A Step-by-Step Guide for the Production of Recombinant Fluorescent TAT-HA-Tagged Proteins and their Transduction into Mammalian Cells
Christer Abou Anny, Christer Abou Anny, Sébastien Nouaille, Sébastien Nouaille, Régis Fauré, Régis Fauré, Céline Schulz, Céline Schulz, Corentin Spriet, Corentin Spriet, Isabelle Huvent, Isabelle Huvent, Christophe Biot, Christophe Biot, Tony Lefebvre, Tony Lefebvre
DNA molecular assembly
fluorescent protein
fusion protein
Ni-NTA-based protein purification
protein transduction
recombinant protein production
tagged protein
Abstract
Investigating the function of target proteins for functional prospection or therapeutic applications typically requires the production and purification of recombinant proteins. The fusion of these proteins with tag peptides and fluorescently derived proteins allows the monitoring of candidate proteins using SDS-PAGE coupled with western blotting and fluorescent microscopy, respectively. However, protein engineering poses a significant challenge for many researchers. In this protocol, we describe step-by-step the engineering of a recombinant protein with various tags: TAT-HA (trans-activator of transduction-hemagglutinin), 6×His and EGFP (enhanced green fluorescent protein) or mCherry. Fusion proteins are produced in E. coli BL21(DE3) cells and purified by immobilized metal affinity chromatography (IMAC) using a Ni-nitrilotriacetic acid (NTA) column. Then, tagged recombinant proteins are introduced into cultured animal cells by using the penetrating peptide TAT-HA. Here, we present a thorough protocol providing a detailed guide encompassing every critical step from plasmid DNA molecular assembly to protein expression and subsequent purification and outlines the conditions necessary for protein transduction technology into animal cells in a comprehensive manner. We believe that this protocol will be a valuable resource for researchers seeking an exhaustive, step-by-step guide for the successful production and purification of recombinant proteins and their entry by transduction within living cells. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : DNA cloning, molecular assembly strategies, and protein production
Basic Protocol 2 : Protein purification
Basic Protocol 3 : Protein transduction in mammalian cells
INTRODUCTION
Over the years, the interest in innovative approaches for the expression, purification, and characterization of recombinant proteins has increased. It empowers scientists to create, engineer, and produce proteins with diverse functions, characteristics, and applications (Assenberg et al., 2013). Thus, production of recombinant proteins represents a cornerstone for various applications in drug discovery (Andersen & Krummen, 2002; Overton, 2014), therapeutic proteins (Burnett & Burnett, 2020; Wurm, 2004) or vaccine production (Farzaneh et al., 2017; Joung et al., 2016). Recombinant DNA encoding proteins are typically generated through the use of recombinant DNA technology from historical methods (e.g., amplifications and use of restriction enzymes) to recently developed sophisticated technologies (e.g., Gibson assembly method for DNA cloning) (Bordat et al., 2015; Casini et al., 2015; Cohen, 2013; Ferro et al., 2019; Gibson et al., 2009; Jang & Magnuson, 2013; Kostylev et al., 2015; Li et al., 2019; Li et al., 2020; Li et al., 2018; Norris et al., 2015; Roberts, 2019; Rudenko & Barnes, 2018; Sands & Brent, 2016; Thomas et al., 2015; Wang et al., 2015). One of the most significant breakthroughs in recombinant protein technology has been the development of expression systems for the efficient production of complex proteins by selecting an appropriate method in microbial hosts (Brondyk, 2009), such as Escherichia coli (Baneyx, 1999; Baneyx & Mujacic, 2004; Chen, 2012; Gopal & Kumar, 2013; Rosano & Ceccarelli, 2014; Sørensen & Mortensen, 2005), yeast (Cregg et al., 2000; Mattanovich et al., 2012; Porro et al., 2005), Streptomyces spp. (Anné et al., 2012), mammalian cells (Andersen & Krummen, 2002; Barnes et al., 2003; Hopkins et al., 2012; Jäger et al., 2015; Sunley & Butler, 2010) or plants (Anné et al., 2012; Newman et al., 2011; Nienhaus & Nienhaus, 2014; Noguchi & Matsumoto, 2006). These systems provide researchers with the ability to produce proteins that may be challenging to obtain by fastidious protein purification from various biological sources (Brondyk, 2009). Among these recombinant proteins, fluorescent proteins have proven to be invaluable tools for monitoring and visualizing cellular processes in real-time (Ai et al., 2014; Nienhaus & Nienhaus, 2014; Wang et al., 2008; Wiedenmann et al., 2009). Fluorescent proteins are derived from naturally occurring proteins with intrinsic fluorescence, such as the Green Fluorescent Protein (GFP) originally isolated from Aequorea victoria (Jakobs et al., 2000; McRae et al., 2005; Nolte et al., 2001) or the red fluorescent protein mCherry, derived from DsRed isolated from Discosoma sea anemones (Shen et al., 2017). These color-differentiated fluorescent proteins can be used together to perform multicolor imaging experiments in molecular and cellular biology research in order to label specific cellular structures, organelles, or proteins for visualization and tracking within living cells (Helmuth et al., 2009; Kirchhausen, 2009; McDonald et al., 2002; Newman et al., 2011; Rizzuto et al., 1995; Salomonnson et al., 2012; Subach et al., 2009; Yin et al., 2013).
This protocol is designed to provide a step-by-step guide to successfully express recombinant fluorescent proteins in bacteria and subsequently visualize them in animal cells (Fig. 1). The successful execution of this protocol involves several steps, starting with the design and construction of recombinant DNA vectors using the Gibson assembly method to express the target protein fused with the HIV-1 trans-activator peptide called TAT-tag (Trans-Activator of Transduction), a fluorescent protein, and a TEV protease recognition sequence to eliminate the downstream 6×His-tag. This method is used for the purpose of gene cloning and expression, through constructing plasmids for protein expression. These assembled vectors are then transformed into a suitable host organism, typically E. coli BL21(DE3), to produce recombinant fluorescent proteins. Recombinant protein production is carried out through transcription induction by L-arabinose thanks to the PBAD bacterial promoter (Greenblatt & Schleif, 1971; Schleif, 2000; Schleif et al., 1973). The protein purification is performed via immobilized metal affinity chromatography (IMAC) thanks to the 6×His-tag (Block et al., 2009; Gaberc-Porekar & Menart, 2001; Porath, 1992; Sun et al., 2005). The TAT-tag, due to its transduction property, enables the fused protein to enter animal cells (Becker-Hapak et al., 2001; Eguchi et al., 2001; Fittipaldi & Giacca, 2005; Ford et al., 2001; Gump & Dowdy, 2007; Gump et al., 2010; Guo et al., 2012; Kabouridis, 2003; Kaplan et al., 2005; Leifert et al., 2002; MacKay & Szoka, 2003; Noguchi & Matsumoto, 2006; Pawson & Nash, 2000; Schwarze et al., 2000; Shokolenko et al., 2005; Wadia & Dowdy, 2002; Zhang et al., 2012). Once inside the living cells, the recombinant protein can be monitored and tracked according to its localization and behavior by its fluorescent signal.
