positioning of AirN and AirC on each bait to find a combination with optimal biotin ligase activity and bait protein function. 2.The length and flexibility of the linker between AirN/AirC and the bait protein(s) will impact the efficiency of complementation, the labeling radius, and bait protein function. Alter the linkers as necessary to optimize these characteristics (e.g., decrease linker length to increase labeling specificity, use glycine and serine residues to increase linker flexibility, etc.). 3.Adding different epitope tags to AirN and AirC may be useful for differentiating the two when co-expressing AirN- and AirC-fused bait(s), but this is not mandatory.
Assess AirID biotin ligase function
As in the protocol for full-length AirID detailed above, biotin ligase function must be validated to confirm that splitAirID biotinylation activity remains intact despite fusion with the bait protein(s) (Fig. 3B). This procedure assesses whether AirN and AirC interact with one another and reconstitute a functional biotin ligase when fused to the bait protein(s).
2.Perform the assay “Assess AirID biotin ligase function” from Basic Protocol 1 as described above (see steps 5-9), with the following set of conditions:
-
Unfused bait protein (bait without AirID) as a negative control.
-
Subcellular localization control condition (AirID without bait) as a positive control for biotin ligase activity (if previously validated, bait fused to full-length AirID may also be used).
-
Bait fused with AirN.
-
Bait fused with AirC.
-
Co-expression of AirN-fused bait and AirC-fused bait.
Percentages of each plasmid in the transfectant for each condition may require optimization to achieve similar levels of expression across conditions, with particular attention paid to the co-expression of AirN- and AirC- fused baits.
Assay bait protein function
As described above for full-length AirID in Basic Protocol 1, if an established functional assay for the bait protein is available, evaluate fusion protein activity as compared to unfused bait protein activity to determine whether bait function is significantly altered by fusion with the splitAirID fragments.
Assess subcellular localization by immunofluorescence assays
Use different fluorophores to visualize co-localization of the splitAirID fusion proteins with biotinylated proteins (Fig. 3C). For example, we use streptavidin conjugated to Alexa Fluor 488 and anti-V5 antibody conjugated to Alexa Fluor 594, which are compatible with our microscope excitation and emission settings and provide the spectral separation needed for imaging. If different epitope tags were appended to AirN and AirC fusions, co-localization of the two components can also be directly assessed in the co-expression condition by using different fluorophore-conjugated antibodies to detect each epitope.
3.Perform immunofluorescence assays as described above for full-length AirID in Basic Protocol 1 (see steps 10-20), with the following set of conditions:
-
Empty vector negative control to establish background signal.
-
Wild-type bait protein (bait without AirID) to demonstrate normal localization.
-
Subcellular localization control (AirID without bait) as a positive control for biotinylation and to confirm that this control is localizing as intended.
-
Bait fusion with AirN.
-
Bait fusion with AirC.
-
AirN-fused bait co-expressed with AirC-fused bait.
Percentages of each plasmid in the transfectant for each condition may require optimization to achieve similar levels of expression across conditions, with particular attention paid to the co-expression of AirN- and AirC- fused baits.
Support Protocol: WESTERN BLOT FOR BIOTINYLATED PROTEINS
This protocol describes the western blotting procedure for detecting biotinylated proteins and evaluating AirID fusion protein expression levels and stability.
Materials
-
2× Laemmli sample buffer (see recipe)
-
Precast SDS-PAGE gels (BioRad, cat. no. 4561036)
-
Nitrocellulose membrane (Bio-Rad, cat. no. 1620146)
-
1× BSA blocking solution (see recipe)
-
Fluorophore-conjugated streptavidin, IRDye 800CW Streptavidin (LI-COR, cat. no. 925-32230)
-
Antibodies specific to AirID fusion protein or epitope tag (e.g., anti-V5, Bethyl Laboratories, Inc., cat. no. A190-220A)
-
Antibody for housekeeping gene (e.g., anti-GAPDH, Proteintech, cat. no. 600004-1-Ig)
-
Secondary antibodies to detect chosen primary antibodies
-
TBS-Tween (0.1%) (see recipe)
-
Heat block or thermocycler
-
SDS-PAGE electrophoresis unit (e.g., Mini-PROTEAN II Electrophoresis Cell, Bio-Rad)
-
Semi-dry transfer cell (e.g., Trans-Blot Turbo, Bio-Rad)
-
Western blot imaging system (e.g., LI-COR)
NOTE : Exact amount of lysate and dilutions of antibodies and streptavidin will depend on your specific reagents. Start with manufacturer-recommended conditions and optimize further as needed.
Perform western blotting for biotinylated proteins
1.Denature clarified lysate. Add 10 μl of lysate to 10 μl of 2× Laemmli sample buffer. Mix. Heat 10 min at 95°C. Centrifuge 60 s at 12,000 × g to collect the sample at the bottom of the tube.
2.Load 10 μl of denatured sample (i.e., 5 μl of lysate) per lane of a 15-well 10% SDS-PAGE gel. Run PAGE at 200 V until the dye front reaches the bottom of the gel.
3.Transfer proteins to nitrocellulose membrane under semi-dry conditions per manufacturer instructions.
4.Block membrane(s) in 1× BSA blocking solution, rocking 1 hr at room temperature.
5.Use one of the replicate membranes to probe for biotin using streptavidin conjugated to an infrared (IR) fluorophore. Dilute the conjugated streptavidin 1:5000 in fresh 1× BSA blocking solution. Incubate the membrane in conjugated streptavidin overnight (∼16 hr), rocking, at 4°C, protected from light.
6.Wash the membrane 3 times with TBS-Tween (0.1%) for ∼5 min per wash.
7.Use a LI-COR or other appropriate western blot imaging system to visualize signal on the blot.
8.Use the other replicate membrane to probe for the bait protein, or corresponding epitope tag, and a housekeeping protein of your choice with appropriate primary antibodies. Dilute antibodies in 1× BSA blocking solution. Incubate the membrane in primary antibodies overnight (∼16 hr), rocking, at 4°C.
9.Wash the membrane 3 times with TBS-Tween (0.1%) for ∼5 min per wash.
10.Dilute appropriate secondary antibodies in TBS-Tween (0.1%). Incubate the membrane with secondary antibodies for 1-4 hr, rocking, at room temperature.
11.Wash the membrane 3 times with TBS-Tween (0.1%) for ∼5 min per wash.
12.Use a LI-COR or other appropriate western blot imaging system to visualize signal on the blot.
Basic Protocol 2: BIOTINYLATION, ENRICHMENT, AND IDENTIFICATION OF PROTEIN INTERACTORS
With validated AirID fusion constructs in-hand from Basic (or Alternate) Protocol 1, the full-scale proximity biotinylation experiment may be performed. AirID fusion proteins are transiently expressed and given time to label proteins in proximity with the bait. Biotinylated proteins are purified from cell lysates and identified and quantified by mass spectrometry (Fig. 1). Ideally, the proximity biotinylation experiment is performed in biological triplicate. This protocol requires techniques in DNA transfection, western blotting, and affinity purification. The validation experiments from Basic Protocol 1 should have already resulted in optimization of conditions such as the proportion of each plasmid in the transfection, duration of incubation after transfection, and the level of exogenous biotin supplementation. If your experiment requires an exogenous stimulus such as interferon induction, infection, drug treatment, etc., optimization of the timing of AirID fusion construct transfection and cell stimulation will also be necessary. This protocol is the same for both the full-length AirID and splitAirID systems.
