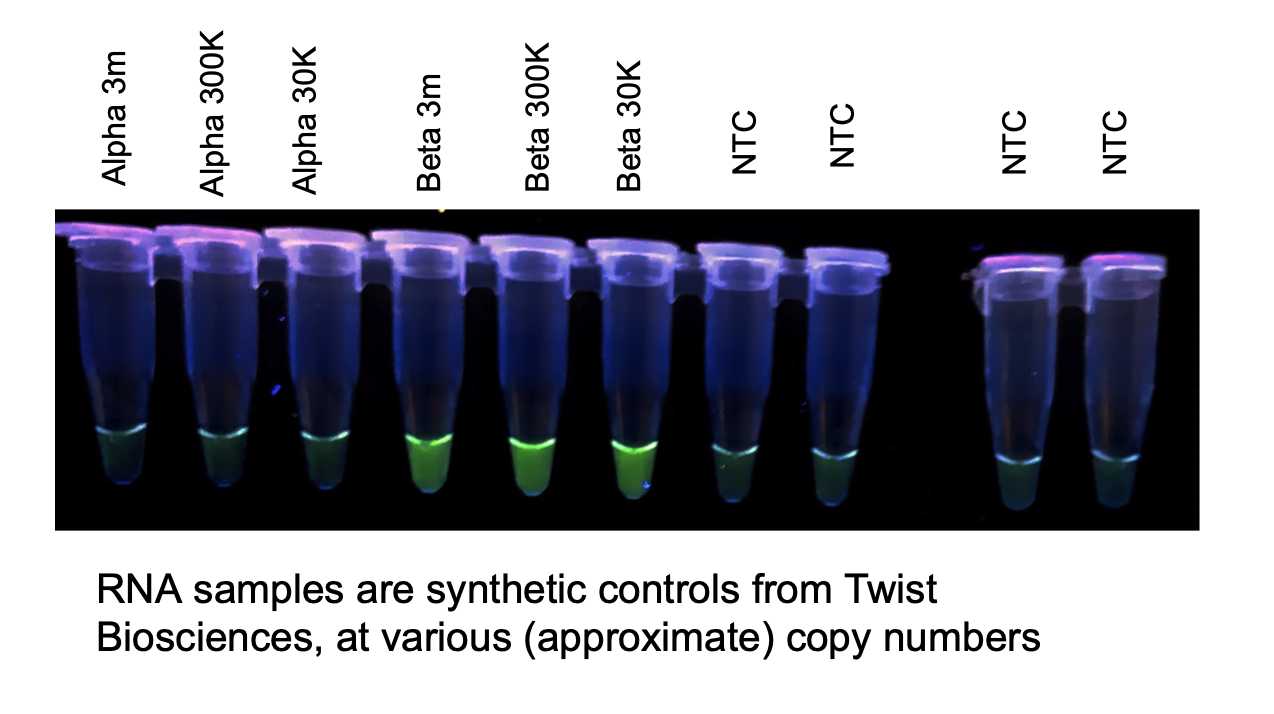
Variant (E484K) ALERT - Ligation-Dependent Loop-Mediated Isothermal Amplification
Ali Bektas, Anitha Jayaprakash
Abstract
A 2-hour, 2-temperature protocol, using RNA templated DNA ligation, for the visual detection of the E484K mutation of concern pertaining to SARS-CoV-2.
N.B. This is the first version of this protocol, stay tuned for increased sensitivity and multiplexing (A.B.)
Steps
Ligation Primers
Oligonucleotides used for RNA templated DNA ligation using
All sequences shown 5' to 3. Underlined segments are complementary to the target and bold nucleotide is specific to the G>A substitution at position 23012 resulting in the E484K amino acid change.
E484K-Donor
/5Phos/ AACACCATTACAAG CTACGTACATGACATAATCCAAACTCATAAATTCTCGCATTTTAGATCCGTCTCCTTACAGGACACATCATCC
E484-Acceptor
CATCTCTAACTCTACTAAGACTTCCATTTATCAACAATAGCTGACATGTTCTAGCAGCGACAGGACACACGACAGG AGGAAAGTAACAATTAAAACCTTT T
Ligation
Prepare ligation mix to use with previously isolated RNA. A standard Trizol/Chloroform method or column purification method for RNA isolation works well.
X1 Ligation mix (multiply for number of samples processed)
2µL E484K-Donor (100nM)
2µL E484K-Acceptor (100nM)
1µLSplintR Buffer (NEB)
0.2µL
1.8µL molecular biology grade water
- add
3µLRNA per7µLof reaction mix for a10µLreaction volume
Incubate at 37°C for 0h 20m 0s followed by a 0h 20m 0s at 65°C
inactivation step.
Place ligation reactions on ice.
LAMP Primers
Oligonucleotides used for Loop-Mediated Isothermal Amplification
all sequences shown 5' to 3'
FIP
CTTTCCTCCTGTCGTGTGTCCTCCATTTATCAACAATAGCTGAC
BIP
ACCTTTAACACCATTACAAGCTACGACGGATCTAAAATGCGAGAA
F3
CATCTCTAACTCTACTAAGACT
B3
GGATGATGTGTCCTGTAAGG
LAMP
Prepare LAMP mix.
X1 LAMP mix (multiply for number of samples processed)
2.5µL Isothermal Buffer 1 (NEB)
0.5µL
3.5µL dNTPs (10µM)
0.8µL FIP (50µM)
0.8µL BIP (50µM)
0.5µL F3 (10µM)
0.5µL B3 (10µM)
0.1µL
1µL
11.8µLMolecular Biology Grade water
- add
3µLof the ligation product from the Ligation step to22µLof LAMP mix for a25µLreaction.
Incubate at 65°C for 0h 50m 0s