single-cell Micro-C protocol
Honggui Wu, Longzhi Tan
Abstract
Here we present a single-cell 3D genome mapping assay with improved spatial resolution with the help of micrococcal nuclease (MNase), termed single-cell Micro-C (scMicro-C). To achieve scMicro-C, we made three improvements. First, we titrated MNase digestion to reduce DNA loss and produce proper DNA fragments. Second, we solubilized chromatin with an ionic detergent, sodium dodecyl sulfate (SDS), which dramatically improved ligation efficiency. Third, we adopted our high-coverage transposon-based whole-genome amplification method, META ( 19 ), using Tn5 ( 20 ), to enhance the detection of chromatin “contacts.” scMicro-C retains the high signal-to-noise and high-resolution characteristics of bulk Micro-C, as indicated the accurate detection chromatin loops and other fine-scale chromatin structures. Furthermore, scMicro-C retains nucleosome and other chromatin-bound proteins (e.g., transcription factor) occupancy information. With our previously developed Dip-C algorithm, we confirmed that scMicro-C enables the reproducible reconstruction of 3D genome structures in single-cell at 5 kb resolution. With such kilobase 3D structures, we were able to explore the fine-scale chromatin folding in individual cells.
Steps
Crosslink Cells for Micro-C
Prepare 1 x BSA in PBS for Washing.
- Add
50µL10% BSA (MACS 130-091-376, final 1x) to50mL1x PBS.
Prepare 1% Paraformaldehyde (1mL for 1 million cells), recover to room temperature.
1mL32% PFA (EMS 15714, final 1%)31mL1x PBS
Prepare 3 mM DSG (1mL for 1 million cells).
- Dissolve 50 mg DSG (Thermo Fisher 20593) with
511µLDMSO to a final concentration of 300 mM. - Add
300µL300 mM DSG to29.7mL1 x PBS to a final concentration of 3 mM, protected from light.
Harvest cells.
- Collect cell growth with appropriate density and high viability.
- Wash cells once with 1x PBS.
Fixation.
- Resuspend cell pellets in 1% PFA, and incubate at room temperature with rotation for 10 min.
- At the end of incubation, add 2 M Tris-HCl ph 7.5 to a final concentration of 0.75 M, and incubate at RT for 5 min.
- Centrifuge at 3000 x g for 5 min in a swing bucket centrifuge.
Remove the supernatant, then wash twice with 5mL ice-cold 1 x BSA in PBS.
Crosslink with DSG.
- Resuspend in 3 mM DSG, and incubate at RT for 45 min with rotation.
- At the end of incubation, add 2 M Tris-HCl ph 7.5 to a final concentration of 0.75 M, and incubate at RT for 5 min.
- Centrifuge at 3000 x g for 5 min in a swing bucket centrifuge.
Count cells and aliquot.
- Remove the supernatant, then wash twice with
5mLice-cold 1 x BSA in PBS. - Count cell number, then aliquot 1 million cells each to 1.5 mL tube.
- Store at -80°C (up to a year).
Prepare Micro-C Buffers
Prepare Micro-C Buffers
10mL Prepare Micro-C Buffer 1 (300µL)
50µL5 M NaCl (Invitrogen AM9760G; final 50 mM)50µL1 M Tris pH 7.5 (Invitrogen 15567027; final 10 mM)25µL1 M MgCl2 (Invitrogen AM9530G; final 5 mM)5µL1 M CaCl2 (Sigma 21115; final 1 mM)50µL5% Digitonin (Sigma D141-100MG, final: 0.05%)50µL100 x protease inhibitor (Roche 4693159001) (final: 1 X)4.77mLnuclease-free H2O
Prepare Micro-C Buffer 2 (10mL)
-
100µL5 M NaCl (Invitrogen AM9760G; final 50 mM) -
100µL1 M Tris pH 7.5 (Invitrogen 15567027; final 10 mM) -
100µL1 M MgCl2 (Invitrogen AM9530G; final 10 mM) -
9.7mLnuclease-free H2O
Prepare Micro-C Buffer 3 (10mL)
500µL1 M Tris pH 7.5 (Invitrogen 15567027; final 50 mM)100µL1 M MgCl2 (Invitrogen AM9530G; final 10 mM)9.4mLnuclease-free H2O
MNase Digestion
Dilute MNase.
