Workflow for human fallopian tube and uterine endomyometrium bulk ATACseq
Scott Lindsay-Hewett, Valentina Stanley, Louise Laurent, Mana Parast
Abstract
Described here is the workflow used by the Female Reproductive Tissue Mapping Center at UCSD to generate bulk ATACseq data from human fallopian tube and uterine endomyometrium.
Steps
Tissue preparation
As soon as possible after sterilization (salpingectomy or tubal ligation), prepare fallopian tube tissue according to the following protocol:
At the time of C-section, prepare uterine endomyometrium tissue according to the following protocol:
For this protocol, use tissue that has been snap-frozen.
Nuclei isolation
Isolate nuclei from 4 samples at a time according to the following protocol:
Nuclei isolation from snap-frozen human placental tissue for bulk ATACseq
Proceed to tagmentation immediately.
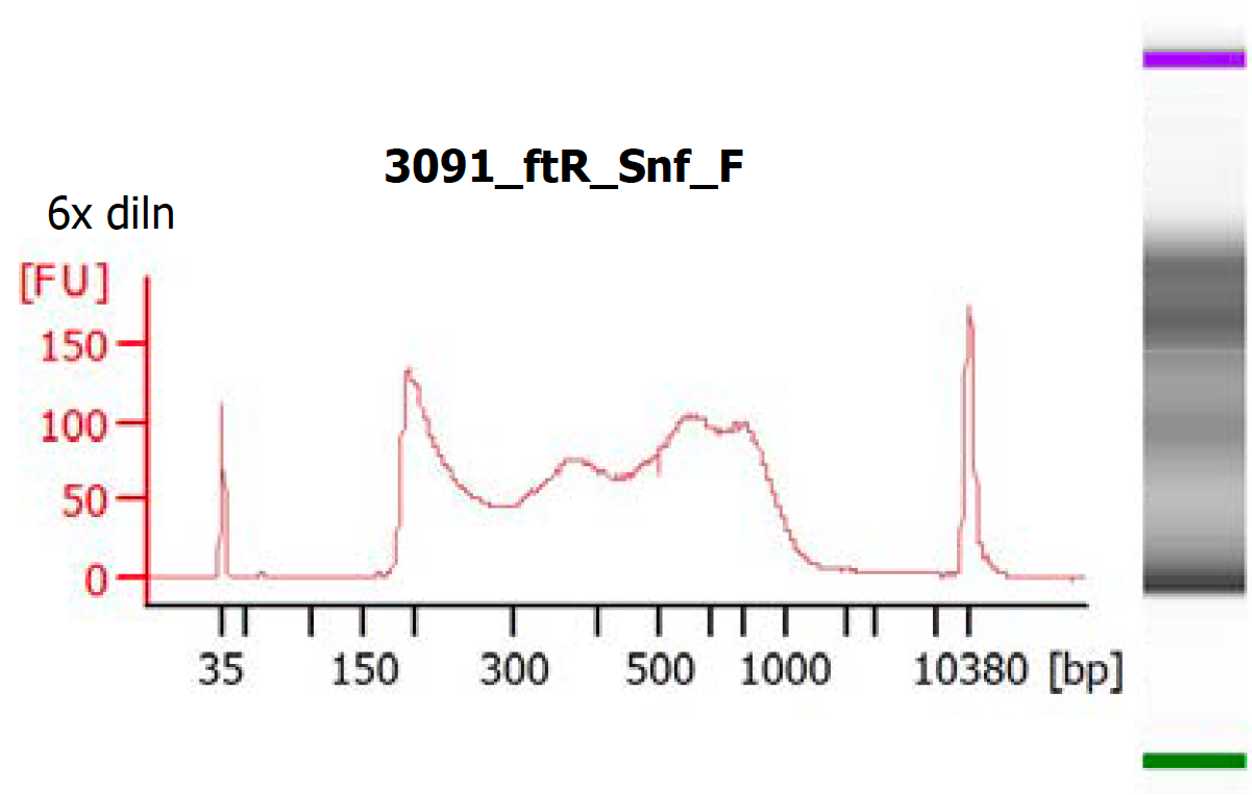
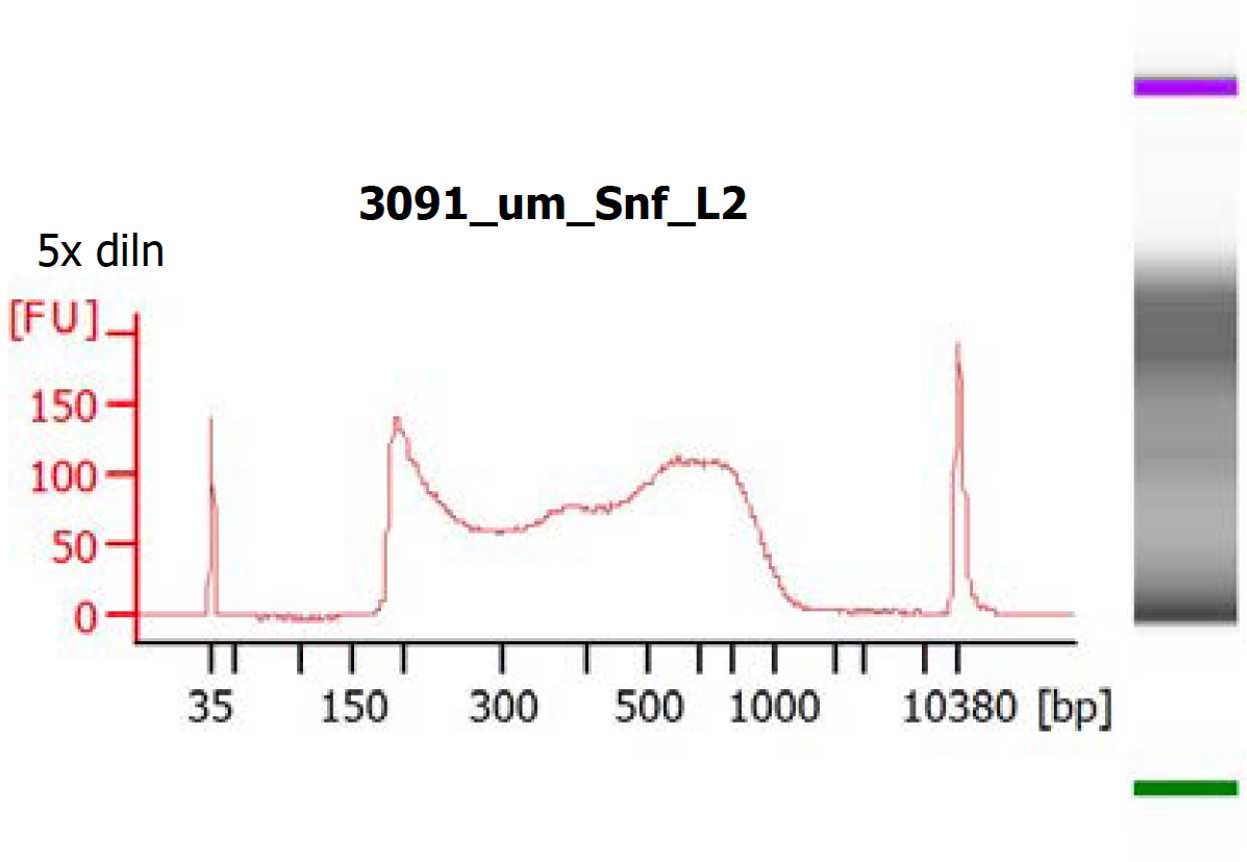
Tagmentation and library generation
Perform tagmentation and library generation according to the following protocol:
Tagmentation and library generation for human placental bulk ATACseq
After passing quality control, proceed to sequencing.
Sequencing
For HuBMAP bulk ATACseq samples, the multiplexed pool was sequenced on a NovaSeq 6000 S4 lane using a 100bp paired-end run configuration. Alignment and peak-calling were performed using the ATAC-seq pipeline within the bcbio Python package.

