SARS-CoV-2 DNA library preparation using an adapted version of Illumina DNA prep protocol v.1.1
Emmanuel Kagning Tsinda, Masahiro Sakamoto, Michiko Okamoto, Clyde Dapat, Mariko Saito, Mayuko Saito, Hitoshi Oshitani
Disclaimer
Funder had no role in the conception and design of this protocol.
Abstract
This protocol describes the steps to prepare DNA libraries from PCR amplicons using the Illumina DNA prep library kit. The current library preparation protocol was adapted from the original Illumina DNA Prep Reference Guide (document #1000000025416 v09) using low input DNA samples as the starting material.
We omitted laboratory equipment such as the Eppendorf 96-well PCR plate , microseal adhesive seals. In addition, we replaced 96-well plate magnetic stand with a magnetic stand suitable for 1.5 ml tubes .
The added value of this protocol is that PCR reactions happen in 0.2 ml PCR tubes, and it can be implemented without a separate purchase of 96-well PCR plates or a magnetic stand for PCR plates. Using individual 0.2 ml tubes increases the user flexibility when running few samples for library preparation. The other advantage of using our adapted protocol is the reduced library preparation cost when running few specimens. For instance, implementing the current protocol might be cheaper than the original Illumina protocol when using the Illumina® DNA Prep, (M) Tagmentation (24 Samples) catalog number 20018704.
The protocol has proven effective for processing hundreds of DNA libraries from tiled virus amplicons such as Sapovirus and SARS-CoV-2 submitted to public repositories such as GISAID and GenBank.
Before start
Please follow steps 1 to 25 of the protocol below ARTIC protocol below.COVID-19 ARTIC v3 Illumina library construction and sequencing protocol
- After the multiplex PCR using the ARTIC protocol (https://www.protocols.io/view/covid-19-artic-v3-illumina-library-construction-an-bgxjjxkn), please pool P1 and P2 amplicons into a single 1.5 ml tube; then clean-up the amplicons using magnetic beads following the manufacturer's protocol.
- Quantify the cleaned DNA product Qubit DNA High-sensitivity kit (important).
- Dilute the genomic DNA of each sample to 1ng/ul in 30 µl of RNAse/DNAse free water.
Steps
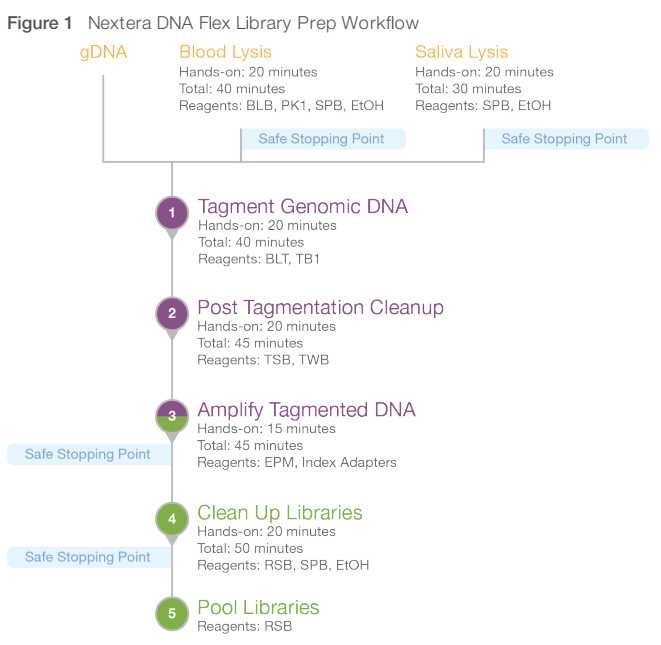
Overview of the library prep workflow
DNA tagmentation
This step uses the Nextera Transposome to tagment genomic DNA. Tagmentation is a process that fragments DNA and then tags the fragmented DNA with adapter sequences in a single reaction.