- Make MNase Dilution Buffer (10 mM Tris-HCl ph 7.5, 50 mM NaCl, 1 mM EDTA, 50% Glycerol).
- Aliquot 10,000 Unit MNase (NEB M0247S, 2,000 U/µL) to
500µLMNase Dilution Buffer to a final concentration of 20 U/µL. - Store at -80°C for several month.
Thaw 5 tubes of cell pellets on ice.
Resuspend in 100µL Micro-C Buffer 1, incubate on ice for 20 min, and aspiration every 5 min.
Centrifuge at 800 x g at 4°C for 5 min.
Take 5 ul to check digestion efficiency, remaining centrifuge at 800 x g at 4°C for 5 min. Carefully discard the supernatant, and wash once with 500µLMicro-C Buffer 2. Store at -80°C (stable for up to several weeks) or proceed to End Repair.
Check Digestion Efficiency
- Take
5µLabove sample to check digestion efficiency. - Add 85 ul 1 x PBS, 5 ul 10% SDS (Sigma, 71736-100ML), and 5 ul 20 mg/mL ProteaseK (QIAGEN, 19131).
- incubate at 65°C for 2 hr.
- Purify with DNA clean column (ZYMO, D4014).
- Run capillary electrophoresis (Fragment Analyzer/TepeStation/Bioanalyzer) to analyze digestion efficiency.
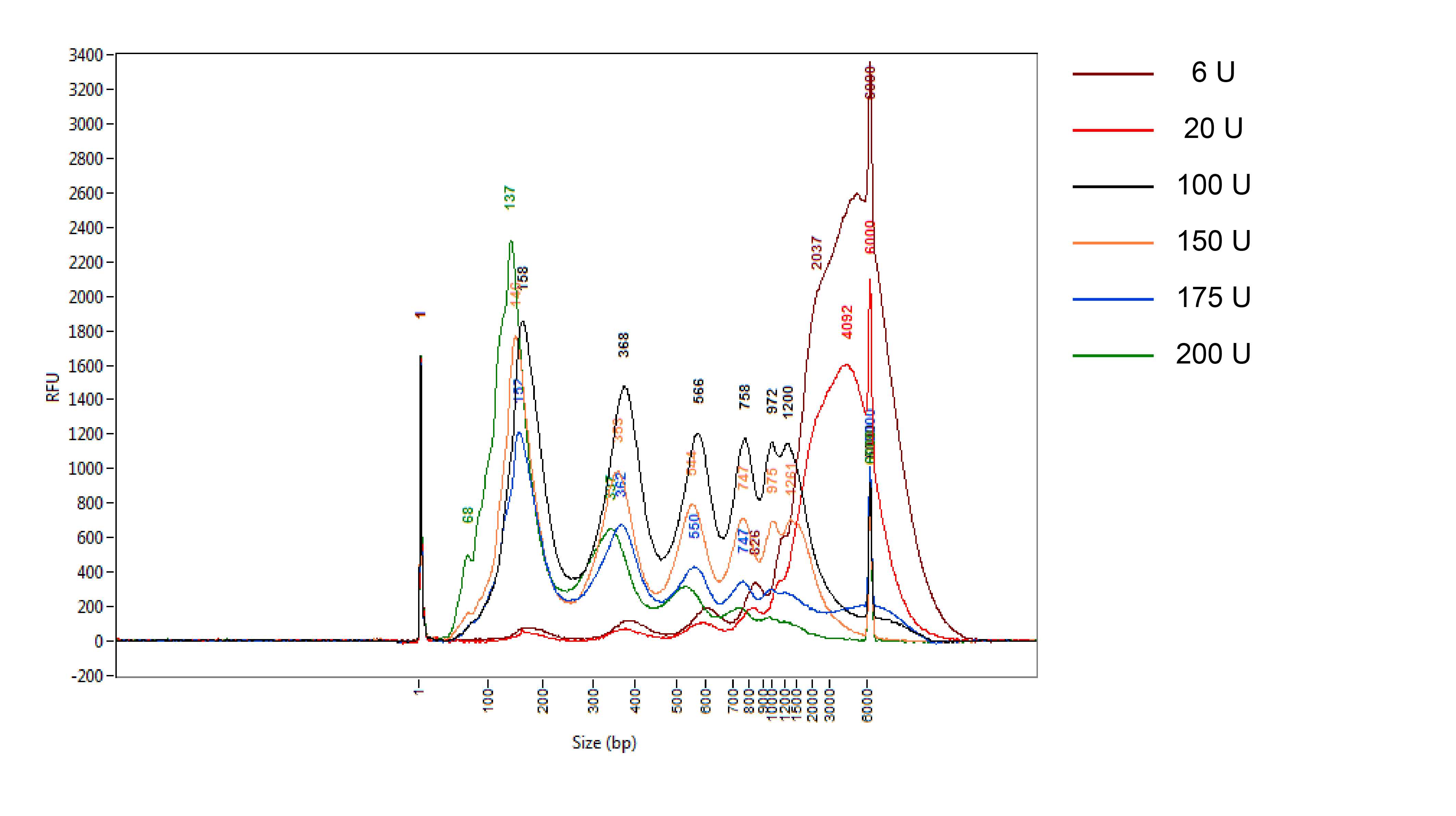
An exemplified MNase titration experiment: fragment size distribution with different MNase concentrations.
End Repair
Thaw the cell pellet (with the most appropriate fragment size) on ice.
