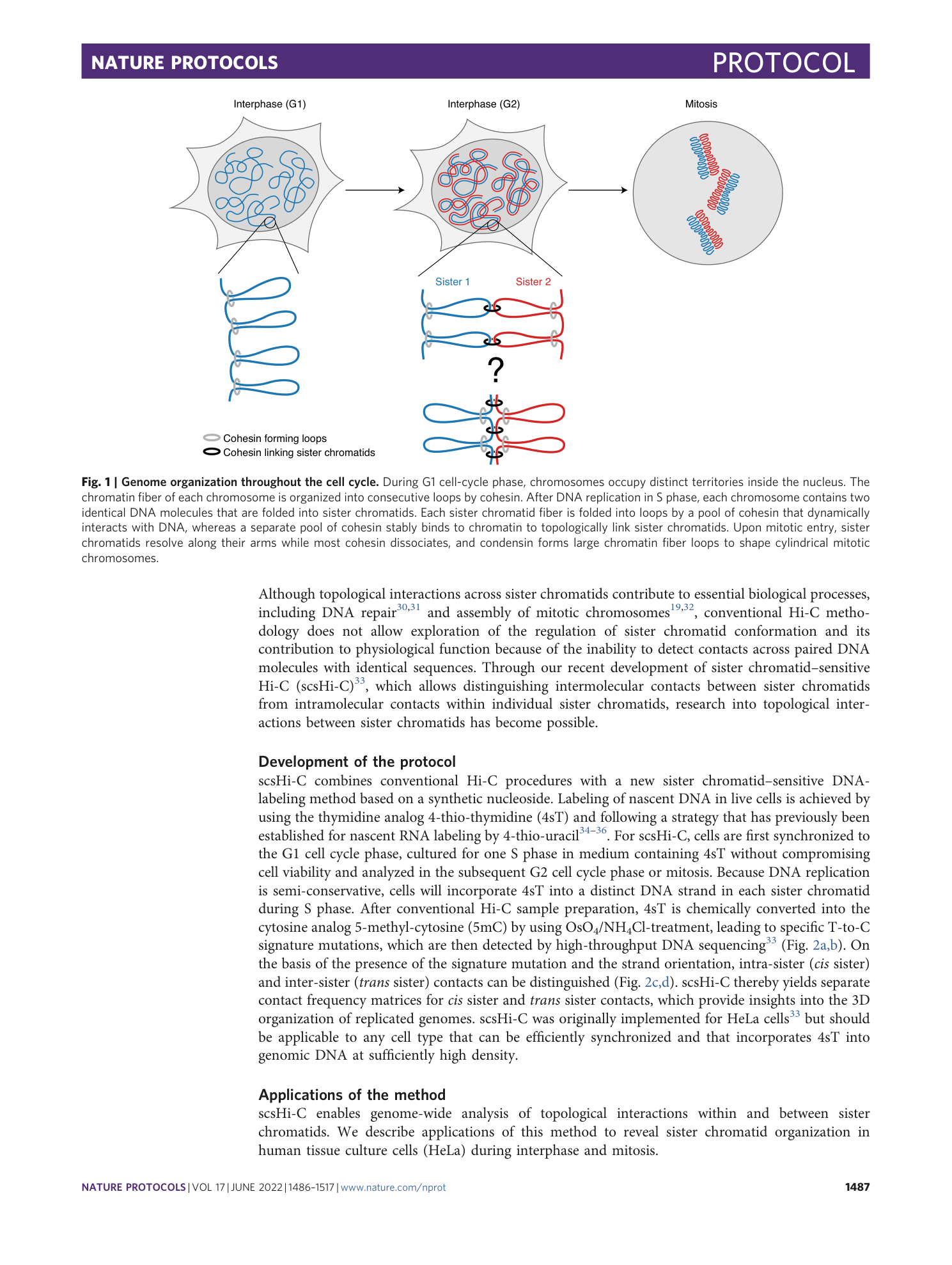
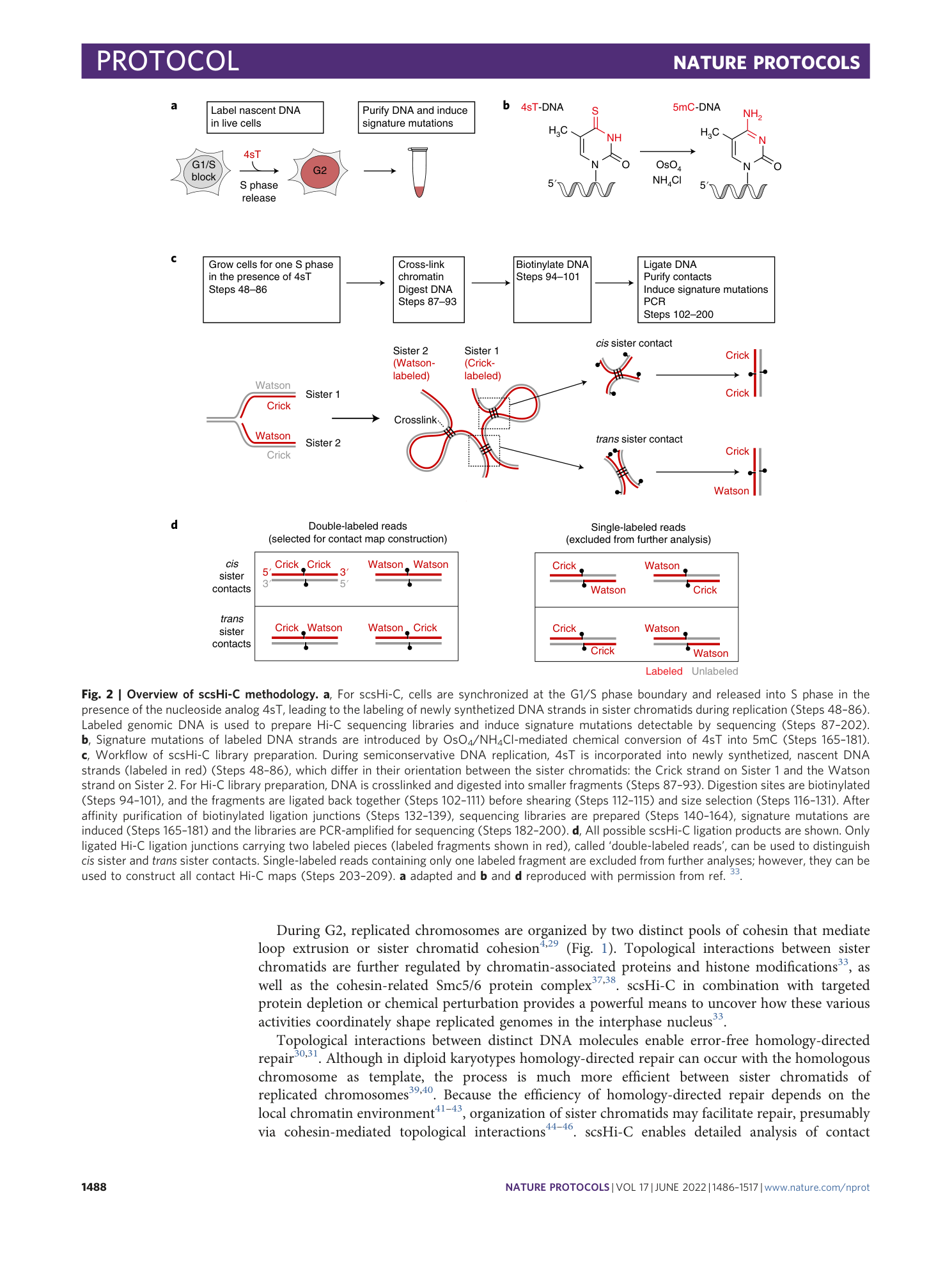
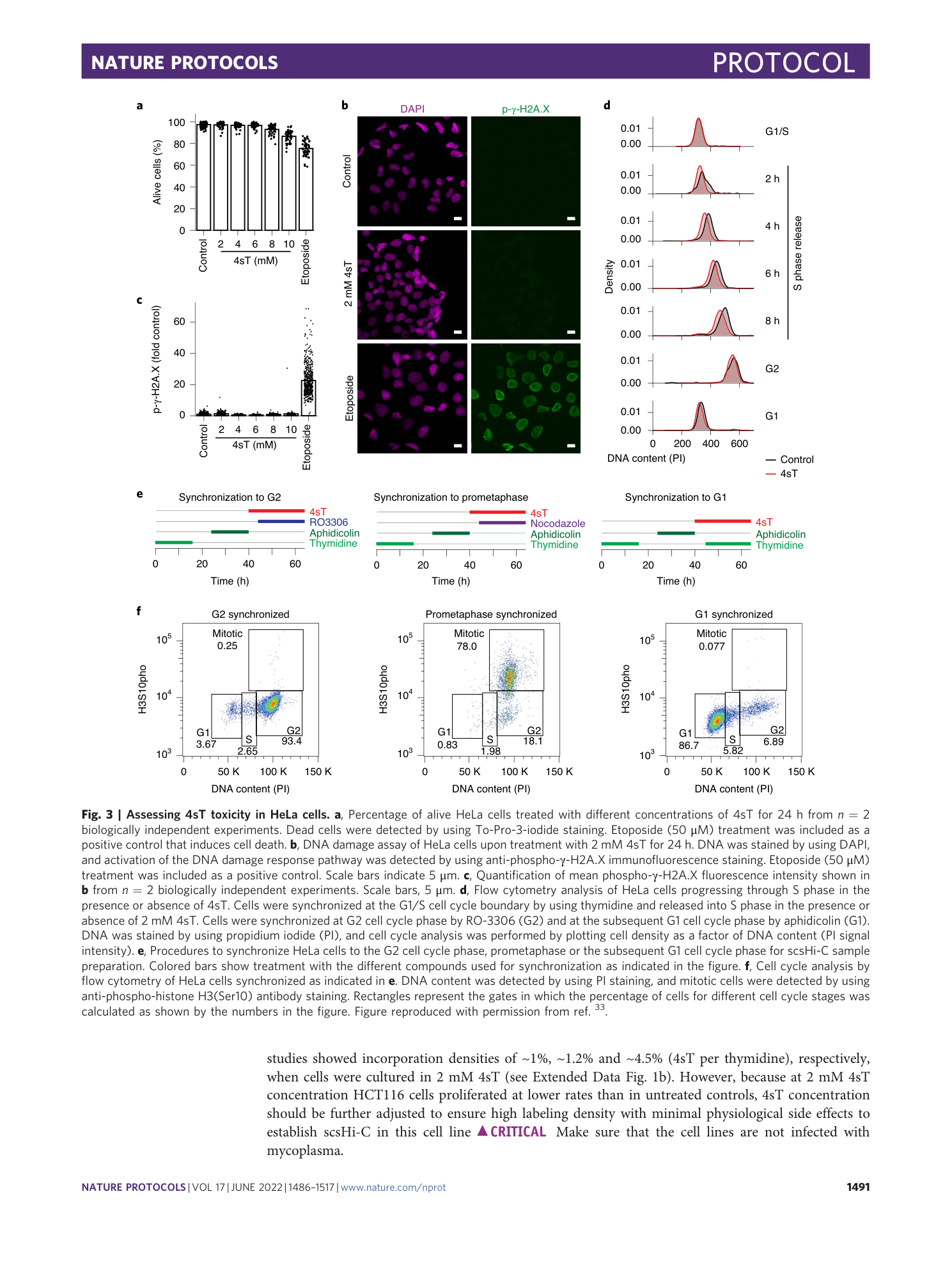
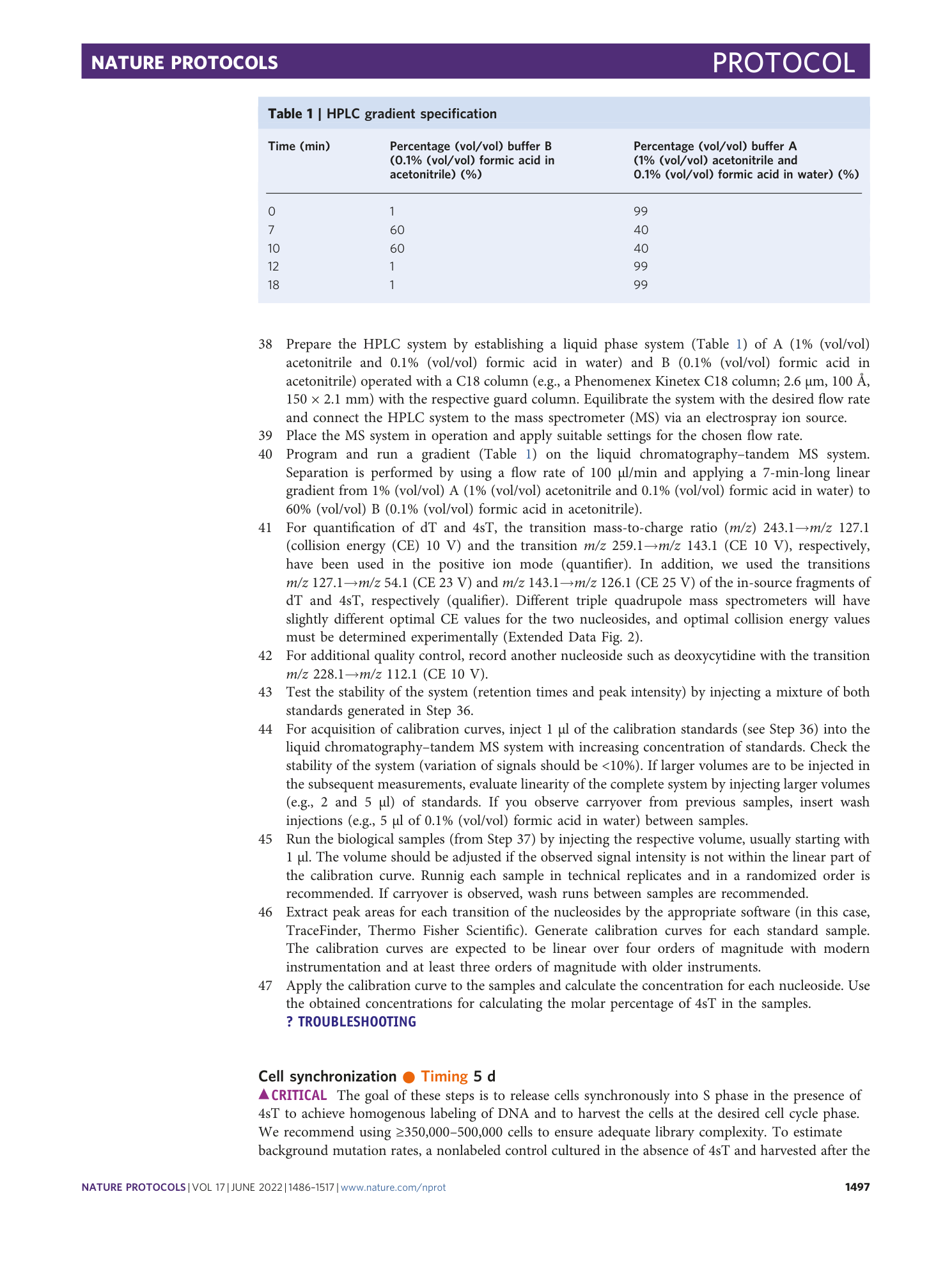
Sister chromatid–sensitive Hi-C