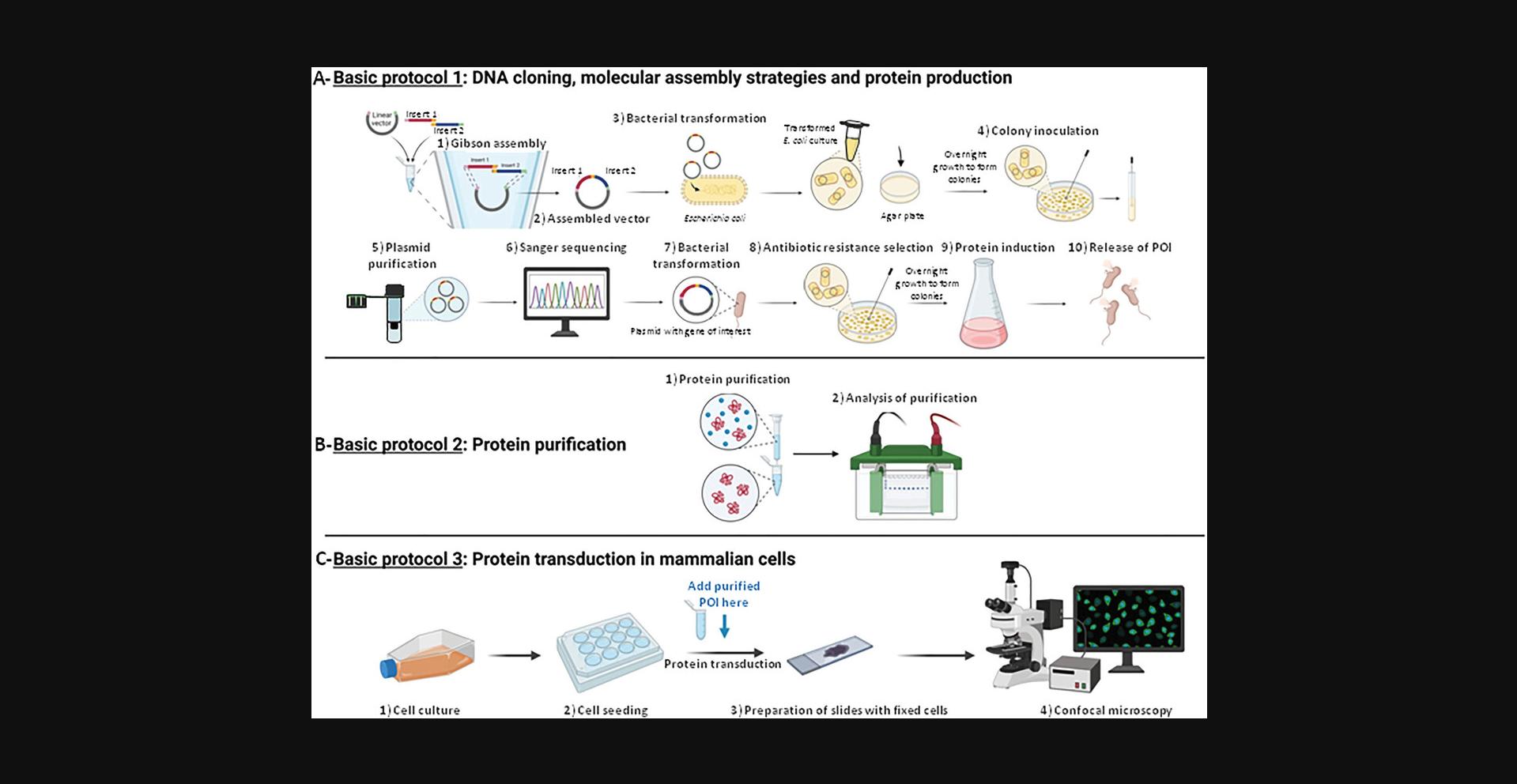
The production of proteins in bacteria and their delivery to animal cells by a penetration peptide has the advantage of working with well-defined quantities of proteins, obtaining a homogeneous distribution of the latter throughout the cell population and to be a vector-free method. The transient transfection of vectors, e.g., plasmids, makes it impossible to control the quantity of protein expressed within the cell and very often leads to heterogeneous expression within the cell population, as well as being cell type dependent.
We offer the reader, with a range of readily accessible techniques and devices found in research laboratories or with a simple Ni-NTA column, the opportunity to produce fluorescent proteins using three distinct but complementary protocols. Basic Protocol 1 details, thanks to Gibson-type molecular assembly, the construction of custom plasmid vectors making it possible to express a specific protein of interest (POI) in phase with any fluorescent protein and various tags, and to produce it in E. coli bacteria. Basic Protocol 2 presents the purification of the POI by taking advantage of a poly-His-tag by using a preparative chromatography system with a fast protein liquid chromatography (FPLC) Ni-nitrilotriacetic acid (NTA) agarose column, and its quality control by SDS-PAGE coupled to western blotting. Lastly, Basic Protocol 3 exposes a very simple experimental strategy for protein transduction into animal cells using a cell penetration peptide. We use direct fluorescence microscopy to avoid long, complex, and often costly indirect immunofluorescence steps.
Basic Protocol 1: DNA CLONING, MOLECULAR ASSEMBLY STRATEGIES, AND PROTEIN PRODUCTION
In this protocol, we describe a procedure to construct a specific DNA sequence encompassing the different blocks needed for expression of the final protein containing the protein of interest (POI) moiety (Fig. 1A and Fig. 2). For that purpose, the choice of the gene encoding the POI, the plasmid vector and its features highly depend on specific needs of the experiment, including the type of host organism (e.g., E. coli) and the size of the DNA to be inserted. For the expression in E. coli , the desired level of gene expression is dependent on the choice of the appropriate promoter (e.g., PBAD promoter, used here). These choices greatly influence the success of gene expression studies (for detailed information see Tables 1–3). The plasmids undergo construction through Gibson molecular assembly of purified PCR fragments. The advantage with this approach is that it is possible, with a whole set of constructions either available in the laboratory or obtained elsewhere, to recover a fragment of interest (comparable to a block or a brick from a game assembly) and nesting it with other fragments to custom manufactured plasmids expressing the POI in phase with any desired tag or fluorescent protein.