Prepare 0.5% SDS.
- Add
25µL10% SDS to475µLMicro-C Buffer 2 to a final concentration of 0.5%.
Solubilize Chromatin.
- Resuspend the cell pellets in
50µL0.5% SDS. - Incubate at 62°C for 10 min.
- At the end of incubation, add
50µL5% Triton X-100 to quench SDS. Incubate at 37°C for 15 min.
Centrifuge at 800 x g for 5 min, carefully remove supernatant.
Prepare End Repair Mix 1 (45µL each sample).
5µL10 X NEBuffer 2.1 (NEB B7202S)1µL100 mM ATP (Thermo R0441)2.5µL100 mM DTT (final: ~5 mM)2.5µL10 U/uL T4 PNK (NEB M0201S) (final: ~0.5 U/uL)34µLnuclease-free H2O
Resuspend in 45µL End Repair Mix1, and incubate at 37°C for 15 min.
At the end of incubation, add 5µL 5 U/uL Klenow Fragment (NEB M0210S), incubate at 37°C for another 15 min.
Prepare End Repair Mix2 (25µLeach sample)
2.5µL10 X T4 DNA Ligase Buffer (NEB B0202S)0.5µL10 mM (each) dNTP (NEB N0447S)0.125µL20 mg/mL BSA (NEB B9000S)21.875µLnuclease-free H2O
After Klenow incubation, add 25µL End Repair Mix2 to each tube, incubate at 23°C for 45 min (shake 30 s and pause 2 min).
Add 5 ul 0.5 M EDTA (Invitrogen AM9260G).
Incubate at 65°C for 20 min.
Wash once with 1mL Micro-C Buffer 3.
Proximity Ligation
Prepare Ligation Mix (250µL each sample).