Materials
-
Expression plasmids for AirID fusion protein(s) and subcellular localization control (user-generated from Basic/Alternate Protocol)
-
Cell line of choice and appropriate culture medium (HEK 293T, 1× DMEM with 10% FBS)
-
jetPRIME transfection reagent and buffer (PolyPlus cat. No. 101000015)
-
100 mM biotin (see recipe)
-
1× DPBS (Corning, cat. No. 21-031-CV)
-
Streptavidin-conjugated magnetic beads, Dynabeads MyOne Streptavidin T1 (Invitrogen, cat. No. 65602)
-
Co-IP lysis buffer (see recipe) or other lysis buffer
-
Protease inhibitor cocktail (Millipore, cat. no. P2714-1BTL)
-
RIPA buffer (see recipe)
-
1 M KCl (see recipe)
-
0.1 M Na2CO3 (see recipe)
-
2 M urea (see recipe)
-
Biotin elution buffer (see recipe)
-
50% MeOH (see silver stain solutions recipe)
-
5% MeOH (see silver stain solutions recipe)
-
33 μM DTT (see silver stain solutions recipe)
-
Silver nitrate solution (see silver stain solutions recipe)
-
Developer solution (see silver stain solutions recipe)
-
Citric acid monohydrate
-
4 M NaCl (see silver stain solutions recipe)
-
10 cm tissue culture dishes
-
Cell scrapers
-
15 ml conical tubes
-
Tabletop centrifuge
-
Microcentrifuge tubes
-
Microcentrifuge
-
Nutator
-
Magnetic rack
-
Heat block
-
Clean glass or plastic storage container
CAUTION : Use personal protective equipment as needed to reduce keratin contamination (e.g., gloves, lab coat, disposable sleeves, hair net).
Perform proximity biotinylation
Perform the proximity biotinylation experiment at full scale. The overall workflow is the same as that of assessing biotin ligase function described in Basic Protocol 1 above: transfect plasmids encoding the fusion construct into cells, allow time for protein expression and biotinylation to occur, then harvest cell lysate. Biotinylation and AirID fusion protein expression are confirmed by western blot before proceeding to enrichment. The protocol presented here reflects timings and cell numbers that may be used as a starting point but should be adjusted per your own optimizations to suit your biological process of interest.
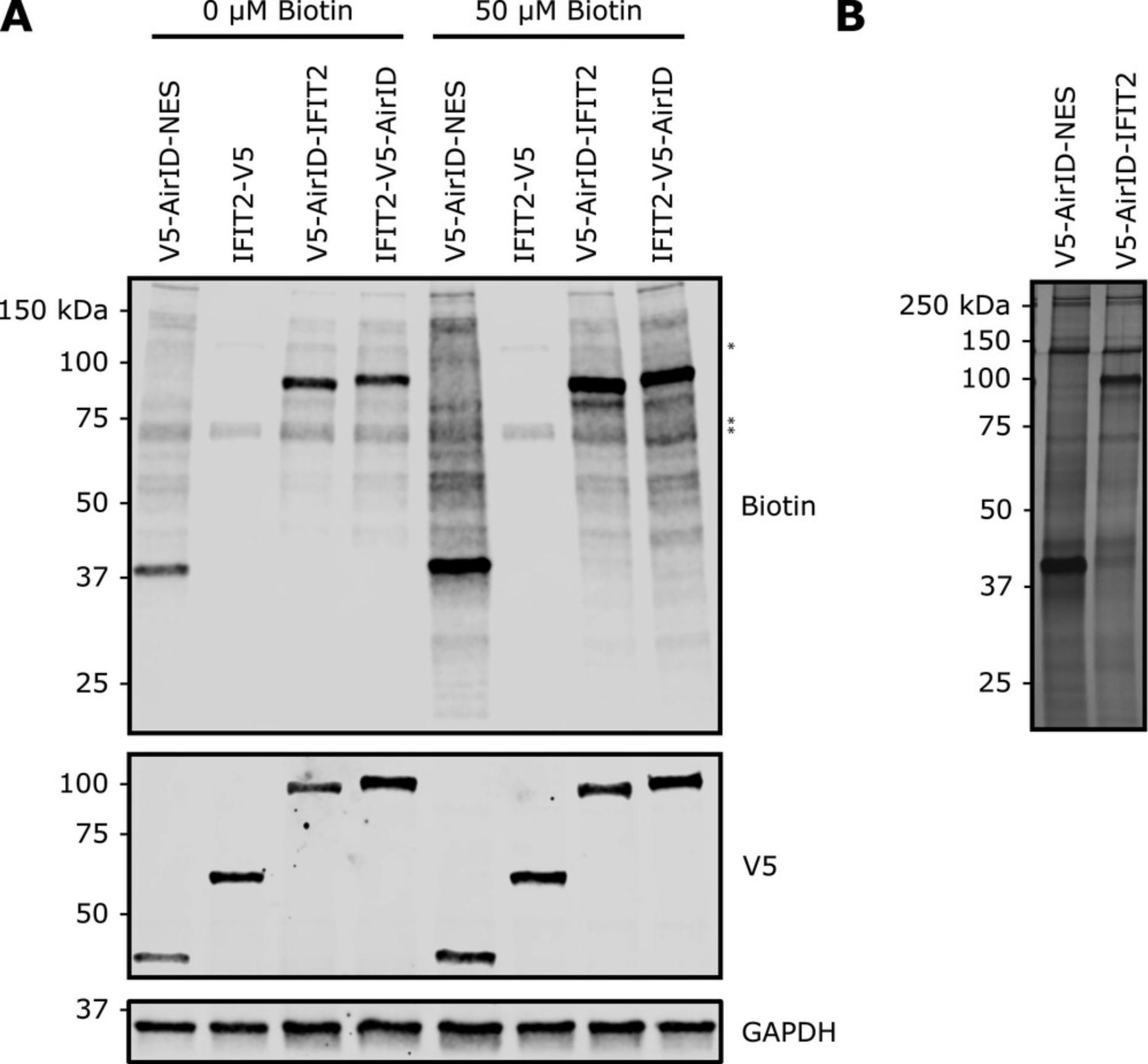
1.Seed 10 cm tissue culture dishes (2 dishes per condition) with 4 × 106 HEK 293T cells per dish in DMEM with 10% FBS. Allow cells to grow to ∼80% confluency. For example, to generate data like that shown in Figure 4B, we set up a subcellular localization control condition (V5-AirID-NES) and a condition with our bait protein of interest fused to AirID (V5-AirID-IFIT2).