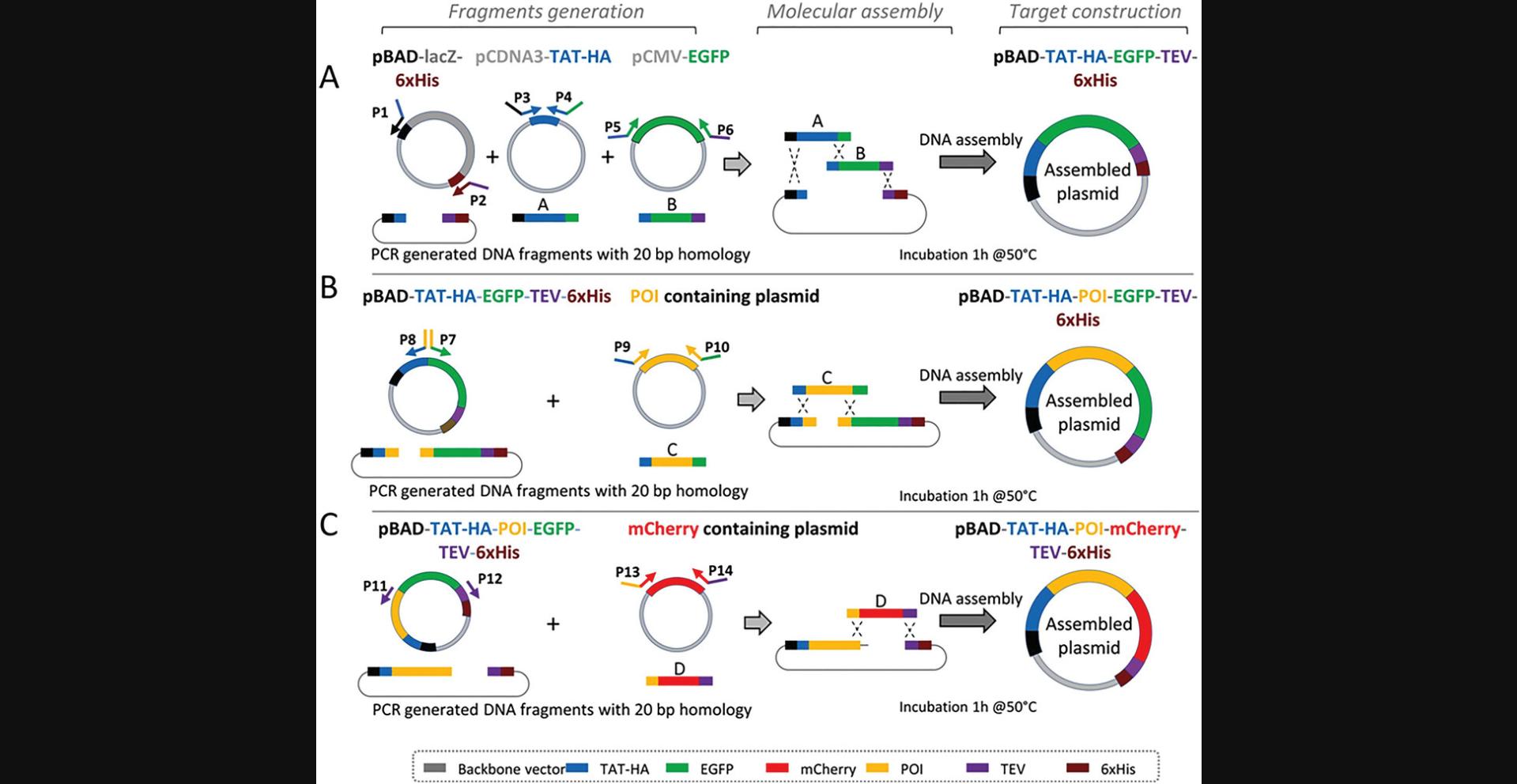
| Plasmid DNA source | Primer pair | Amplicon size (bp) |
|---|---|---|
| pBAD-LacZ-X-6×His | P1-P2 | 4047 |
| pCDNA3-Y-TAT-HA | P3-P4 | 122 |
| vCMV-EGFP-POI | P5-P6 | 745 |
| pBAD-TAT-HA-EGFP-TEV-6×His | P7-P8 | 4873 |
| vCMV-EGFP-POI | P9-P10 | 2360 |
| pBAD-TAT-HA-POI-EGFP-TEV-6×His | P11-P12 | 6479 |
| pCMV-mCherry | P13-P14 | 745 |
| Strain or Plasmid | Characteristics | Source |
|---|---|---|
| Strain | ||
| Competent E. coli (High Efficiency) | DH5-alpha | New England BioLabs |
| Competent E. coli (High Efficiency) | BL21(DE3) | New England BioLabs |
| Plasmid | ||
| pBAD-LacZ-6×His | E. coli expression vector, ampr | Life Science |
| pcDNA3-Y-TAT-HA | E. coli expression vector, ampr | UGSF, Lille (France) |
| vCMVp-EGFP-POI | E. coli expression vector, kanr | e-Zyvec, Loos (France) |
| pBAD-TAT-HA-EGFP-TEV-6×His | E. coli expression vector, ampr | This study |
| pBAD-TAT-HA-POI-EGFP-TEV-6×His | E. coli expression vector, ampr | This study |
| pCMV-mCherry | E. coli expression vector, kanr | UGSF, Lille (France) |
| Primer | Sequence (5’ → 3’) | Construct |
|---|---|---|
|
P1-TEV-6×His P2-TAT-HA-pBAD P3-pBAD-TAT-HA P4-EGFP-TAT-HA P5-EGFP-TAT-HA P6-TEV-EGFP |
GAAAACCTGTACTTCCAGGGTAATAGCGCCGTCGACCATCAT AGCCAAGCTTTGGATCCATGGTTAATTCCTCCTGT CATGGATCCAAAGCTTGGCTACGGCCGCAAGAAA CCTTGCTCACGGCCATGGAGCCAGCATAGTCTG CTCCATGGCCGTGAGCAAGGGCGAGGAGCTGTTC ACCCTGGAAGTACAGGTTTTCCTTGTACAGCTCGTCCATGCCGAG |
pBAD-TAT-HA-EGFP-TEV-6×His |
|
P7-POI-EGFP P8-POI-TAT-HA P9-TAT-HA-POI P10-EGFP-POI |
NNNNNNNNNNGTGAGCAAGGGCGAGGAGCTGTTC NNNNNNNNNNGGCCATGGAGCCAGCATAGTCTG CTCCATGGCCNNNNNNNNNNNNNNNNNNNNNN CCTTGCTCACNNNNNNNNNNNNNNNNNNNNNN |
pBAD-TAT-HA-POI-EGFP-TEV-6×His |
|
hP11-POI Rv P12-5’TEV P13-mcherry P14-mcherry |
CAGGTCAGTATCAAACCAGGCCAGC GAAAACCTGTACTTCCAGGGTAATAGCG CCTGGTTTGATACTGACCTGGTGAGCAAGGGCGAGGAGGATAACAT CCCTGGAAGTACAGGTTTTCCTTGTACAGCTCGTCCATGCCGCC |
pBAD-TAT-HA-POI-mcherry-TEV-6×His |
- a
For clarity the color code used for sequences is the same as that used in Figure 2 for fragments.
