Well-tempered Metadynamics protocol
Vidya Niranjan, Akshay Uttarkar
Abstract
Metadynamics is a technique in which the potential for one or more chosen variables ("collective variables") is modified by periodically adding a repulsive potential of Gaussian shape at the location given by particular values of the variables. These repulsive Gaussians eventually fill up the well that is being sampled, and force the calculation to sample elsewhere. At certain points in the simulation, the sum of the Gaussians and the free-energy surface (FES) becomes flat, and therefore the sum of Gaussians is the negative image of the FES.
The present protocol is running the well-tempered metadynamics module of Desmond (Free academic license) and the analysis of the results
Steps
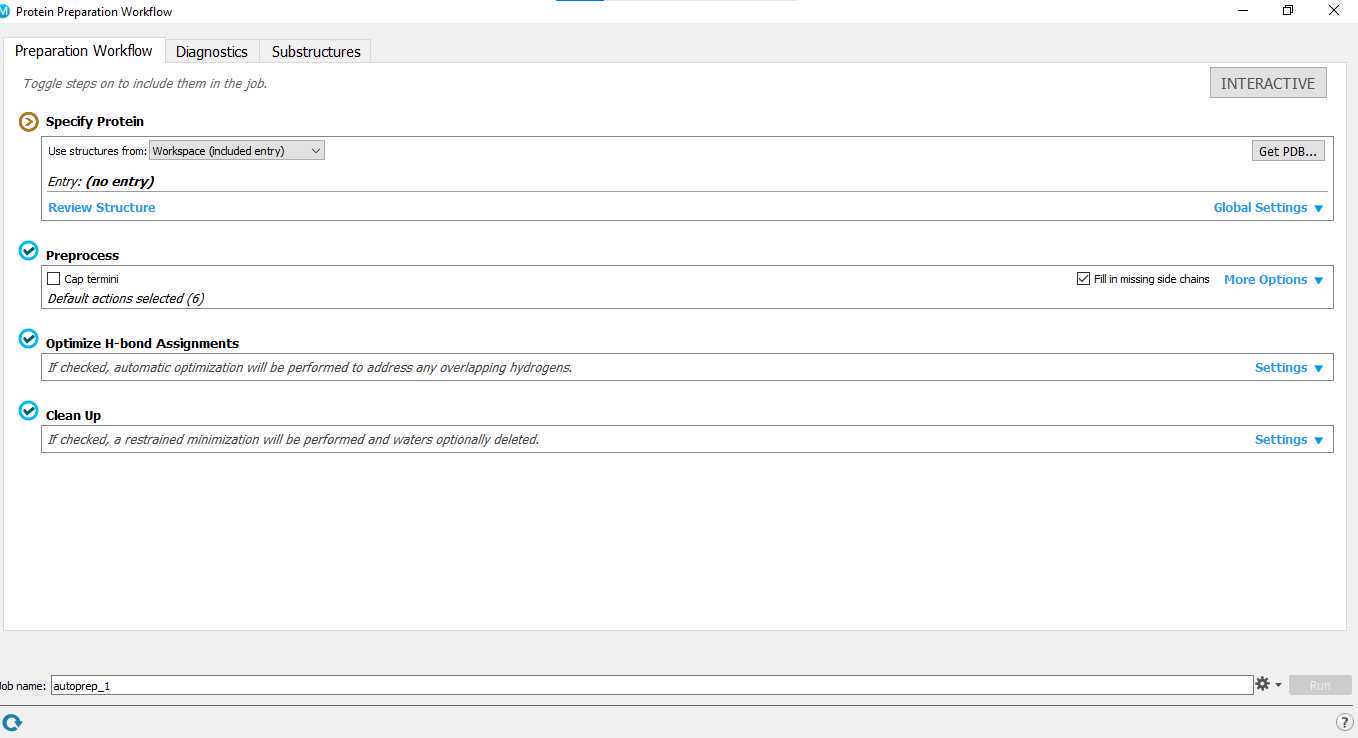
Protein preparation.
The crystal structure is imported into Maestro GUI. The Protein Preparation Wizard panel44 is used to add hydrogen atoms, patch end groups, add missing side chains and missing loops, assign protonation states of histidine, aspartate and glutamate at pH 7.045 and optimize the polar hydrogen orientations in crystal water molecules and
proteins.
Any problems related to your structure can be viewed in "Diagnostics Tab".
Simulation system setup .
In the solvation tab
The complex is solvated in an orthorombhic box with the dimensions 10Å additional to the total size of the receptor complex.
Click on minimize volume to obtain the optimal box dimensions.
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.b5fyq3pw/SS.png" alt="The "Solvation tab" of simulation system builder" loading="lazy" title="The "Solvation tab" of simulation system builder"/>
In the Ions tab
Place ions based on the requirement of neutral, acidic or basic simulation environment.