25µL10 X T4 DNA Ligase Buffer (NEB B0202S)1.25µL20 mg/mL BSA (NEB B9000S)12.5µL400 U/uL T4 DNA Ligase (NEB M0202S)211.25µLnuclease-free H2O
Resuspend the cell pellet in 250µL Ligation Mix.
Take 25µL as ligation efficiency control.
Check Ligation Efficiency
-
Take
25µLligation product above sample to check ligation efficiency. -
Add 65 ul 1 x PBS, 5 ul 10% SDS (Sigma, 71736-100ML), and 5 ul 20 mg/mL ProteaseK (QIAGEN, 19131).
-
incubate at 65°C for 2 hr.
-
Purify with DNA clean column (ZYMO, D4014).
-
Run capillary electrophoresis (Fragment Analyzer/TepeStation/Bioanalyzer) to analyze ligation efficiency.
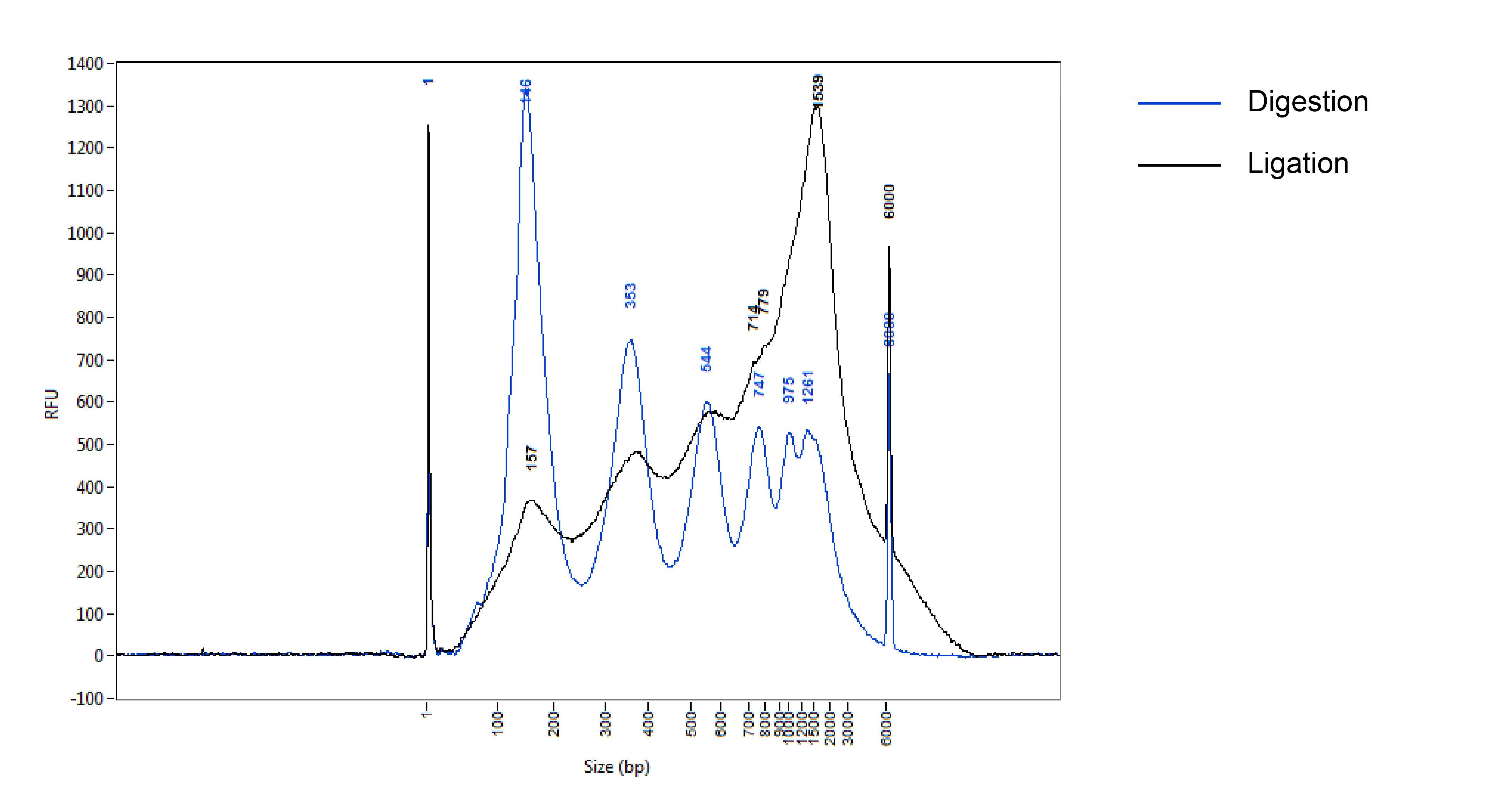
A typical fragment size distribution after digestion and ligation.