Materials
-
Plasmids, or other source of DNA insert (see Table 2 for detailed information)
-
Custom primers (see Table 3 for primers used in this study)
-
Q5 high-fidelity DNA polymerase, 2000 U/ml (New England BioLabs, cat. no. M0491S)
-
Q5 reaction buffer, 5× (New England BioLabs, cat. no. B9027S)
-
Deoxynucleotide (dNTP) solution mix, 10 mM (New England BioLabs, cat. no. N0447S)
-
Ultrapure DNase-free H2O
-
Agarose powder
-
Tris-acetate-EDTA (TAE) buffer, 0.5× (Euromedex)
-
Gel loading buffer (New England Biolabs, cat. no. B7025)
-
1 kb Plus DNA ladder, 1000 µg/ml (New England BioLabs, cat. no. N3200S)
-
100 bp DNA ladder, 500 µg/ml (New England BioLabs, cat. no. N3231S)
-
Ethidium bromide (Thermo Fisher Scientific, cat. no. 15585011)
-
QIAquick gel extraction kit (Qiagen, cat. no. 28706)
-
NEBuilder HiFi DNA assembly master mix (New England BioLabs, cat. no. E2621S)
-
Competent 5-alpha E. coli (New England BioLabs, cat. no. C2987H)
-
SOC medium (New England BioLabs, cat. no. B9020S)
-
LB agar plates: LB medium plus 15 g/L agar
-
LB medium: 10 g/L tryptone, 10 g/L NaCl, and 5 g/L yeast extract supplemented with the appropriate antibiotics, e.g., 100 mg/ml ampicillin (stock solution) diluted at 1/1000
-
QIAprep spin miniprep kit (Qiagen, cat. no. 27104)
-
Competent BL21(DE3) E. coli (New England BioLabs, cat. no. C2527H)
-
L-arabinose (Sigma-Aldrich, cat. no. A3256)
-
Complete EDTA-free protease inhibitor cocktail tablets (Sigma-Aldrich, cat. no. 11873580001)
-
Buffer A (see recipe)
-
DNase I (3000 U/mg) 20,000 U (Euromedex, cat. no. 1307)
-
0.2-ml PCR tubes (Dutsher, cat. no. 010207)
-
Benchtop centrifuge
-
Thermal cycler (Eppendorf Mastercycler)
-
Microwave oven or hotplate
-
Agarose gel electrophoresis system
-
E-Box gel documentation imaging system (Vilber Lourmat, France)
-
Benchtop UV transilluminator machine
-
Scalpel
-
NanoDrop ND-1000 spectrophotometer
-
Petri dishes (TRP, cat. no. 067002)
-
1.5-ml microcentrifuge tubes (Thermo Fisher Scientific, cat. no. 69715)
-
Ice
-
Heat block (Eppendorf ThermoMixer) or water bath
-
Shaking incubator
-
Sterile toothpicks
-
1-L glass Erlenmeyer flasks
-
Centrifuge Beckman-Coulter Avanti J-26S XP (with a rotor adapted for 1-L volumes)
-
50-ml sterile conical tubes (Falcon, cat. no. 352070)
-
Sonicator
-
Centrifuge, rotor 19776H, angle rotor, for six 50-ml culture tubes
-
0.45-µm filter (Dutsher, cat. no. 033942)
CAUTION : Ethidium bromide is a putative carcinogen compound (the dye intercalates between the stacked bases of DNA fragments) and must be handled with gloves. Do not use ethidium bromide powder to avoid irritation by inhalation and never use a microwave oven to heat solutions containing ethidium bromide. It is preferrable to use the less toxic SYBR safe stain gel to visualize DNA fragments in agarose gels. When using the transilluminator to visualize DNA bands, it is essential to protect yourself from aggressive UV rays with plastic glasses, gloves and sleeves that must cover the integrity of the arms.
Amplification of the various PCR products
1.In PCR tubes, mix the Q5 reaction buffer (adjusted with DNase-free ultrapure water) with the DNA templates, primers (designed as P1 to P14 in Table 1 and Fig. 2), dNTP and the Q5 high-fidelity DNA polymerase at desired concentrations (Table 4). Mix and pulse-centrifuge to ensure the total liquid is located at the bottom of the tube.
| Composition of PCR mixture | |||
|---|---|---|---|
| Component | Stock solution | Volume (µl) | Final concentration |
| Q5 reaction buffer | 5× | 5 µl | 1× |
| dNTP mix | 10 mM each | 0.5 µl | 0.2 mM each |
| Primers (10 µM) | 10 µM | 1.25 µl for each primer | 0.5 µM each |
| Plasmid matrix | Variable | Variable | ∼ 4 pg/µl |
| Q5 polymerase | 2 U/µl | 0.3 µl | 0.024 U/µl |
| Ultrapure water | To 25 µl | ||
| PCR Thermocycling conditions | |||
| Step | Temperature (°C) | Time | Cycle |
| Denaturation | 98 | 30 s | 1 |
| Denaturation | 98 | 10 s | |
| Annealing | 60 | 20 s | ×30 |
| Elongation | 72 | 30 s/kb | |
| Elongation | 72 | 3 min | 1 |
| Cooling | 14 | ∞ | - |
2.Place the PCR tubes in a thermal cycler and apply the corresponding PCR amplification program (Table 4).
3.Prepare the gel by mixing agarose at a concentration of 1% (w/v) in 0.5× TAE. Heat the solution in a microwave oven or on a hotplate until the agarose has dissolved.
4.Pour the gel in the electrophoresis casting tray and let it solidify. Remove the comb and put the gel in the electrophoresis chamber containing 0.5× TAE as migration buffer.
5.Mix the 5 µl PCR reaction to 2 µl gel loading buffer, load on the gel along with 5 µl DNA size marker to monitor DNA size after migration. Migrate at 100 V for 30 min.
6.After migration, incubate the gel for 10 min in water containing 0.5 µg/ml ethidium bromide. Wash the excess of ethidium bromide by incubation in water for 5 min.
7.Visualize the PCR products (Fig. 3) by exposition of the gel to a UV transilluminator, such as the E-box gel documentation imaging system.