2.Transfect each well with appropriate DNA for each condition using jetPRIME per manufacturer's instructions.
3.At 18 hr post-transfection, carefully remove the spent media and replace with media supplemented with biotin to 50 μM.
4.24 hr later (at 42 hr post-transfection), carefully remove the media from each dish. Wash gently 5 times with cold DPBS to remove excess biotin and end the labeling reaction. Scrape cells into conical tubes (the duplicate dishes may be pooled at this point) and pellet the cells by centrifugation at 500 × g for 5 min. Remove the supernatant and freeze the cell pellets at −80°C.
5.Lyse cells in co-IP lysis buffer supplemented with the protease inhibitor cocktail by resuspending each cell pellet in 1200 μl co-IP lysis buffer. Transfer to microcentrifuge tubes and nutate for 20-30 min at 4°C, then clarify by centrifuging 15 min at 15,000 × g , 4°C. Transfer supernatants to fresh tubes. Reserve 800 μl of each clarified lysate for subsequent enrichment.
6.Use a portion of the remaining lysate to perform western blotting according to Support Protocol to detect biotinylated proteins in the lysates and to confirm expression of the AirID fusion proteins. Store the remaining lysate for additional analysis as needed.
Enrich for biotinylated proteins
Next, enrich the samples for biotinylated proteins using streptavidin-coated magnetic beads to pull down biotinylated proteins from each clarified lysate. This section of the protocol yields a concentrated solution of biotinylated proteins that may then be identified by mass spectrometry. The entire protocol should be performed carefully and with sufficient personal protective equipment to reduce keratin contamination from the user.
7.Prepare 200 μl of bead slurry per 800 μl of clarified lysate. Pellet the beads on a magnetic rack and remove the supernatant. Wash the beads in 1.5 volumes of co-IP lysis buffer by nutation. Pellet the beads on a magnetic rack and remove the supernatant. Resuspend the beads in another 1.5 volumes of co-IP lysis buffer to perform a second wash. After the second wash, remove the supernatant and resuspend the beads in 200 μl co-IP lysis buffer per sample.
8.Add 200 μl of the washed bead suspension to each of the 800 μl samples (the reserved clarified cell lysates from step 5). Nutate overnight (∼16 hr) at 4°C.
9.Pellet the beads on a magnetic rack. Collect the supernatant and save this flow-through for downstream western blotting (keep samples on ice if performing the western blot same-day or freeze for later use).
10.Perform the following series of washes (adapted from Cho et al., 2020b). For each wash, resuspend the beads in the wash solution by shaking vigorously by hand, then incubate at room temperature for the time indicated. Pellet the beads on a magnetic rack, remove the supernatant, and proceed to the next wash step. Care should be taken to closely follow the times indicated for each wash.
-
Wash once in 1 ml lysis buffer (co-IP or other buffer used in steps 5 and 7).
-
Wash once in 500 μl lysis buffer + 500 μl RIPA buffer.
If RIPA was used as the cell lysis buffer, this step may be omitted.
-
Wash in 1 ml RIPA buffer. Incubate for 2 min. Repeat this step.
-
Wash once in 1 ml 1 M KCl. Incubate for 2 min.
-
Wash once, briefly, in 1 ml 0.1 M Na2CO3. Perform this step as quickly as possible.
-
Wash once, briefly, in 1 ml 2 M urea in 10 mM Tris·HCl (pH = 8). Perform this step as quickly as possible.
-
Wash in 1 ml RIPA buffer. Incubate for 2 min. Repeat this step. Remove the final wash completely.
11.Elute the biotinylated proteins from the beads by adding 40 μl biotin elution buffer. Fully resuspend the beads. Boil at 95°C for 5 min.
12.Centrifuge 60 s at 12,000 × g to collect sample at the bottom of the tube, then pellet the beads on a magnetic rack. Quickly collect the eluate into a fresh tube.
13.Repeat steps 11-12 to perform a second elution. Combine the first and second eluates.
Visualize proteins by silver staining
Visualize proteins in the eluates by silver staining (Fig. 4B). A commercial silver stain kit may be used as an alternative to the protocol below.
14.Perform denaturing PAGE with 5 μl of each eluate. Mix 5 μl of eluate with an equal volume of 2× Laemmli sample buffer. Heat at 95°C for 10 min. Load the entire 10 μl of each denatured sample on a 10% SDS-PAGE gel. Run PAGE at 200 V until the dye front reaches the bottom of the gel.
15.Carefully transfer the gel to a clean container.
16.Soak the gel in 50% MeOH for 20 min, rocking, at room temperature.
17.Decant. Soak in 5% MeOH for 20 min, rocking, at room temperature.
18.Decant. Soak in 33 μM DTT for 20 min, rocking, at room temperature.
19.Decant. Rinse once, quickly, in water.
20.Rinse quickly with silver nitrate solution, then incubate in fresh silver nitrate solution for 20 min, rocking, at room temperature.
21.Remove the silver nitrate solution. Rinse 3 times, as quickly as possible, with water.
22.Rinse with a minimal volume of developer (just enough to cover the gel) until the solution turns yellowish in color, then decant and add fresh developer solution. Protein bands will become visible and darken in color as the gel develops. Continue to develop the gel until as dark as desired.
23.Add solid citric acid monohydrate to stop development. Approximately 5 g of citric acid monohydrate is needed for every 10 ml of developer used in step 22.
24.Store gel in water until ready to examine.
25.Image the gel (use a scanner, photograph over a light source, etc.).
To monitor the efficiency of biotin capture, assess the depletion of biotinylated proteins from cell lysates by blotting input lysates (use a portion of the remaining lysate from step 6) and flow-through (from step 9) according to Support Protocol. Blot input and flow-through on the same membrane or otherwise ensure that they are visualized under identical exposure conditions such that they can be directly compared with one another.
Identify protein interactors by mass spectrometry
If the mass spectrometry will be done by a core facility or other collaborator, discuss the experiment and desired quantitative mass spectrometry method ahead of time in case any additions or modifications to the sample preparation protocol are required to fit into their workflows. A detailed discussion of mass spectrometry parameters is outside the scope of this protocol.
26.Submit samples for quantitative mass spectrometry.
27.Identify significant PPIs using a data analysis method appropriate for the type of mass spectrometry performed.
REAGENTS AND SOLUTIONS
Use ultrapure water in all recipes.
Biotin, 100 mM
Dissolve 24.4 mg biotin (Millipore, cat. no. B4501-500MG) in 1 ml of DMSO (Corning, 21-031-CV) for use in tissue culture or in 1 ml alkaline water for use in making elution buffer. Filter-sterilize (pore size = 0.2 μm). Store up to 8 weeks at −20°C.
Biotin elution buffer
- 25 mM biotin (from 100 mM stock of biotin in water)
- 4% SDS
- 0.125 M Tris (pH 6.8)
- Prepare fresh immediately before use
BSA blocking solution, 1×
Dilute 10% (w/v) BSA solution (Thermo Scientific, cat. no. 37520) 1:10 with 1× TBS-Tween (0.1%). Store up to 3 months at 4°C.