to map the conformation of replicated genomes
Michael Mitter, Zsuzsanna Takacs, Thomas Köcher, Ronald Micura, Christoph C. H. Langer, Daniel W. Gerlich

























Extended
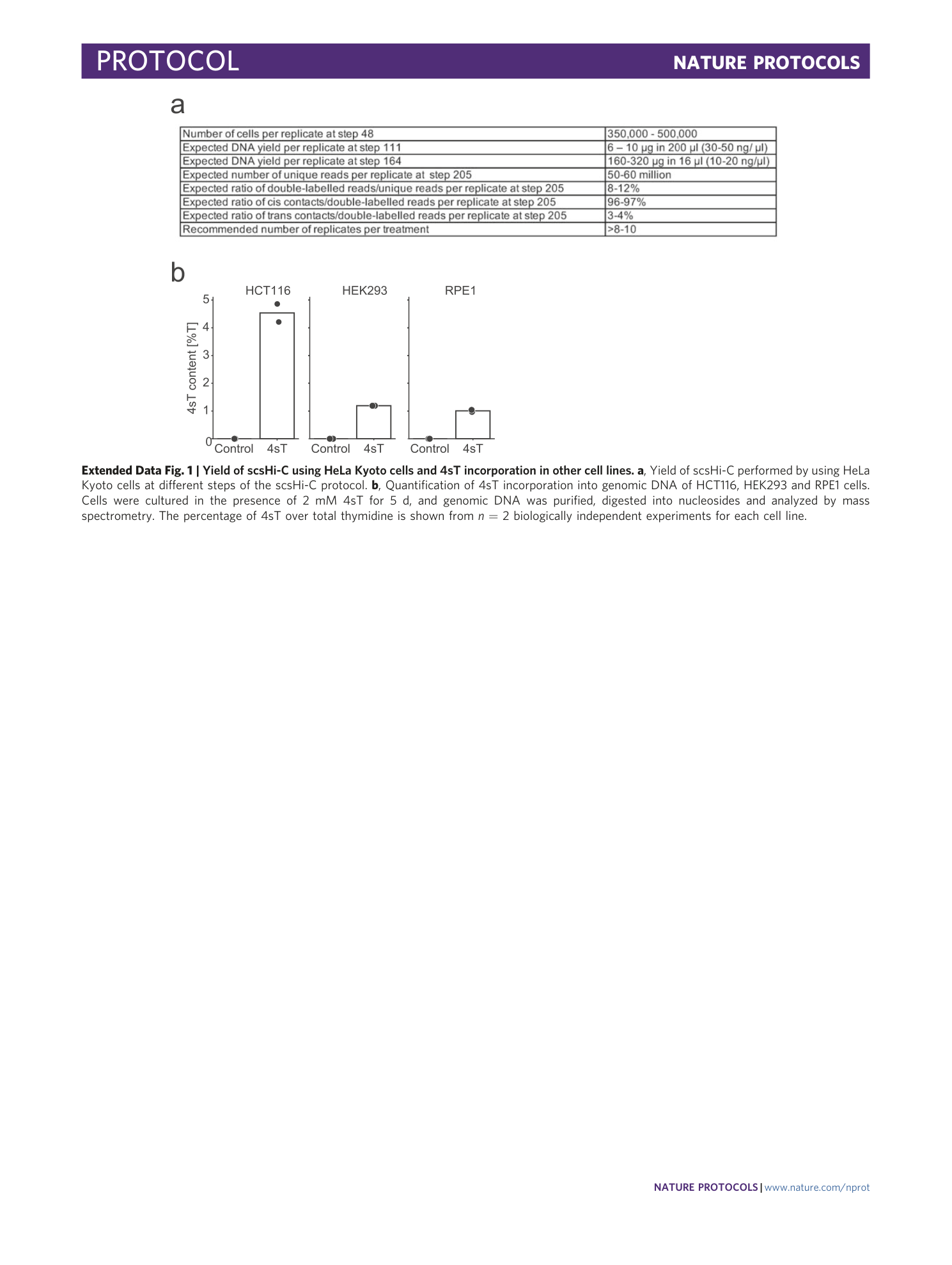
Extended Data Fig. 1 Yield of scsHi-C using HeLa Kyoto cells and 4sT incorporation in other cell lines.
a , Yield of scsHi-C performed by using HeLa Kyoto cells at different steps of the scsHi-C protocol. b , Quantification of 4sT incorporation into genomic DNA of HCT116, HEK293 and RPE1 cells. Cells were cultured in the presence of 2 mM 4sT for 5 d, and genomic DNA was purified, digested into nucleosides and analyzed by mass spectrometry. The percentage of 4sT over total thymidine is shown from n = 2 biologically independent experiments for each cell line.