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.b5fyq3pw/SS%20ions.png" alt="The "Ions tab" of simulation system builder" loading="lazy" title="The "Ions tab" of simulation system builder"/>
The solvents are composed of water molecules and neutralizing agents (i.e Na or Cl).
Preparation of metadynamics input files.
The solvated complex is loaded into the Metadynamics panel. The CV distance is defined.
The distance for the set of atoms has to be defined by selection or by using "Define" option present in Maestro GUI
This can be done via two modes
-
Maestro GUI > Select > Define > " Molecule list"
-
Maestro GUI > Select > More Objects > "Your selection"
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.b5fyq3pw/meta.png" alt=""Metadynamics panel" for selection of CV atoms and defining related parameters" loading="lazy" title=""Metadynamics panel" for selection of CV atoms and defining related parameters"/>
The parameters that control the accuracy of the simulation are the height and width of the Gaussian potential and the interval at which the Gaussians are added. The accuracy is not very sensitive to the ratio of the height to the interval; however, smaller values of this ratio increase the accuracy somewhat. The width of the Gaussian should be roughly 1/4 to 1/3 of the average fluctuations of the collective variable during a free MD run.
A wall at the CV value equal to the sum of the largest dimension of the complex.
The time interval between which a Gaussian is injected is set to 0.09 picoseconds (ps). The simulation temperature and pressure are set to 310 K and 1.01325 bar, respectively. The simulation time is set to the desired time (25-50ns)
Click on "Write file" under the action buttons to generate a the following files
This step generates three input files (cfg, msj and cms) and a command file (sh)
Changes in parameters for Well-tempered metadynamics
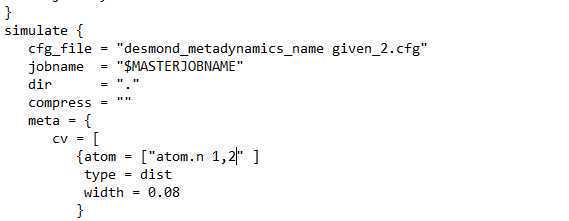
Add the bias factor keyword kTemp and its value to the meta block of the .msj file (generated in previous step)
kTemp = 2.4
(Boltzmann's constant times temperature ΔT in units of kcal/mol – In this case, kTemp corresponds to 1200 K.)
During well-tempered metadynamics, the dynamics of the system are effectively accelerated (without heating up the system) up to T + ΔT, where T is the chosen MD simulation temperature.
There are two limiting cases for ΔT (the bias factor kTemp):
ΔT → 0: The well-tempered metadynamics run is identical to a standard MD simulation at temperature T.
ΔT → ∞: The well-tempered metadynamics run is identical to a standard/non-well-tempered metadynamics run.
The choice of the bias factor value is guided by the highest barrier in the simulation system which the well-tempered metadynamics run should overcome.
Here are some suggestions, assuming that the initial hill height ω0 (height parameter in the Metadynamics panel) has been set to 0.3 kcal/mol:
| A | B |
|---|---|
| 3 | 1.7 |
| 6 | 3.4 |
| 10 | 5.6 |
| 15 | 8.4 |
| 20 | 11.2 |
Column A is Max. barrier height [kcal/mol] and Column B is kTemp [kcal/mol]
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.b5fyq3pw/Ktemp%20added.png" alt="Meta block of .msj file with bias factor "Ktemp" added" loading="lazy" title="Meta block of .msj file with bias factor "Ktemp" added"/>
Save the .msj file
Run the simulation from the control panel using the .sh file generated.
Multisim run.
Each run undergoes multiple stages as instructed in the msj file as follows. (a) Brownian dynamics of NVT ensemble: temperature 10 K, small timesteps, restraints on solute heavy atoms and 100 ps. (b) Berendsen dynamics of NVT ensemble: temperature 10 K, small timesteps, restraints on solute heavy atoms and 12 ps. (c) Berendsen dynamics of NPT ensemble: temperature 10 K, restraints on solute heavy atoms and 12 ps. (d) Berendsen dynamics of NPT ensemble: temperature 310 K, restraints on solute heavy atoms and 12 ps. (e) Berendsen dynamics of NPT ensemble: temperature 310 K, no restraints and 500 ps. (f) Production. Martyna-Tobias-Klein dynamics of NPT ensemble with metadynamics is performed at a temperature of 310 K without restraints up to the targeted simulation time. This generates a kerseq file recording all the Gaussian functions infused into the system along the simulation time, the trajectory file and other output files.
Metadynamics analysis
Open the Metadynamics analysis panel from Maestro GUI
Input the "-out.cms" file and run the analysis.
Save the plot
Apart from this, by default a .fes file is generated by the module which contains corresponding FES values for each CV.