Co-|P lysis buffer
- 50 mM Tris-HCl, pH 7.4
- 150 mM NaCl
- 0.5% Nonidet-P40 substitute (US Biological, cat. no. N3500)
- Store up to 3 months at 4°C
KCl, 1 M
- Dissolve 3.73 g of KCl up to 50 ml water. Prepare fresh immediately before use.
Laemmli sample buffer, 2×
- 2× Laemmli Sample Buffer with bromophenol blue (BioRad, cat. no. 1610737)
- 5% (v/v) 2-mercaptoethanol (Sigma cat. no. M6250, add immediately before use)
Na2CO3, 0.1 M
- Dissolve 53 mg of Na2CO3 up to 50 ml water. Prepare fresh immediately before use.
RIPA buffer
- RIPA (1×)
- 100 mM Tris-HCl, pH 7.5
- 150 mM NaCl
- 0.2% (w/v) SDS
- 1% (w/v) DOC
- 2% (v/v) Nonidet-P40 substitute
- Store up to 3 months at 4°C
Silver stain solutions
- 50% methanol (v/v) (Fisher Chemical, cat. no. CAS 67-56-1) in water. Shelf stable.
- 5% methanol (v/v) in water. Shelf stable.
- 33 μM DTT: combine 8.3 μl of 1 M DTT in 250 ml of water. Prepare fresh immediately before use.
- AgNO3 6 mM: dissolve 0.25 g of silver in 250 ml of water. Prepare fresh immediately before use.
- Developer: dissolve 7.5 g Na2CO3 in 125 μl of 37% formaldehyde (Millipore, cat. no. 47608-250ML-F) up to 250 ml of water. Prepare fresh immediately before use.
- NaCl 4 M: dissolve 234 g NaCl up to 1 L of water. Shelf stable.
TBS-Tween (0.1%), 1×
- Prepare a 10× solution as:
- 0.5 M Tris-HCl, pH 7.4
- 1.5 M NaCl
- 1% Tween 20 (v/v) (Fisher bioreagents, BP337-500)
- Dilute 1:10 to obtain the 1 × 50 mM Tris pH 7.4, 150 mM NaCl, 0.1% (v/v) Tween 20
- Store up to 3 months at room temperature
Urea, 2 M
- Dissolve 6 g of urea up to 50 ml in water buffered with 0.5 ml of 1 M Tris-HCl (pH 8) to obtain 2 M Urea 10 mM Tris pH 8. Prepare fresh immediately before use.
COMMENTARY
Background Information
Proximity biotinylation is a powerful technique for defining PPIs (Qin, Cho, Cavanagh, & Ting, 2021). Unlike traditional affinity purification, proximity labeling methods capture stable interactions as well as transient and/or non-covalent interactions, yielding a more comprehensive dataset (Liu, Salokas, Weldatsadik, Gawriyski, & Varjosalo, 2020; Sears, May, & Roux, 2019). Labeling occurs in living cells, prior to cell lysis, precluding identification of irrelevant protein interactions that form post-lysis (Bosch et al., 2021). Furthermore, the exceptionally strong interaction between biotin and streptavidin enables stringent washing to markedly increase the specificity of protein capture (Samavarchi-Tehrani, Samson, & Gingras, 2020).
Biotinylation is achieved enzymatically by a promiscuous biotin ligase (e.g., AirID, BioID, BioID2, BASU, TurboID, miniTurbo) or by an engineered peroxidase (e.g., HRP, APEX, APEX2) fused to the bait (Cho et al., 2020b). Peroxidase-based methods are very fast, capable of completing labeling within one minute, but require hydrogen peroxide treatment and are toxic to cells (Cho et al., 2020b; Lam et al., 2015; Lobingier et al., 2017; Rhee et al., 2013). BioID and its relatives are generally slower acting, but better tolerated by cells, and thus better suited to applications in live cells (Cho et al., 2020b).
BioID is the original Escherichia coli BirA derivative used for proximity labeling (Roux, Kim, Raida, & Burke, 2012). BioID2, derived from a biotin ligase in Aquifex aeolicus , is smaller than BioID, making it less impactful on bait subcellular localization (Kim et al., 2016). BASU is an alternative biotin ligase derived from Bacillus subtilis (Ramanathan et al., 2018). BioID, BioID2, and BASU exhibit similar labeling kinetics, requiring >16 hr of labeling time to produce sufficient signal (Branon et al., 2018). TurboID was produced by directed evolution of BirA, creating a highly active, fast enzyme that biotinylates proteins within 10 min (Branon et al., 2018). A smaller version of TurboID, miniTurbo, exhibits lower activity but greater specificity than TurboID (Branon et al., 2018). AirID is a relatively recent addition to the BioID family of enzymes that was created via computational reconstruction of an ancestral BirA (Kido et al., 2020). Requiring at least 1-3 hr of labeling time, AirID is not as highly active as TurboID, but it is significantly faster than the older BioID enzymes (Kido et al., 2020). AirID also exhibits favorable labeling specificity compared to TurboID (Kido et al., 2020).
Each of these biotin ligases respond to exogenous biotin supplementation in the cell culture medium to differing degrees. While the relatively small amount of biotin typically present in serum-containing cell culture media can sometimes sustain low-level activity, most of these enzymes require additional biotin supplementation to produce robust biotinylation. BioID2 requires less exogenous biotin than original BioID, while TurboID is highly biotin-consumptive (Branon et al., 2018; Kim et al., 2016). The activity of miniTurbo can be tightly controlled via addition of exogenous biotin to biotin-depleted media (Branon et al., 2018). The exceptional activity of TurboID comes at the cost of toxicity as it competes for and depletes biotin from normal cellular processes. AirID, on the other hand, does not require exogenous biotin supplementation, though addition of exogenous biotin can improve signal strength (Kido et al., 2020). Of the fast-acting ligases, AirID is the least toxic to cells, enabling its use over prolonged periods of time, on the order of hours to days (Kido et al., 2020).
In addition to use in cell culture systems, proximity biotinylation has also been applied in vivo. BioID has been successful in plants, protists, eukaryotic parasites, and mice (Batsios, Meyer, & Gräf, 2016; Chen et al., 2015; Conlan, Stoll, Gorman, Saur, & Rathjen, 2018; Dingar et al., 2015; Khan, Youn, Gingras, Subramaniam, & Desveaux, 2018; Khosh-Naucke et al., 2018; McAllaster et al., 2015; Morriswood et al., 2013; Tu et al., 2020; Uezu et al., 2016). BioID2 has also been used in a protist model (Pitzen, Askarzada, Gräf, & Meyer, 2018). TurboID has been used in plants, yeast, and Drosophila melanogaster , though the duration of TurboID expression must be limited to avoid excessive toxicity (Branon et al., 2018; Larochelle, Bergeron, Arcand, & Bachand, 2019; Mair, Xu, Branon, Ting, & Bergmann, 2019; Shinoda, Hanawa, Chihara, Koto, & Miura, 2019; Zhang et al., 2019). The low toxicity of AirID has allowed for its stable expression in transgenic lines of D. melanogaster (personal communication, M. Harrison).