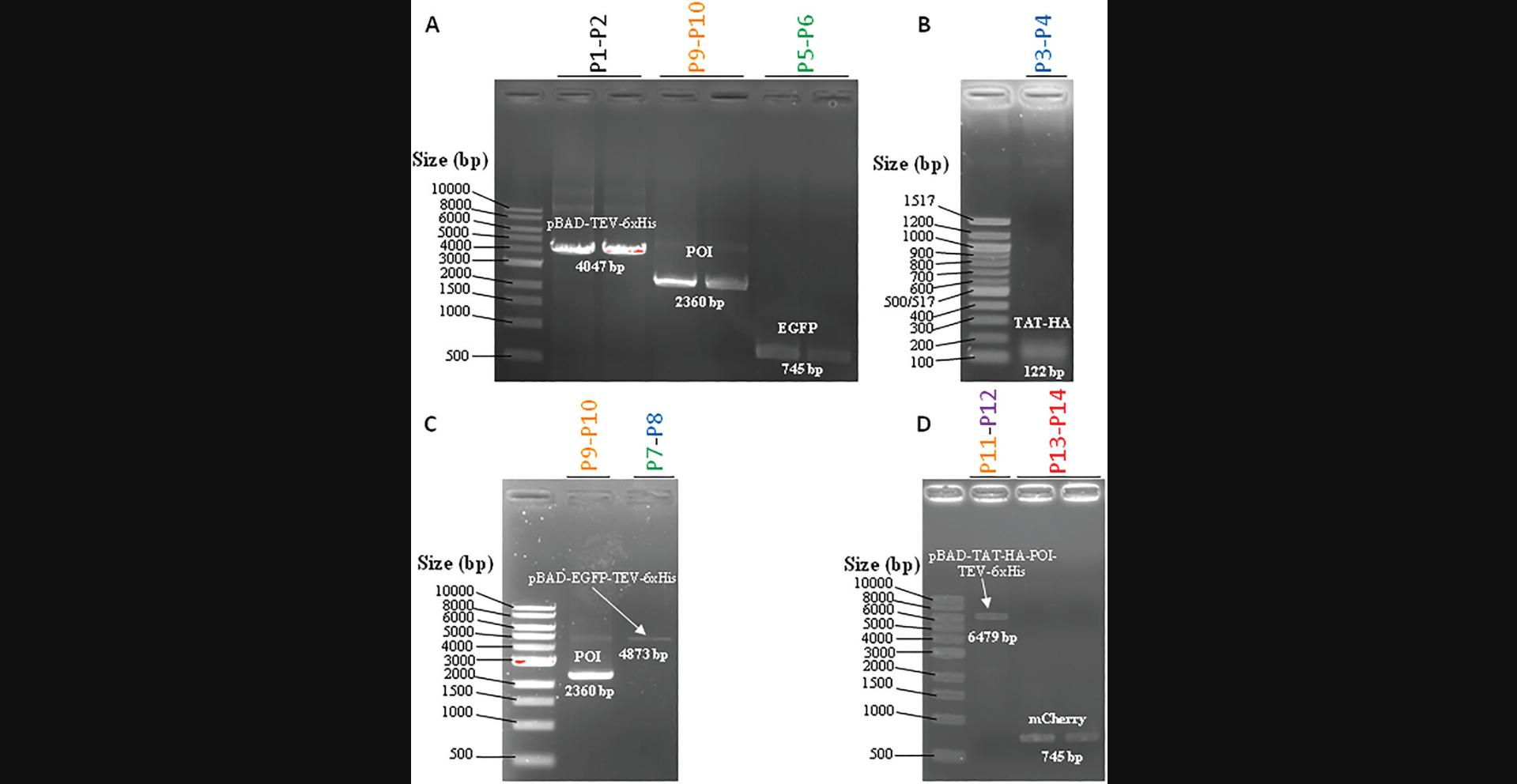
8.To purify the amplicon of interest, cut out the UV-detected DNA bands using a scalpel blade under a UV transilluminator instrument.
9.Weigh the DNA containing the gel slice and add 3 volumes (v/w) of QG buffer. Purify by using QIAquick gel extraction kit following the manufacturer's instructions. Elute with 50 µl elution buffer. Quantify the DNA using a NanoDrop ND-1000 spectrophotometer.
DNA molecular assembly
10.Mix each purified PCR fragment, i.e, TAT-HA, POI, pBAD-TEV-6×His, and EGFP or mCherry for the construction of pBAD-TAT-HA-POI-EGFP-TEV-6×His and pBAD-TAT-HA-POI-mCherry-TEV-6×His, respectively (Fig. 2), with NEBuilder HiFi DNA assembly 2× master mix at a ratio (1:1) (Tables 5 and 6). Complement with water to a final volume of 10 µl. Incubate the mixture at 50°C for 1 hr in a thermocycler.
| Components | Stock solution | Volume | Final concentration |
|---|---|---|---|
| Purified PCR | Variable | Variable | 5-15 ng/µl |
| T4 polynucleotide kinase PNK | 10 U/µl | 1 µl | 0.5 U/µl |
| T4 DNA ligase Buffer with ATP | 10× with 10 mM | 1 µl | 0.5× with 0.5 mM |
| T4 DNA ligase | 400 U/µl | 1 µl | 20 U/µl |
| Ultrapure water | To 20 µl |
| Component | Volume (µl) |
|---|---|
| Purified PCR fragments | x µl |
| Enzymes, 2× Hi-Fi DNA assembly master mix | 5 µl |
| Water | Up to 10 µl |
| DNA assembly thermocycling conditions | |
| 50°C for 1 hr | |
| 14°C/∞ | |
11.Use 5 µl assembly reaction for bacterial transformation in E. coli 5-alpha chemically competent cells (Froger & Hall, 2007). Mix DNA to 50 µl cells in 1.5-ml microcentrifuge tubes and mix gently by pipetting up and down or flicking the tube 4 to 5 times (do not vortex).
12.After 20 min on ice, perform heat shock using a heat block or water bath set to 42°C for 30 s and immediately transfer cells on ice for 3 min.
13.Add 450 µl SOC medium and incubate the mixture under shaking (150 rpm) for 1 hr at 37°C.
14.Spread 10 and 100 µl on LB agar plates containing the appropriate antibiotic and incubate at 37°C overnight.
15.Pick single colonies using toothpicks and cultivate to saturation in 5 ml LB medium containing antibiotic (37°C, 150 rpm) overnight.
16.Extract plasmid DNA using a QIAprep spin miniprep kit following the manufacturer's protocols. Elute the plasmid DNA in 50 µl elution buffer and quantify with a NanoDrop ND-1000 spectrophotometer. Check and confirm the fidelity of the construction by DNA sequencing (Eurofins).
Transformation and selection procedure
17.Perform the transformation with the plasmid expressing the POI by using the heat shock method described in step 11.Thaw E. coli BL21(DE3) competent cells on ice.
18.Add 0.1 µg purified DNA plasmid to 50 µl competent cells and mix gently by pipetting up and down or flicking the tube 4 to 5 times (do not vortex).
19.After 20 min on ice, perform heat shock at 42°C for 45 s and immediately transfer cells on ice for 3 min.
20.Add 450 µl SOC medium and incubate the mixture under shaking (150 rpm) for 1 hr at 37°C.
21.Spread 10 and 100 µl of the transformation mixture on separate LB agar plates (Petri dishes) supplemented with the antibiotic.
22.Incubate overnight at 37°C.
Recombinant protein expression
23.Pick single colonies and cultivate to saturation in 5 ml LB containing antibiotic (37°C, 150 rpm) overnight.