Thaw EPM and indexes on ice.
Bring the following reagents at room temperature: BLT (beads linked transposomes), TB1(Tagmentation buffer), TSB (Tagment Stop buffer), and TWB (Tagment Wash Buffer).
Vortex BLT and TB1 to mix before use.
Pre-heat the lid of the Thermocycler (by Running the " TAG " program, and press "Pause")
The TAG program is set as follows:
- Set the reaction volume to
50µL 55°Cfor0h 15m 0s15 min- Hold at
10°C
Vortex BLT vigorously for 0h 0m 10s seconds to resuspend. Do not centrifuge BLT.
Prepare Tagmentation master mix by adding the following to a separate tube for the master mix.
- BLT:
11µL - TB1:
11µLMultiply the volume of BLT and TB1 by the number of samples.
Vortex tagmentation master mix thoroughly to resuspend BLT in tagmentation buffer.
Distribute 20µL into each 0.2 tubes containing 30µL of DNA samples (1ng/μl ) and gently pipette 10 times to resuspend. Gentle pipetting prevents liquid drops splits on the walls of tubes. Do not spin down.
Close caps and place tubes into the pre-heated thermocycler and resume the " TAG " program.
Immediately proceed to the next step --> Post-Tagmentation cleanup.
Post tagmentation cleanup
This step washes the adapter-tagged DNA on the BLT before PCR amplification.
Prepare the following reagents:
-Tagment Stop Buffer (TSB): if precipitates are observed, heat at 37°C for 10 min (using a heat-block) and vortex to dissolve the precipitate.
-Tagment Wash Buffer (TWB)
-DynaMag magnetic stand
-0.2 ml PCR tubes
Run the PTC program below on the thermocycler and press "pause".
- Choose the preheat lid option and set it to
100°C - Set the reaction volume to 60 μl
37°Cfor0h 15m 0smin- Hold at
10°C
Add 10µL of TSB to the tagmentation reaction and slowly pipette each well 10 times to resuspend the beads.
Place tubes in the pre-programmed thermal cycler, and run the "PTC" program.
Transfer content of each PCR tube (60µL ) to a 1.5 ml low adhesion microcentrifuge tube.
Equipment
| Value | Label |
|---|---|
| 1.5 ml graduated microcentrifuge tube Flat Top Cap | NAME |
| Tube | TYPE |
| Quality Scientific Plastics | BRAND |
| 509-GRD-Q | SKU |
| https://www.fishersci.co.uk/gb/en/brands/IG2AR7AN/mbp-molecular-bioproducts.html | LINK |
| certified RNase and DNase free | SPECIFICATIONS |
Place tubes on the magnetic stand and wait until liquid is clear (~3 minutes).
Using a P100 pipette, carefully remove and discard supernatant.
Washing steps:
Remove from the magnetic stand and use a deliberately slow pipetting technique to add 100 µl TWB directly onto the beads. The deliberately slow pipetting technique minimizes the potential of TWB foaming to avoid incorrect volume aspiration and incomplete mixing.
Mix at 1400 rpm for 2 min at room temperature (or pipette mix slowly 10 times until beads are re-suspended).
Place the tubes on the magnetic stand and wait until the liquid is clear (~3 minutes).
Using a P200 pipette, remove and discard the supernatant.
Repeat the washing steps 15.1 to 15.3 above.
Keep tubes on the magnetic stand until step 22 of the section Amplify Tagmented DNA section Amplify Tagmented DNA section below. The TWB remains in the tube to prevent overdrying of the beads.
Amplify tagmented DNA
This step amplifies the tagmented DNA through a limited-cycle PCR program. The PCR step adds Index 1 (i7) adapters, Index 2 (i5) adapters, and sequences required for sequencing cluster generation.