For defining interactors of dimeric protein complexes, split biotin ligase complementation systems are an elegant approach. Split biotin ligases have proven successful in the forms of split-BioID, Contact-ID, and split-TurboID (Cho et al., 2020a; De Munter et al., 2017; Kwak et al., 2020; Schopp et al., 2017). Each of these split proteins come with the underlying advantages and limitations of their parent full-length enzyme. We made a split version of AirID, splitAirID, to bring the low toxicity and high specificity of AirID to a split system.
The variety of characteristics displayed by these enzymes means that there is likely an appropriate enzyme available no matter the experimental situation. The labeling speed and dependence on exogenous biotin provision make miniTurbo, for example, ideally suited to experiments in which strict temporal control over labeling is desired. AirID is well suited to experiments of longer duration or situations in which maintaining exogenous biotin supplementation is challenging. The very high activity of TurboID may make it ideal for detecting low abundance interactions. Ultimately, the choice of which biotin ligase to use relies on balancing enzymatic activity, specificity, and toxicity, each of which may be of greater or lesser importance depending on the experimental question being addressed. Note that while the protocols presented here were optimized for AirID, they may be easily modified for another biotin ligase as the overall workflow is the same for all of the BioID derivatives. Several excellent protocols are also available for BioID, TurboID, and split-TurboID (Chastney, Lawless, & Humphries, 2020; Cho et al., 2020b; Le Sage, Cinti, & Mouland, 2016; Roux, Kim, & Burke, 2013).
Critical Parameters
As noted in the protocol, when designing the AirID fusion constructs, several factors should be taken into consideration. Appending AirID to the N- or C-terminus of the bait may yield different interaction results and differently impact fusion protein stability and activity. The length and flexibility of the linker between the AirID and bait domains may also impact protein stability and function. The longer the linker, the greater the effective AirID labeling radius, so linkers should be sufficiently long and flexible to maximize protein function while remaining short enough to preserve labeling specificity. Optimization of these factors may require generation of multiple versions of AirID fusion proteins to assess their functionalities prior to selecting one or more constructs for use in a full-scale experiment.
While AirID does not require exogenous biotin supplementation to function effectively, supplementing the medium with biotin may still be of use for enhancing the signal:noise ratio. We recommend, at a minimum, comparing biotinylation signal with and without biotin supplementation to 50 μM in the media as part of your early optimization efforts (Fig. 4A).
Because AirID does not require exogenous biotin to function, the enzyme will be actively labeling proteins for the duration of its expression. Depending on your experimental design, this may or may not be advantageous. If tight control over the timing of AirID expression is required, consider generating stable cell lines capable of inducible AirID fusion protein expression. The protocols herein rely on transient overexpression by transfection but could easily be modified for the inducible expression format.
To preserve physiologic relevance, AirID fusion protein expression should be optimized to closely match endogenous bait protein expression levels occurring in the biological context of interest. An inducible expression system is again an alternative option if transient overexpression proves overly problematic.
Time is another important consideration. How long should labeling be allowed to occur If treating with an exogenous stimulus, how long should cells be treated? Should treatment occur prior to or after expression of the AirID fusion construct? Answers to these questions will be specific to the experimental question(s) and relevant biological context. These questions should be addressed prior to performing the full-scale proximity biotinylation experiment.
Collecting a pool of biotinylated proteins of sufficient quantity and purity is important for generating biologically relevant mass spectrometry data (i.e., “garbage in, garbage out”). Two parameters impacting sample abundance and purity are the number of cells used in the full-scale proximity biotinylation experiment and the amount of streptavidin-conjugated beads used in the enrichment step. Either or both of these factors may require optimization to ensure collection of an accurately representative sample of bait-interacting proteins for identification by mass spectrometry. In our hands, two 10 cm dishes of HEK 293T cells per condition yield more than enough protein for analysis, but your experiment may require greater or fewer cells depending on the cell line and level of AirID fusion protein expression. When enriching cell lysates for biotinylated proteins, enough streptavidin-conjugated beads should be used to fully sample the population of biotinylated proteins but not so many as to promote non-specific binding. An optimization experiment titrating the amount of streptavidin resin used for the enrichment step should be performed to identify conditions that significantly deplete biotinylated proteins from the input lysate. See “Understanding Results” below for further discussion of lysate:bead volume optimization.
Another consideration is the type of mass spectrometry to be performed. The most statistically robust approaches that provide the highest-confidence lists of interactors involve quantitative mass spectrometry. Label-free quantification (LFQ), stable isotope labeling by amino acids in cell culture (SILAC), and isobaric labeling methods like tandem mass tag (TMT) proteomics are all demonstrated, viable options for proximity biotinylation experiments (Bersuker et al., 2018; Branon et al., 2018; Roux et al., 2012). Samples must be measured individually in LFQ, making it time-intensive. However, LFQ offers high sensitivity and is easy to incorporate into experimental workflows as it does not require an additional labeling procedure (Li et al., 2012). TMT enables multiplexing and with rigorous statistical analyses can yield high-confidence interactomes (Cho et al., 2020b). Sample loss during TMT labeling may require additional starting material or result in identification of fewer peptides than LFQ (Li et al., 2012). SILAC offers high accuracy and does not require extensive processing prior to mass spectrometry, but the number of possible experimental conditions is limited by the number of stable isotopes used. Note that if a SILAC approach is desired, the cell culture conditions in Basic Protocol 2 must be modified to incorporate heavy isotope labeling. The choice of quantitative proteomics method then dictates the appropriate data analysis pipelines.
Finally, prior to launching the full-scale proximity biotinylation experiment, controls and replicates should already be carefully planned. To acquire high-quality, reproducible data that identify meaningful interactions, it is recommended to generate three biological replicates in which cells are transfected on different days for each replicate and samples from each replicate are enriched for biotinylated proteins in separate batches (Choi et al., 2012). The subcellular control is key to ultimately distinguishing true positives from spurious interactions. This control consists simply of the AirID enzyme targeted to the same subcellular compartment as the bait. This “naked” AirID without fusion to the bait biotinylates proteins that interact with AirID at baseline in that cellular compartment. This establishes a background level of biotinylation against which bait-directed biotinylation is compared. If your experiment involves multiple treatment conditions, the subcellular localization control should be included in each individual treatment condition.