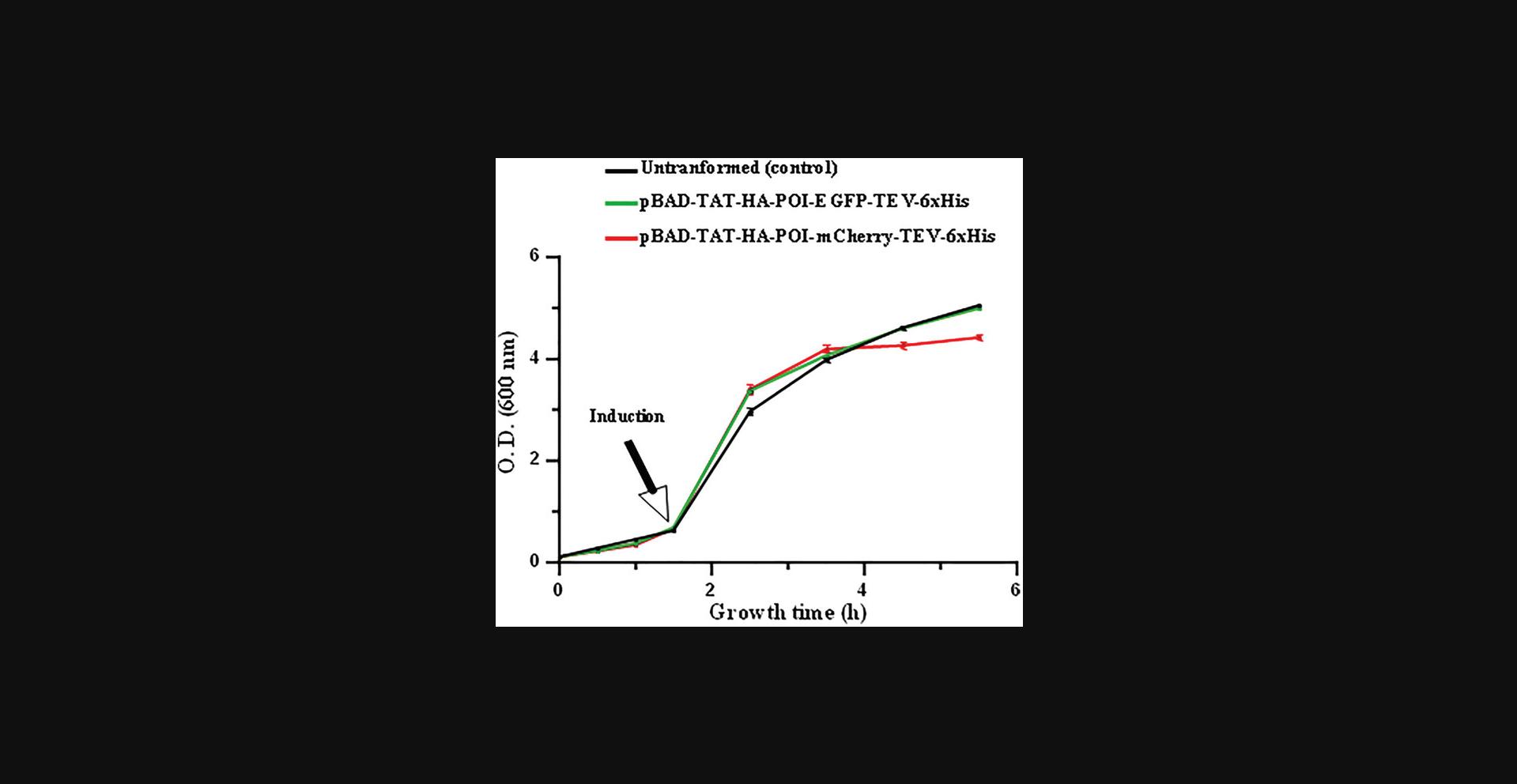
24.Add pre-culture of transformed E coli BL21(DE3) to 1 L of LB medium in 1-L glass Erlenmeyer flasks containing the antibiotic to reach a starting optical density at 600 nm (OD600) of 0.1. Monitor the OD600 until 0.6; then induce the recombinant POI expression (Table 7) for 4 h by adding 0.1% (w/v) L-arabinose (final concentration).
| Recombinant protein | Profiling amino acid sequence |
|---|---|
| TAT-HA- POI- EGFP-TEV-6×His | MDPKLGYGRKKRRQRRRGGSTMSGYPYDVPDYAGSMA[POI]VSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYKENLYFQGNSAVDHHHHHH |
| TAT-HA- POI- mCherry-TEV-6×His | MDPKLGYGRKKRRQRRRGGSTMSGYPYDVPDYAGSMA[POI]VSKGEEDNMAIIKEFMRFKVHMEGSVNGHEFEIEGEGEGRPYEGTQTAKLKVTKGGPLPFAWDILSPQFMYGSKAYVKHPADIPDYLKLSFPEGFKWERVMNFEDGGVVTVTQDSSLQDGEFIYKVKLRGTNFPSDGPVMQKKTMGWEASSERMYPEDGALKGEIKQRLKLKDGGHYDAEVKTTYKAKKPVQLPGAYNVNIKLDITSHNEDYTIVEQYERAEGRHSTGGMDELYKENLYFQGNSAVDHHHHHH |
- a
For clarity the color code used for sequences is the same as that used in Figure 2 for fragments.
25.To ensure that the conditions for bacterial growth are optimal for the expression of the recombinant protein, monitor the bacterial growth by measuring the OD600 during the culture and at the end of the expression phase (4 hr post-induction) (Fig. 4).
26.For cell harvesting, centrifuge the 1 L of LB culture medium 30 min at 6000 × g , 4°C, using the Avanti J-26S XP centrifuge (Fig. 5A).
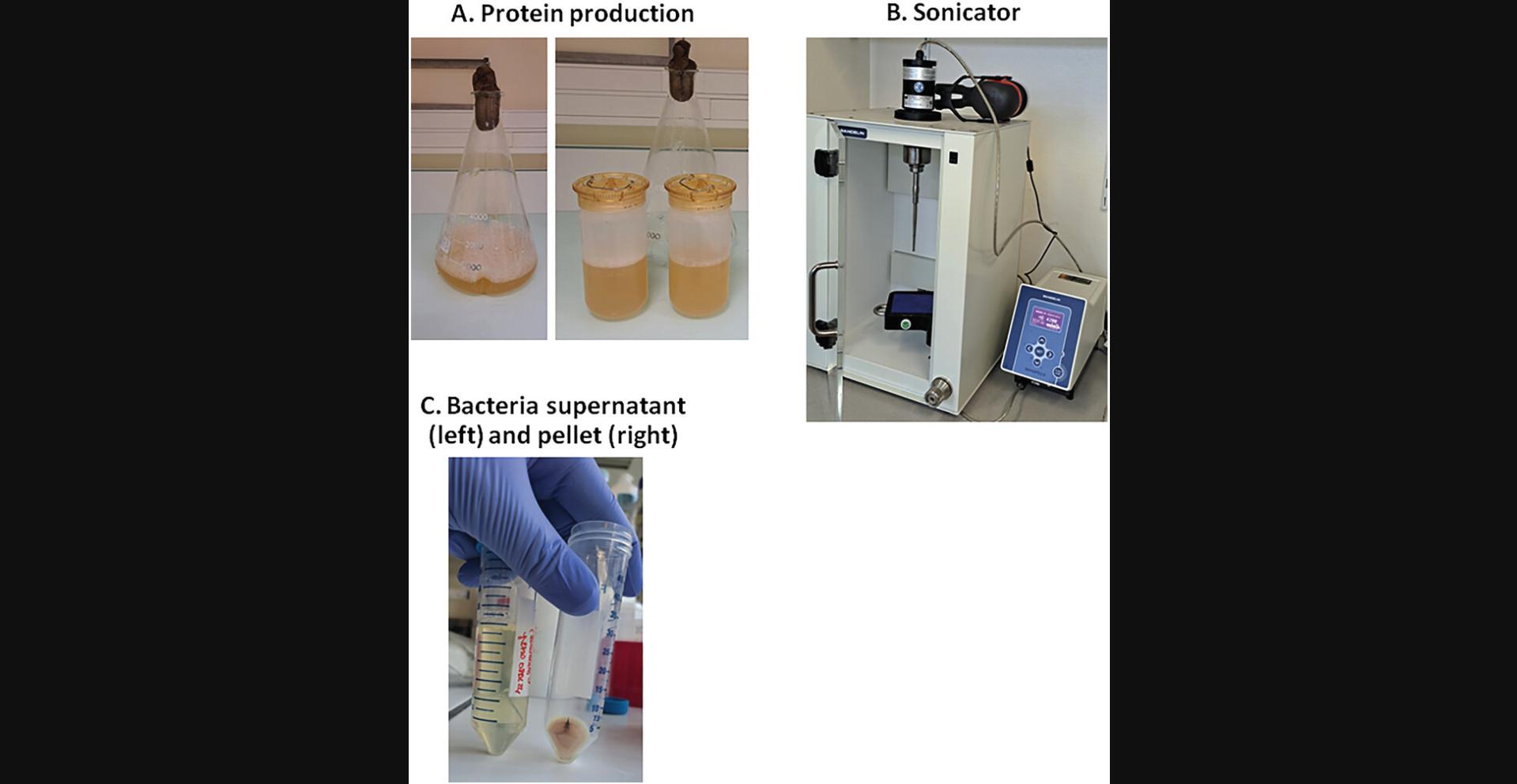
27.Discard the supernatant LB and add an anti-protease complete EDTA-free protease inhibitor cocktail tablet to the bacterial pellet (one tablet per L culture medium).