Fill the table below including index information.
| A | B | C | D | E | F | G | H | I |
|---|---|---|---|---|---|---|---|---|
| H503 | H505 | H506 | H517 | |||||
| H705 | sample 1 | sample 2 | sample 3 | |||||
| H706 | ||||||||
| H707 | ||||||||
| H710 | ||||||||
| H711 | ||||||||
| H714 | ||||||||
Note: Index names may vary depending on the purchased Illumina index kit.
Pre-heat the thermocycler by running the " BLT " program below and press "Pause".
Choose the preheat lid option and set it to 100°C. set thermocycler:
- 68°C for 3 minutes
- 98°C for 3 minutes
- (6) cycles of:98°C for 45 seconds, 62°C for 30 seconds, 68°C for 2 minutes
- 68°C for 1 minute
- Hold at 10°C
Label 0.2 ml PCR tubes with the sample number.
Combine the following volumes to prepare the PCR master mix. Multiply each volume by the number of samples being processed. Reagent overage is included in the volume to ensure accurate pipetting
- EPM :
22µL - Nuclease-free water
22µL
Briefly vortex then centrifuge for 10 seconds.
With the 1.5 ml tubes on the magnetic stand, use a P300 pipette to carefully remove and discard the supernatant.
Foam that remains on the well-walls does not adversely affect the library.
Remove tubes from the magnetic stand.
Immediately add 40µL of PCR master mix directly onto the beads in each sample tube, then gently pipette to mix until the beads are fully re-suspended and transfer all the contents into labeled PCR tubes. Gentle pipetting avoids bubbles and liquid drop splits on the walls of the tube.
Add the 5µl of I5 index adapters and 5µl of I7 index adapters to each sample, using 1 pair of gloves per index adapters to minimize contamination risk.
After adding each index, mix by gently pipetting up and down 5 times. Careful pipetting ensures the absence of bubbles and liquid splits on the tube walls.
Place on the thermal cycler and run the "BLT" PCR program.
Cleaning the library
This step uses a double-sided bead purification procedure to purify the amplified libraries.
Items to prepare :
- Sample Purification Beads (SPB): bring at room temperature 30 min before use, vortex to resuspend the beads, and invert to mix.
- Freshly prepared 80% ethanol (EtOH). use milli-Q water to prepare the 80% ethanol.
- RSB (Resuspension Buffer): thaw and bring to room temperature; vortex to mix.
- 1.5 ml low binding microcentrifuge tubes; label two tubes per sample.
- Nuclease-free water.
Transfer the contents to a 1.5 ml tube.
Spin down for 10 seconds to collect contents at the bottom of tube.
Place the tubes on the magnetic stand and wait until the liquid is clear (~5 minutes).
Transfer 45 µl supernatant from each tube to new 1.5 ml tubes.
Vortex and invert SPB multiple times to resuspend.
The starting DNA was a small PCR amplicon input. Perform the following steps:
Add 81 µl SPB to each 1.5 ml tube containing supernatant.
Mix using a plate shaker at 1600rpm,0h 0m 0s for 1 minute.
Incubate caped tubes at room temperature for 5 minutes.
Place on the magnetic stand and wait until the liquid is clear (~5 minutes).
Without disturbing the beads, remove and discard supernatant.
Wash two times as follows:
With the plate on the magnetic stand, add 200 µl fresh 80% EtOH without mixing.
Incubate for 30 seconds.
Without disturbing the beads, remove and discard supernatant.
Use a 20 µl pipettor to remove and discard residual EtOH.
Air-dry on the magnetic stand for 5 minutes, then remove from the magnetic stand.
Add 32 µl RSB to the beads and gently pipette 5 times to resuspend.
Incubate at room temperature for 2 minutes.
Place the tube on the magnetic stand and wait until the liquid is clear (~2 minutes).
Transfer 30 µl supernatant to a new low binding 0.5 ml tube. This supernatant contains double stranded DNA library.
Pool and dilute library
Library quality control
Check the concentration of library using
Dilute Libraries to the starting concentration

Dilute the library using RSB to 2nanomolar (nM) (10µL) into a low binding 0.6 ml tubes.