Single-cell Amplification
Primers
- META 16 sequence
| A | B |
|---|---|
| META-16-1 | GGCACCG AAAA |
| META-16-2 | CTCGGCGA TAAA |
| META-16-3 | GGTGGAGC ATAA |
| META-16-4 | CGAGCGCA TTAA |
| META-16-5 | AGCCCGGT TATA |
| META-16-6 | TCGGCACC AATA |
| META-16-7 | GCCTGTGG ATTA |
| META-16-8 | GCGACCCT TTTA |
| META-16-9 | GCATGCGG TAAT |
| META-16-10 | GCGTTGCC ATAT |
| META-16-11 | GGCCGCAT TTAT |
| META-16-12 | ACCGCCTC TATT |
| META-16-13 | CCGTGCCA AAAT |
| META-16-14 | TCTCCGGG AATT |
| META-16-15 | CCGCGCTT ATTT |
| META-16-16 | CTGAGCTCG TTTT |
-
META Transposon | A | B | | --- | --- | | META Transposon | 5'-[META sequence]-AGATGTGTATAAGAGACAG-3' | | 19 bp ME | 5'-/phos/-CTGTCTCTTATACACATCT-3' |
-
First Step PCR Primers
| A | B |
|---|---|
| First PCR primers | 5'-[META sequence]-AGATGTGTATAAG-3' |
Here we use 12 x 8 barcode combinations to distinguish individual cells.
-
META16-ADP1 cell barcode | A | B | | --- | --- | | 1-1 | GATATG | | 1-2 | ATACG | | 1-3 | CCGTCTG | | 1-4 | TGCG | | 1-5 | GAACTCG | | 1-6 | ATGTAG | | 1-7 | CCCG | | 1-8 | TATGT | | 1-9 | GAGTAAG | | 1-10 | ATCG | | 1-11 | CCTAG | | 1-12 | TGACCG |
-
META16-ADP2 cell barcode
| A | B |
|---|---|
| 2-1 | ACTCTA |
| 2-2 | AGAGCAT |
| 2-3 | GGTATG |
| 2-4 | TCGATGC |
| 2-5 | CTACTAG |
| 2-6 | TATGCA |
| 2-7 | CACACGA |
| 2-8 | GTCGAT |
Second Step Barcoded PCR Primers
| A | B |
|---|---|
| META16-ADP1 | 5'-CTTTCCCTACACGACGCTCTTCCGATCT-[cell barcode]-[META sequence]-AGATGTGTATAAG-3' |
| META16-ADP2 | 5'-GAGTTCAGACGTGTGCTCTTCCGATCT-[cell barcode]-[META sequence]-AGATGTGTATAAG-3' |
Prepare Reagents.
Prepare 60 mg/mL Protease.
- Dissolve 1 vial QIAGEN protease (QIAGEN 19155) in 2.78 mL 50% glycerol.
- Aliquot and store at -80°C (stable for up half a year).
Cell Triton Lysis Buffer (1mL).