28.Resuspend the bacterial pellet in 20 ml buffer A and store the 50-ml tube at −20°C overnight.
29.Thaw the frozen bacterial pellet by placing the 50-ml tube containing the bacterial pellet in a water bath at room temperature.
30.Add 100 µl DNase I for every 1 L bacterial culture to degrade DNA that will be released by the bacterial lysis.
31.Perform bacterial lysis by sonication on ice, employing cycles of 30 s ON and 15 s OFF during 5 min, with the probe operating at 70% of maximal power (Fig. 5B).
32.Using a centrifuge and rotor 19776H, centrifuge the lysate 15 min at 12,000 × g , 4°C (Fig. 5C).
33.Filter the supernatant through a 0.45-µm filter. The supernatant containing the POI is ready for protein purification (Basic Protocol 2).
Basic Protocol 2: PROTEIN PURIFICATION
In this protocol, we describe the procedure of production and purification of recombinant protein isoforms (Fig. 1B). The remaining supernatant (from Basic Protocol 1, step 33) is then subjected to IMAC thanks to the 6×His-tag. Subsequent wash steps remove impurities and then the POI is eluted using a buffer that disrupts the binding interaction. Further purification steps may include size-exclusion chromatography and/or ion-exchange chromatography to achieve higher purity. Throughout the protocol protein concentration is monitored and quality is assessed using SDS-PAGE and a brilliant Coomassie blue staining and by western blot. If conducted properly, the final purified POI should exhibit the expected molecular weight, high purity, and biological activity.
Materials
-
Buffer A (see recipe)
-
Buffer B (see recipe)
-
Ultrapure H2O
-
Desalting buffer: Dulbecco's phosphate-buffered saline (DPBS), w/o Mg, w/o Ca (Dutscher, cat. no. L0615-500)
-
Laemmli buffer, 5× (see recipe)
-
Polyacrylamide gels, 8% separating and 4% stacking (see recipes)
-
SDS-PAGE buffer: 10% (v/v) Tris-glycine-SDS 10× (Euromedex, cat. no. EU0510) in distilled H2O
-
PageRuler prestained protein ladder (Thermo Fisher Scientific, cat. no. SM0671)
-
Quick Coomassie stain (NeoBiotech, cat. no. NB-45-00078-1litre)
-
Transfer buffer: 10% (v/v) Tris-glycine 10× (Euromedex, cat. no. EU0550) in distilled water, 20% (v/v) methanol (Carlo Erba, cat. no. 524102)
-
Ponceau S red solution (Sigma-Aldrich, cat. no. P7170)
-
TBS-Tween buffer (see recipe)
-
Blocking buffer (see recipe)
-
Mouse monoclonal antibodies:
- Anti-HA-tag (F-7) (Santa Cruz Biotechnology, cat. no. sc-7392)
- His-Tag (HIS. H8 / EH158) (Covalab, cat. no. mab90001-P)
- Anti-POI
- Anti-EGFP (Roche, cat. no. 45-11814460001)
-
ECL anti-mouse IgG (from sheep), horseradish peroxidase-linked whole secondary antibodies (Fisher Scientific, cat no. NA931V)
-
SuperSignal West Pico PLUS (Thermo Scientific, cat. no.34577) or SuperSignal West Femto, maximum sensitivity substrate (Thermo Scientific, cat. no.34095).
-
Preparative chromatography system (e.g., ÄKTA start protein purification system associated with UNICORN start control software)
-
HisTrap fast flow, 1- or 5-ml, Cytiva (Dutscher, cat. no. 17-5255-01)
-
HiTrap desalting, 5-ml, Cytiva (Dutscher, cat. no. 17-1408-01)
-
Vivaspin 6, centrifugal concentrator, 50,000 MWCO PES (Grosseron, cat. no. VS0632)
-
Benchtop centrifuge
-
Heat block (Eppendorf ThermoMixer)
-
Buffer tank lid
-
Electrode assembly
-
Combs electrophoresis Cell, 10-well, 1.5-mm
-
Hamilton syringe, 50-µl with needles
-
Power supply generator
-
Rotating platform
-
Gel cassette
-
Gel holder cassette and foam pads
-
0.45-µm pore nitrocellulose membrane (Amersham, cat. no. 10600016)
-
Whatman paper sheets, 6 × 9.5 cm, and sponges
-
Electroblotting system
-
Fusion Solo machine (Vilber Lourmat) for western blot detection
NOTE : It is possible that the POI aggregates and forms inclusion bodies that are difficult to solubilize and extract from the bacteria. In that case we recommend to process via mechanical rupture of the bacteria by sonication or by using a French press.
Protein fractionation
To purify your recombinant TAT-HA-POI-EGFP/mCherry-TEV-6×His protein by IMAC (exploiting the presence of a C-terminal 6×His-tag), use a simple benchtop nickel affinity column or combine with a preparative chromatography system, such as the ÄKTA Start system associated with a ÄKTA Start Fraction Collector “Frac30” and a UNICORN Start Software (Fig. 6). This can be carried out by employing an automated protein purification workflow with features like automated sample injection, fractions collection, and real-time monitoring caution.

1.Prepare bottles for buffer A, buffer B, ultrapure water, and the column HisTrap fast flow.
2.Set the IMAC purification program to use both buffer A and buffer B.
3.Equilibrate the column with 20 to 35 ml buffer A.
4.Inject your protein sample containing the POI into the equilibrated column (usually done using an automated injection system).
5.Wash the column with 25 ml buffer A to remove unbound proteins and contaminants.
6.Start the imidazole concentration gradient between buffer A and buffer B, up to 100% buffer B, to ensure the controlled release of the POI from the column.
7.Collect 1 ml fractions from the column. Monitor the elution process in real-time by measuring absorbance at a specific wavelength of 280 nm (detection of proteins).
Protein desalting and concentration
8.Use a bottle of 1× DPBS and a HiTrap desalting column.
9.Set the protein desalting program to only use pump A.
10.In the injection loop, re-inject the fraction containing your purified POI -1 ml per 1 ml and re-collect 5 to 10 desalted fractions of 1 ml each.