Equipment
| Value | Label |
|---|---|
| Platinum 0.6 ml tube | NAME |
| Tube | TYPE |
| BMBio | BRAND |
| BM4006 | SKU |
| https://bmbio.com/shop/g/gBM4006/ | LINK |
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| sample | Library concentration [Qubit(ng/ul)] | Molarity(nM) | DNA2nM(ul) | RSB (ul) | total |
| P1743 | 1.65 | 1.65*1E6/660/400=6.25 | 20/6.25=3.2 | 10-3.2=6.8 | 10 |
| P1866 | 0.968 | 0.95*1E6/660/400=3.5985 | 5.57 | 4.43 | 10 |
sample P1743: add 3.2 DNA + 6.8 ul RSB to make 2nM (10ul) DNA library
Library pooling
Label a clean 1.5 ml low binding microcentrifuge tube.
Add 5ul of each 2nM diluted libraries into the 1.5 ml tube to make a pooled 2nM DNA library.
Mix for 2 min 30 seconds at 1400rpm,0h 0m 0s.
Library denaturation before the sequencing using Illumina MiSeq v2
Prepare :
• thaw HT1 ((Hybridization Buffer), RSB, <reagents text="phiX V3 control" label="Illumina, Inc."/> and place on ice.
• prepare 1N NaOH = 10M NaOH (50ul) + MilliQ H2O (450ul)
• prepare fresh [0.2N NaOH] from 1N NaOH stock solution :
○ Mix `800µL`milli-Q water + `200µL`of Stock 1N NaOH
○ Invert tube several times to mix
NB: 0.2N NaOH should be used within 12 hours.
Label a new 1.5 ml low binding microcentrifuge tube and combine the following volumes in the tube:
-
5 μl of pooled library (2nM)
-
5 μl of 0.2N NaOH
Briefly vortex the mixture.
briefly centrifuge for 2- 3 seconds.
incubate for 5 minutes at room temperature.
Add 990µL of pre-chilled HT1 into the tube to obtain 10picomolar (pM) of denatured library (1mL) --> then flick tube to mix.
Please make a 10picomolar (pM)of final solution (600µL). Therefore,
-
transfer
600µLof denatured library into new low binding 1.5 ml tube. -
Invert to mix and spin down
Preparation of PhiX control
PhiX Control v3 is a reliable, adapter-ligated library used as a control for Illumina sequencing runs. The library is derived from the small, well-characterized PhiX genome, offering several benefits for sequencing and alignment.
This is a small, ready-to-use Illumina library with a balanced nucleotide representation. Adding a 2% PhiX spike-in to your library provides additional metrics. For low-diversity libraries, use a 10% spike-in to increase base diversity.
In a 0.2 ml PCR tube, prepare 4nanomolar (nM) Phix from the 10nanomolar (nM) stock.
Mix:
•`2µL` (of 10 nM stock)
• `3µL` of RSB
Store the prepared PhiX control and library pool at -30°C until sequencing using MiSeq flowcell reagents.
In a clean1.5 ml tube, Mix :
• 4 nM PhiX library: `5µL`
• 0.2 N NaOH: `5µL`
Vortex the modified PhiX solution vigorously.
Briefly centrifuge.
Incubate at room temperature for 5 minutes.
Dilute denatured PhiX to 20picomolar (pM)
Mix :
• Modified PhiX solution: `10µL`
• Ice-cooled HT1: `990µL`
The newly prepared 1 mL of PhiX solution has a final concentration of 20 pM. It can be kept frozen for up to 3 weeks.
To prepare 10 pM of PhiX control, mix :
• `240µL` of PhiX solution (20 pM),
• `240µL` of ice cooling HT1.
In a new e-tube, prepare the PhiX 10% solution by mixing :
• PhiX (10 pM): `60µL`
• DNA library (10 pM): `540µL`