10µL1 M Tris pH 8.0 (Invitrogen AM9855G, final 10 mM)4µL5 M NaCl (Invitrogen AM9760G , final 20 mM)2µL0.5 M EDTA (Invitrogen AM9260G, final 1 mM)10µL10% Triton X-100 (Sigma 93443, final 0.1%)25µL1 M DTT (Sigma 646563, final 25 mM)5µL100 µM Carrier ssDNA (final 100 nM)- Add H2O to 1 mL
25µL60 mg/mL QIAGEN protease (QIAGEN 19155, final 1.5 mg/mL) (freshly add when used)
| A | B |
|---|---|
| Carrier ssDNA | TCAGGTTTTCCTGAA |
2 X Transposition Buffer (1mL)
20µL1 M Tris-HCl pH 8.0 (Invitrogen AM9855G, final 20 mM)10µL1 M MgCl2 (Thermo Fisher AM9530G, final 10 mM)- 400 ul 40% (w/w) Polyethylene glycol 8000 (Sigma P1458, final 16% (w/w) )
570µLnuclease-free H2O Store at -20℃ if needed
Pick single cells.
Use mouth pipette or Fluorescence-activated cell sorting (FACS) to pick single cell into single PCR tubes or 96-well plates containing 2µL cell lysis buffer.
Cell lysis.
Incubate as:
50℃, 1 hr
65℃, 1 hr
70℃, 15 min
Stored at -80℃ or proceed to transposon-based amplification.
Transposition (8µLfor each well, below recipe for 1 96-well plate).
- Prepare Transposition Mix
440µL2 X Transposition Buffer- Add META transposome* to a final concentration of 0.3 nM
- Add nuclease-free H2O to
880µL - Transposome: META transposome is optional. Alternatively, Nextera transposome can be used (if using Nextera procedure, please refer to "Dip-C (Part 2: Whole-genome Amplification with Nextera)") on protocol.io.
-
Add
8µLTransposition Mix to each well with a multi-channel pipette, vortex to mix. -
Incubate at 55℃ for 10 min.
Tn5 Stop.
-
Prepare Tn5 Removal Mix (250 mM NaCl, 37.5 mM EDTA, 2 mg/mL QIAGEN protease).
-
Add
2µLTn5 Removal Mix to each tube, and mix thoroughly. -
Incubate as: 50℃, 30 min
70℃, 15 min
Pre-amplification.
Prepare Preamplification Mix (16µL each well, below recipe for 1 96-well plate)
1540µL2 X Q5 High-fidelity Master Mix (NEB M0492L)98.6µL50 µM First Step PCR Primers (final 0.1 µM each)55µL100 mM MgCl266.4µLnuclease-free H2O
Add 16µL Preamplification Mix each well with a multi-channel pipette, and vortex to mix.
Amplified as:
72℃, 5 min
98℃, 30 s
12 cycles of [98℃ for 10 s, 62℃ for 30 s, 72℃, 2 min]
65℃, 5 min.
Purification.
- Pool a 96-well plate to purify.
- Purify with clean column (ZYMO, D4014).
- Quantify with Qubit Fluorometer.
Library preparation.
-
Take 400 ng amplified product for each plate as input.
-
Prepare Library Preparation Mix (
50µL2 X Q5 High-fidelity Mater Mix, template (400 ng),5µL10 µM Indexed i5 primer,5µL10 µM Indexed i7 primer (Vazyeme N321/N322), nuclease-free H2O to100µL) -
Amplified as: 98℃, 30 s
2 cycles of [98℃, 10 s, 68℃, 30 s, 72℃, 2 min] 72℃, 5 min -
Purify with PCR clean column (ZYMO D4014), then purify with 0.75 x AMPUre XP beads to select > 300 bp fragments for sequencing.
Sequencing.
META library only has 20% valid fragments having both P7 and P5 on either side, need to RUN qPCR quantify before loading.