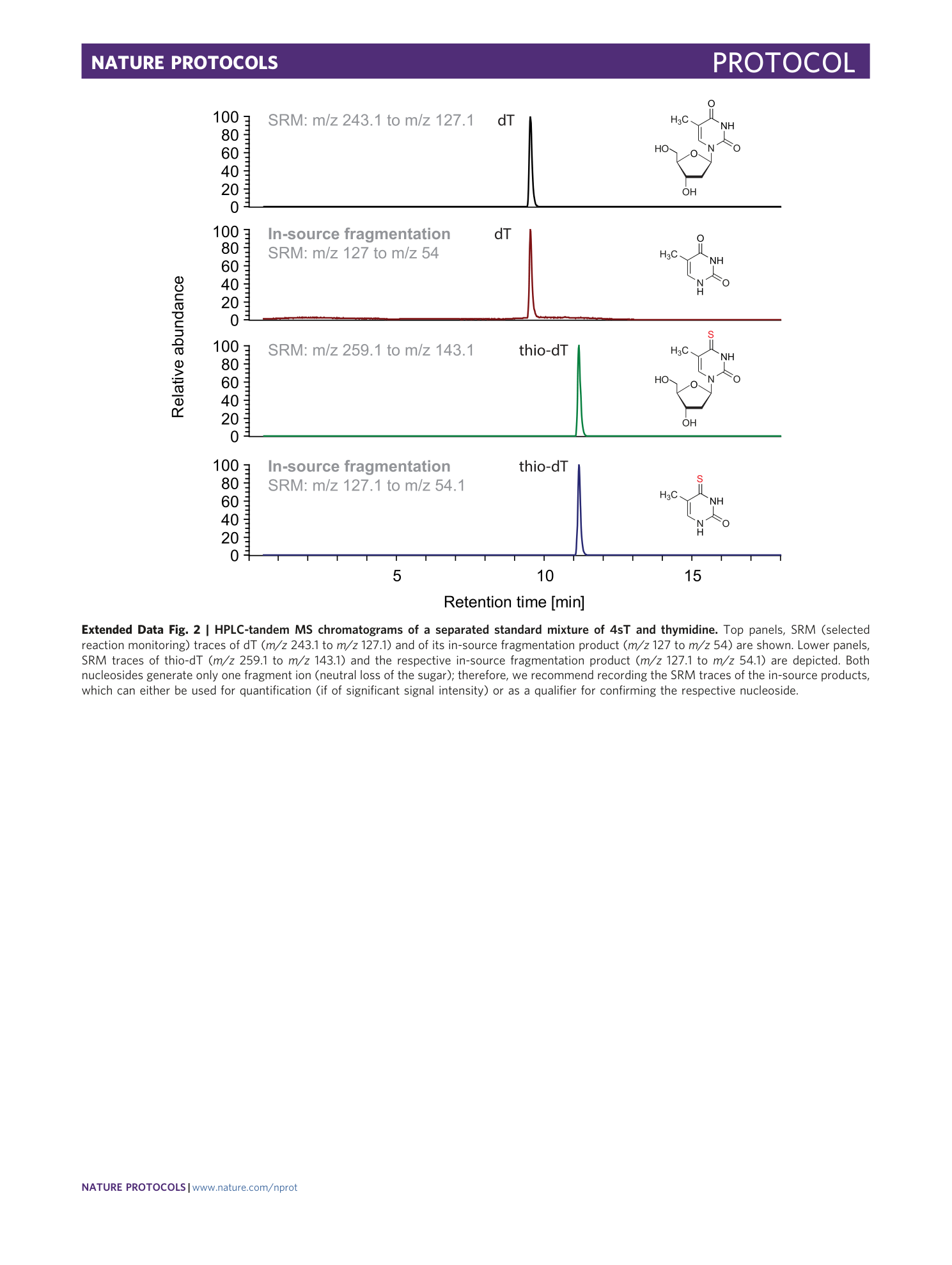
Extended Data Fig. 2 HPLC-tandem MS chromatograms of a separated standard mixture of 4sT and thymidine.
Top panels, SRM (selected reaction monitoring) traces of dT ( m/z 243.1 to m/z 127.1) and of its in-source fragmentation product ( m/z 127 to m/z 54) are shown. Lower panels, SRM traces of thio-dT ( m/z 259.1 to m/z 143.1) and the respective in-source fragmentation product ( m/z 127.1 to m/z 54.1) are depicted. Both nucleosides generate only one fragment ion (neutral loss of the sugar); therefore, we recommend recording the SRM traces of the in-source products, which can either be used for quantification (if of significant signal intensity) or as a qualifier for confirming the respective nucleoside.