Troubleshooting
Common problems encountered while performing the protocols, their possible causes, and potential solutions are described in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| Low signal for biotinylated proteins on western blot | Biotin ligase activity hindered by fusion to bait | Optimize linker length and flexibility; consider fusing the enzyme to a different terminus of the bait |
| Low transfection efficiency | Ensure cells are low-passage, actively growing, and free of mycoplasma; optimize cell density at time of transfection | |
| Insufficient available biotin for robust labeling | Increase concentration of biotin in the cell culture medium | |
| Insufficient labeling time | Increase the duration of AirID fusion overexpression | |
| Poor blotting | Ensure blocking reagent is free of biotin; include commercially available biotinylated markers as a control for biotin detection; expect detection of endogenously biotinylated proteins of 72, 75, and 130 kDa | |
| Bait activity significantly reduced on functional assay | Bait activity hindered by fusion with AirID | Optimize linker length and flexibility; consider fusing the enzyme to a different terminus of the bait |
| Low signal on immunofluorescence assay | Concentration of antibody (or streptavidin reagent) is too low | Optimize concentrations of antibodies and fluorophore-conjugated streptavidin |
| Alteration of epitopes by organic solvent(s) | Fix cells in formaldehyde and permeabilize in Triton X-100 | |
| Biotinylated protein signal in input and flow-through is largely unchanged | Too few beads were used | Use more beads (decrease the cell lysate:bead volume ratio) |
| Free biotin saturates streptavidin resin to prevent capture of biotinylated proteins | Be sure cells and dishes are carefully washed with DPBS to remove free biotin present in culture media | |
| High background banding on silver stain of eluate from biotin pull-down | Too many beads were used | Use fewer beads (increase the cell lysate:bead volume ratio) |
| AirID overexpressed at too high a level | Increase the specificity during labelling by reducing the expression levels of AirID controls and fusions. This may require increasing the total number of cells used to acquire sufficient material. | |
| Low signal on silver stain of eluate from biotin pull-down | Biotinylated proteins remain bound to the beads | Add a third elution step |
| Too few beads were used | Use more beads (decrease the cell lysate:bead volume ratio) | |
| Not enough biotinylated proteins in the cell lysates | Scale up the proximity biotinylation experiment to harvest protein from more cells; express higher amounts of proteins during transfection |
Statistical Analysis
Analyze mass spectrometry data to identify significant interactors. The use of the AirID subcellular localization control establishes background biotinylation that allows for relative quantification of specific interactors with the AirID fusions. Appropriate data analysis methods will be specific to the type of quantitative mass spectrometry performed. Label-free quantification data, for example, may be analyzed using SAINTexpress. SAINTexpress is a computational tool that scores PPIs according to a statistical model that calculates the posterior probability of each interaction being true (Teo et al., 2014). A graphical user interface for SAINTexpress is maintained by the Resource for Evaluation of Protein Interaction Networks (REPRINT) and enables integration of CRAPome controls (see Internet Resources below). Data generated by TMT proteomics may be analyzed using a ratiometric approach with receiver-operator-characteristic-based cutoffs to generate a high-confidence list of interactors (Cho et al., 2020b).
Understanding Results
A fully-validated AirID fusion protein shows evidence of biotin ligase activity and has comparable function and localization to the unfused bait protein. When assessing biotin ligase function, western blots for biotin should detect endogenously biotinylated proteins across all conditions: propionyl CoA carboxylase at 72 kDa, 3-methylcrotonyl CoA carboxylase at 75 kDa, and pyruvate carboxylase at 130 kDa in humans (Chandler & Ballard, 1985). If signal intensity in the AirID-containing samples necessitates short exposure times during imaging, one or more of the endogenous protein bands may not be visible. Samples containing AirID are expected to exhibit multiple additional bands (Fig. 2B). At a minimum, a band corresponding to the AirID fusion protein itself should be visible, representing autobiotinylation of the AirID fusion protein. Should your experiment require expressing AirID without localization signals, epitope tags, or bait fusions, it may not be detected in biotin blots as it has been reported that AirID does not biotinylate itself (Kido et al., 2020). In parallel with detection of biotinylated proteins, be sure to include western blotting for the fusion protein itself to assess protein expression and stability. Subcellular localization will impact the pool of potential interactors, so it is important to confirm that the fusion protein is localizing similarly to the unfused bait protein. Immunofluorescence assays should reveal a similar subcellular localization pattern for AirID-fused and unfused baits. The AirID fusion protein is expected to co-localize with biotin signal (Fig. 2C). Keep in mind that any healthy cell will contain biotinlylated proteins, producing a background level of biotin signal by immunofluorescence. Use the empty-vector control to visualize this background signal and set appropriate brightness and contrast levels such that true, above-background biotinylation levels can be distinguished in AirID-containing samples. Validating bait(s) fused with AirN and AirC follows the same logic, but in the case of splitAirID, biotinylation signal is only expected when AirN and AirC fusion proteins are co-expressed and co-localized (Fig. 3B and 3C). If biotin ligase activity is impaired, localization is perturbed, or bait protein function is significantly attenuated when fused to AirID, consider: changing the position at which the AirID is fused to the bait, moving it from the N-terminus to the C-terminus or vice versa; varying the length and flexibility of the linker between AirID and the bait; or, testing different expression levels of the AirID fusions. Finally, be sure you are using the biotin ligase best suited to your experimental question.
Labeled interactors must be isolated from whole cell lysates such that they may be identified and quantified by mass spectrometry. This is achieved by affinity purification of biotinylated proteins from cell lysates using streptavidin-conjugated beads. Depletion of proteins from cell lysates is assessed when western blotting for biotinylated proteins in the input and flow-through. A gross decrease in biotinylated proteins should be visible when comparing input and flow-through. If too many beads are used, however, this leaves a greater surface area of beads available for non-specific binding, increasing background noise. This may become apparent during silver staining of eluates, where a large number of bands with a similar banding pattern appear across all conditions and bands corresponding to the AirID fusion proteins are less distinctive against the many background bands. The amount of material in the cell lysates and the resin type and manufacturer will impact the amount of beads needed for optimal purification. Use the western blot and silver stain results to optimize amounts of affinity resin accordingly. A simple titration using increasing amounts of beads in parallel captures may be performed to identify conditions where the resin becomes saturated with specific interactors but shows minimal background binders.
Once mass spectrometry data are available and analyzed, the resulting candidate list of interactors can be used to quickly assess the success of the experiment. Compare prey identities with known bait interactors. The presence of previously known interactors in the dataset indicates that the experiment was sufficiently executed to detect true interactors of the bait protein, bolstering its validity and increasing confidence in any novel interactions identified in the AirID-generated interactome. For example, in our work with IFIT2 and IFIT3 as bait proteins, we rediscovered known interactions between IFIT1, IFIT2, and IFIT3, indicating that our proximity labeling experiment was able to capture true PPIs (Fleith et al., 2018; Johnson et al., 2018). Once success of the proximity biotinylation experiment is thus confirmed, novel interactions can be investigated further.
While a list of high-confidence interactors is the endpoint of proximity biotinylation experiments, it is the starting point for functional validation. Independent assays should be used to validate PPIs and confirm their biological significance. Such assays may include: standard co-immunoprecipitation assays, with the acknowledgement that these assays may not detect transient or weak PPIs; bimolecular fluorescence or luciferase complementation assays to detect interactions in living cells; and imaging-based approaches to demonstrate co-localization of interacting proteins in cells (Schaack & Mehle, 2020). Ultimately, functional assays are also needed to confirm the importance of novel PPIs for the activity of your protein of interest.
Time Considerations
The entire protocol can be completed in ∼4-6 months. Generating and validating the AirID fusion constructs (Basic Protocol 1) takes ∼1-2 months, depending on how many constructs need to be cloned and tested. Performing the full-scale proximity biotinylation experiment and isolating biotinylated proteins (Basic Protocol 2) takes 1-2 weeks per biological replicate, followed by another 2-4 weeks for mass spectrometry data acquisition and analysis. More time will be required if lengthy optimization and troubleshooting are needed.
Acknowledgments
The authors thank G. Barrett-Wilt and G. Sabat from the UWBC Mass Spectrometry Core Facility for advice. AM is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease and an H.I. Romnes Faculty Fellow funded by the Wisconsin Alumni Research Foundation (USA). This work is supported by the National Institutes of Health R01AI164690 (AM), T32AI007414 (GS), T32GM140935 (GS), and T32AI055397 (OS).
Author Contributions
Grace A. Schaack : Conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, resources, validation, visualization, writing - original draft, writing-review and editing; Owen M. Sullivan : Conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, validation, visualization, writing-original draft, writing-review and editing; Andrew Mehle : Conceptualization, formal analysis, funding acquisition, project administration, resources, supervision, writing-original draft, writing-review and editing.
Conflict of Interest
The authors declare no conflict of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Open Research
Data Availability Statement
All data are fully presented within the manuscript and associated files.
Supporting Information
Full-length AirID with V5 tag, V5-AirID.gb
SplitAirID components with V5 tags, V5-AirN.gb and V5-AirC.gb
DNA sequences codon-optimized for Homo sapiens
Supporting Information
| Filename | Description |
|---|---|
| cpz1702-sup-0001-SuppMat.gb2.3 KB | Full-length AirID with V5 tag, V5-AirID.gb |
| cpz1702-sup-0002-SuppMat.gb2.6 KB | SplitAirID components with V5 tags, V5-AirN.gb and V5-AirC.gb |
| cpz1702-sup-0003-SuppMat.gb1.6 KB | DNA sequences codon-optimized for Homo sapiens |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Batsios, P., Meyer, I., & Gräf, R. (2016). Proximity-dependent biotin identification (BioID) in dictyostelium amoebae. Methods in enzymology , 569, 23–42. doi: 10.1016/bs.mie.2015.09.007
- Bersuker, K., Peterson, C. W. H., To, M., Sahl, S. J., Savikhin, V., Grossman, E. A., … Olzmann, J. A. (2018). A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Developmental Cell , 44, 97–112.e7. doi: 10.1016/j.devcel.2017.11.020
- Bosch, J. A., Chen, C.-L., & Perrimon, N. (2021). Proximity-dependent labeling methods for proteomic profiling in living cells: An update. Wiley Interdisciplinary Reviews. Developmental Biology , 10, e392. doi: 10.1002/wdev.392
- Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., … Ting, A. Y. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nature Biotechnology , 36, 880–887. doi: 10.1038/nbt.4201
- Chandler, C. S., & Ballard, F. J. (1985). Distribution and degradation of biotin-containing carboxylases in human cell lines. Biochemical Journal , 232, 385–393. doi: 10.1042/bj2320385
- Chastney, M. R., Lawless, C., & Humphries, M. J. (2020). Multiplexed proximity biotinylation coupled to mass spectrometry for defining integrin adhesion complexes. Current Protocols in Cell Biology , 88, e113. doi: 10.1002/cpcb.113
- Chen, A. L., Kim, E. W., Toh, J. Y., Vashisht, A. A., Rashoff, A. Q., Van, C., … Bradley, P. J. (2015). Novel components of the toxoplasma inner membrane complex revealed by BioID. mBio , 6, e02357–14. doi: 10.1128/mBio.02357-14
- Cho, K. F., Branon, T. C., Rajeev, S., Svinkina, T., Udeshi, N. D., Thoudam, T., … Ting, A. Y. (2020a). Split-TurboID enables contact-dependent proximity labeling in cells. Proceedings of the National Academy of Sciences , 117, 12143–12154. doi: 10.1073/pnas.1919528117
- Cho, K. F., Branon, T. C., Udeshi, N. D., Myers, S. A., Carr, S. A., & Ting, A. Y. (2020b). Proximity labeling in mammalian cells with TurboID and split-TurboID. Nature Protocols , 15, 3971–3999. doi: 10.1038/s41596-020-0399-0
- Choi, H., Liu, G., Mellacheruvu, D., Tyers, M., Gingras, A., & Nesvizhskii, A. I. (2012). Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Current Protocols in Bioinformatics , 39, 8.15.1–8.15.23. doi: 10.1002/0471250953.bi0815s39
- Conlan, B., Stoll, T., Gorman, J. J., Saur, I., & Rathjen, J. P. (2018). Development of a rapid in planta BioID system as a probe for plasma membrane-associated immunity proteins. Frontiers in Plant Science , 9, 1882. doi: 10.3389/fpls.2018.01882
- De Munter, S., Görnemann, J., Derua, R., Lesage, B., Qian, J., Heroes, E., … Bollen, M. (2017). Split-BioID: A proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Letters , 591, 415–424. doi: 10.1002/1873-3468.12548
- Dingar, D., Kalkat, M., Chan, P.-K., Srikumar, T., Bailey, S. D., Tu, W. B., … Raught, B. (2015). BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. Journal of Proteomics , 118, 95–111. doi: 10.1016/j.jprot.2014.09.029
- Fleith, R. C., Mears, H. V., Leong, X. Y., Sanford, T. J., Emmott, E., Graham, S. C., … Sweeney, T. R. (2018). IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Research , 46, 5269–5285. doi: 10.1093/nar/gky191
- Johnson, B., VanBlargan, L. A., Xu, W., White, J. P., Shan, C., Shi, P.-Y., … Amarasinghe, G. K. (2018). Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity , 48, 487–499.e5. doi: 10.1016/j.immuni.2018.01.014
- Khan, M., Youn, J.-Y., Gingras, A.-C., Subramaniam, R., & Desveaux, D. (2018). In planta proximity dependent biotin identification (BioID). Scientific Reports , 8, 9212. doi: 10.1038/s41598-018-27500-3
- Khosh-Naucke, M., Becker, J., Mesén-Ramírez, P., Kiani, P., Birnbaum, J., Fröhlke, U., … Spielmann, T. (2018). Identification of novel parasitophorous vacuole proteins in P. falciparum parasites using BioID. International Journal of Medical Microbiology , 308, 13–24. doi: 10.1016/j.ijmm.2017.07.007
- Kido, K., Yamanaka, S., Nakano, S., Motani, K., Shinohara, S., Nozawa, A., … Sawasaki, T. (2020). AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. eLife , 9, e54983. doi: 10.7554/eLife.54983
- Kim, D. I., Jensen, S. C., Noble, K. A., Kc, B., Roux, K. H., Motamedchaboki, K., & Roux, K. J. (2016). An improved smaller biotin ligase for BioID proximity labeling. Molecular Biology of the Cell , 27, 1188–1196. doi: 10.1091/mbc.E15-12-0844
- Kim, D. I., KC, B., Zhu, W., Motamedchaboki, K., Doye, V., & Roux, K. J. (2014). Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proceedings of the National Academy of Sciences , 111, E2453–E2461. doi: 10.1073/pnas.1406459111
- Kwak, C., Shin, S., Park, J.-S., Jung, M., Nhung, T. T. M., Kang, M.-G., … Rhee, H. -W. (2020). Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proceedings of the National Academy of Sciences , 117, 12109–12120. doi: 10.1073/pnas.1916584117
- Lam, S. S., Martell, J. D., Kamer, K. J., Deerinck, T. J., Ellisman, M. H., Mootha, V. K., & Ting, A. Y. (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature Methods , 12, 51–54. doi: 10.1038/nmeth.3179
- Larochelle, M., Bergeron, D., Arcand, B., & Bachand, F. (2019). Proximity-dependent biotinylation mediated by TurboID to identify protein-protein interaction networks in yeast. Journal of Cell Science , 132, jcs232249. doi: 10.1242/jcs.232249
- Le Sage, V., Cinti, A., & Mouland, A. J. (2016). Proximity-Dependent Biotinylation for Identification of Interacting Proteins. Current Protocols in Cell Biology , 73, 17.19.1–17.19.12. doi: 10.1002/cpcb.11
- Li, Z., Adams, R. M., Chourey, K., Hurst, G. B., Hettich, R. L., & Pan, C. (2012). Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ orbitrap velos. Journal of Proteome Research , 11, 1582–1590. doi: 10.1021/pr200748h
- Liu, X., Salokas, K., Weldatsadik, R. G., Gawriyski, L., & Varjosalo, M. (2020). Combined proximity labeling and affinity purification−mass spectrometry workflow for mapping and visualizing protein interaction networks. Nature Protocols , 15, 3182–3211. doi: 10.1038/s41596-020-0365-x
- Lobingier, B. T., Hüttenhain, R., Eichel, K., Miller, K. B., Ting, A. Y., von Zastrow, M., & Krogan, N. J. (2017). An approach to spatiotemporally resolve protein interaction networks in living cells. Cell , 169, 350–360.e12. doi: 10.1016/j.cell.2017.03.022
- Mair, A., Xu, S.-L., Branon, T. C., Ting, A. Y., & Bergmann, D. C. (2019). Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife , 8, e47864. doi: 10.7554/eLife.47864
- McAllaster, M. R., Ikeda, K. N., Lozano-Núñez, A., Anrather, D., Unterwurzacher, V., Gossenreiter, T., … de Graffenried, C. L. (2015). Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Molecular Biology of the Cell , 26, 3013–3029. doi: 10.1091/mbc.E15-04-0219
- Morriswood, B., Havlicek, K., Demmel, L., Yavuz, S., Sealey-Cardona, M., Vidilaseris, K., … Warren, G. (2013). Novel bilobe components in trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryotic Cell , 12, 356–367. doi: 10.1128/EC.00326-12
- Pitzen, V., Askarzada, S., Gräf, R., & Meyer, I. (2018). CDK5RAP2 is an essential scaffolding protein of the corona of the dictyostelium centrosome. Cells , 7, 32. doi: 10.3390/cells7040032
- Qin, W., Cho, K. F., Cavanagh, P. E., & Ting, A. Y. (2021). Deciphering molecular interactions by proximity labeling. Nature Methods , 18, 133–143. doi: 10.1038/s41592-020-01010-5
- Ramanathan, M., Majzoub, K., Rao, D. S., Neela, P. H., Zarnegar, B. J., Mondal, S., … Khavari, P. A. (2018). RNA–protein interaction detection in living cells. Nature Methods , 15, 207–212. doi: 10.1038/nmeth.4601
- Rhee, H.-W., Zou, P., Udeshi, N. D., Martell, J. D., Mootha, V. K., Carr, S. A., & Ting, A. Y. (2013). Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science , 339, 1328–1331. doi: 10.1126/science.1230593
- Roux, K. J., Kim, D. I., & Burke, B. (2013). BioID: A screen for protein-protein interactions. Current Protocols in Protein Science , 74, 19.23.1–19.23.14. doi: 10.1002/0471140864.ps1923s74
- Roux, K. J., Kim, D. I., Raida, M., & Burke, B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. Journal of Cell Biology , 196, 801–810. doi: 10.1083/jcb.201112098
- Samavarchi-Tehrani, P., Samson, R., & Gingras, A.-C. (2020). Proximity dependent biotinylation: Key enzymes and adaptation to proteomics approaches. Molecular & Cellular Proteomics, 19, 757–773. doi: 10.1074/mcp.R120.001941
- Schaack, G. A., & Mehle, A. (2020). Experimental approaches to identify host factors important for influenza virus. Cold Spring Harbor Perspectives in Medicine , 10, a038521. doi: 10.1101/cshperspect.a038521
- Schopp, I. M., Amaya Ramirez, C. C., Debeljak, J., Kreibich, E., Skribbe, M., Wild, K., & Béthune, J. (2017). Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nature Communications , 8, 15690. doi: 10.1038/ncomms15690
- Sears, R. M., May, D. G., & Roux, K. J. (2019). BioID as a tool for protein-proximity labeling in living cells. Methods in Molecular Biology , 2012, 299–313. doi: 10.1007/978-1-4939-9546-2_15
- Shinoda, N., Hanawa, N., Chihara, T., Koto, A., & Miura, M. (2019). Dronc-independent basal executioner caspase activity sustains Drosophila imaginal tissue growth. Proceedings of the National Academy of Sciences , 116, 20539–20544. doi: 10.1073/pnas.1904647116
- Teo, G., Liu, G., Zhang, J., Nesvizhskii, A. I., Gingras, A.-C., & Choi, H. (2014). SAINTexpress: Improvements and additional features in Significance Analysis of INTeractome software. Journal of Proteomics , 100, 37–43. doi: 10.1016/j.jprot.2013.10.023
- Tu, V., Tomita, T., Sugi, T., Mayoral, J., Han, B., Yakubu, R. R., … Weiss, L. M. (2020). The Toxoplasma gondii cyst wall interactome. mBio , 11, e02699–19. doi: 10.1128/mBio.02699-19
- Uezu, A., Kanak, D. J., Bradshaw, T. W. A., Soderblom, E. J., Catavero, C. M., Burette, A. C., … Soderling, S. H. (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science , 353, 1123–1129. doi: 10.1126/science.aag0821
- Zhang, Y., Song, G., Lal, N. K., Nagalakshmi, U., Li, Y., Zheng, W., … Dinesh-Kumar, S. P. (2019). TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nature Communications , 10, 3252. doi: 10.1038/s41467-019-11202-z
Internet Resources
REPRINT/CRAPome/SAINTexpress