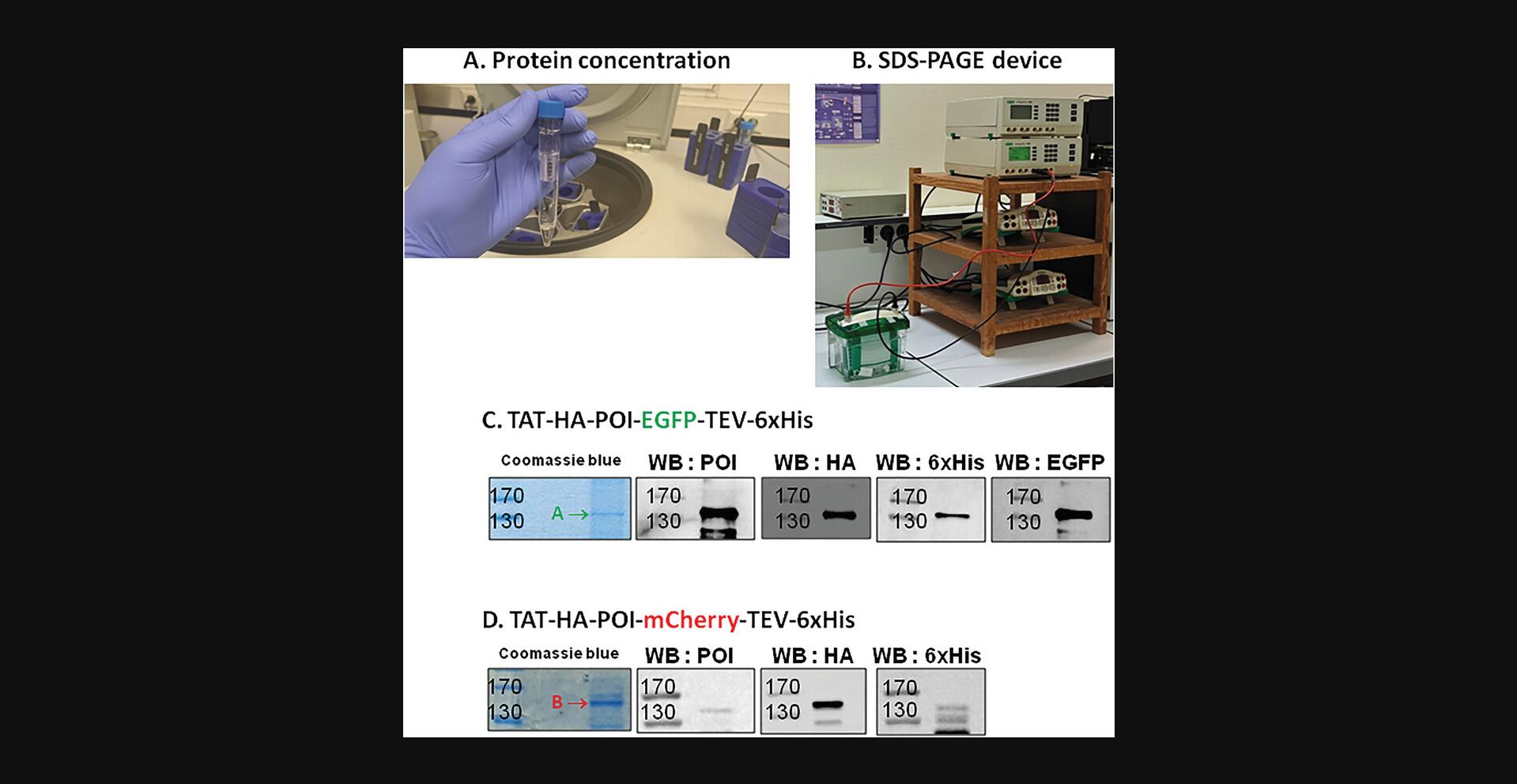
11.Concentrate the proteins (sample of 4 ml POI) using a Vivaspin 50 kDa cut-off through successive centrifugations of 5 min at 3500 × g , 4°C (Fig. 7A). Prior to use, rinse the concentrator column with water and subsequently with DPBS. Gently mix with a pipette before and after each centrifugation.
12.Quantify protein concentration by measuring absorbance at 280 nm (extinction coefficients of 93,170 and 105,660 M-1cm-1 for the EGFP and the mCherry forms of the POI, respectively).
Analysis of recombinant protein expression by SDS-PAGE (Fig. 7B)
13.Load 10 to 20 µg of purified proteins (mixed and boiled 1/5 volume of 5× Laemmli buffer) by using a Hamilton syringe on an 8% SDS-PAGE gel. Also load 5 µl molecular weight markers (typically ranging from 250 to 10 kDa).
14.Using a power supply generator, run at 30 mA in 1× Tris-glycine-SDS buffer until the bromophenol blue line reaches the bottom of the glass plates (∼2 hr).
15.For Coomassie blue staining (Figs. 7C and 7D), after migration carefully transfer the gel to a plastic or glass tray.
16.Cover the gel with Quick Coomassie stain with enough volume to allow it to move freely in the tray.
17.Cover with aluminum wrap and place on a platform rotator overnight at room temperature.
18.Proceed to the gel destaining step by multiple washes with ultrapure water until the band corresponding to the POI is easily visible.
19.For western blotting (Figs. 7C and 7D), transfer the gel onto nitrocellulose and proceed to electro-blotting in a device dedicated for that purpose (e.g., BioRad blotting system).
20.After transfer, check the transfer efficiency (and if necessary, the equal loading of the samples) by incubating the nitrocellulose membrane in the Ponceau S red solution for 1 min.
21.Discard the excess of solution and wash the membrane with ultrapure water until the band(s) is(are) easily visible.
22.Take a picture of the stained membrane for archiving.
23.Destain the membrane by incubation in the TBS-Tween buffer for a few seconds until gentle shaking.
24.Saturate the membrane by incubation in the blocking buffer for 45 min at room temperature (use a rotating platform for gentle shaking).
25.Discard the blocking buffer and incubate the membrane with the appropriate antibodies (mouse polyclonal anti-HA, mouse monoclonal anti-6×His, mouse monoclonal anti-POI at the appropriate dilution according to the manufacturer's instructions in blocking buffer).
26.Place the membrane on a rotating platform (gentle shaking) overnight at 4°C.
27.Wash the membranes with TBS-Tween 3 times for 7 min each and then incubate with an anti-mouse horseradish peroxidase-labeled secondary antibody (usually at a dilution of 1/10,000) for 1 hr.
28.Wash the membranes with TBS-Tween 3 times for 7 min each.
29.Add a mixed volume of buffers A and B (1:1) of SuperSignal West Pico Plus onto a parafilm sheet placed on the bench.
30.Position the nitrocellulose membrane so that the proteins directly face the revelation liquid. Wait for 5 min in the dark.
31.Transfer the membrane onto a tray for protein detection and place the tray into the Fusion Solo machine.
32.For capturing images using enhanced chemiluminescence, use the Fusion software (chemiluminescence and fluorescence imaging).
Protein storage
33.According to protein stability, store the POI at −80°C in 10% (v/v) sterile glycerol until use.
Basic Protocol 3: PROTEIN TRANSDUCTION IN MAMMALIAN CELLS
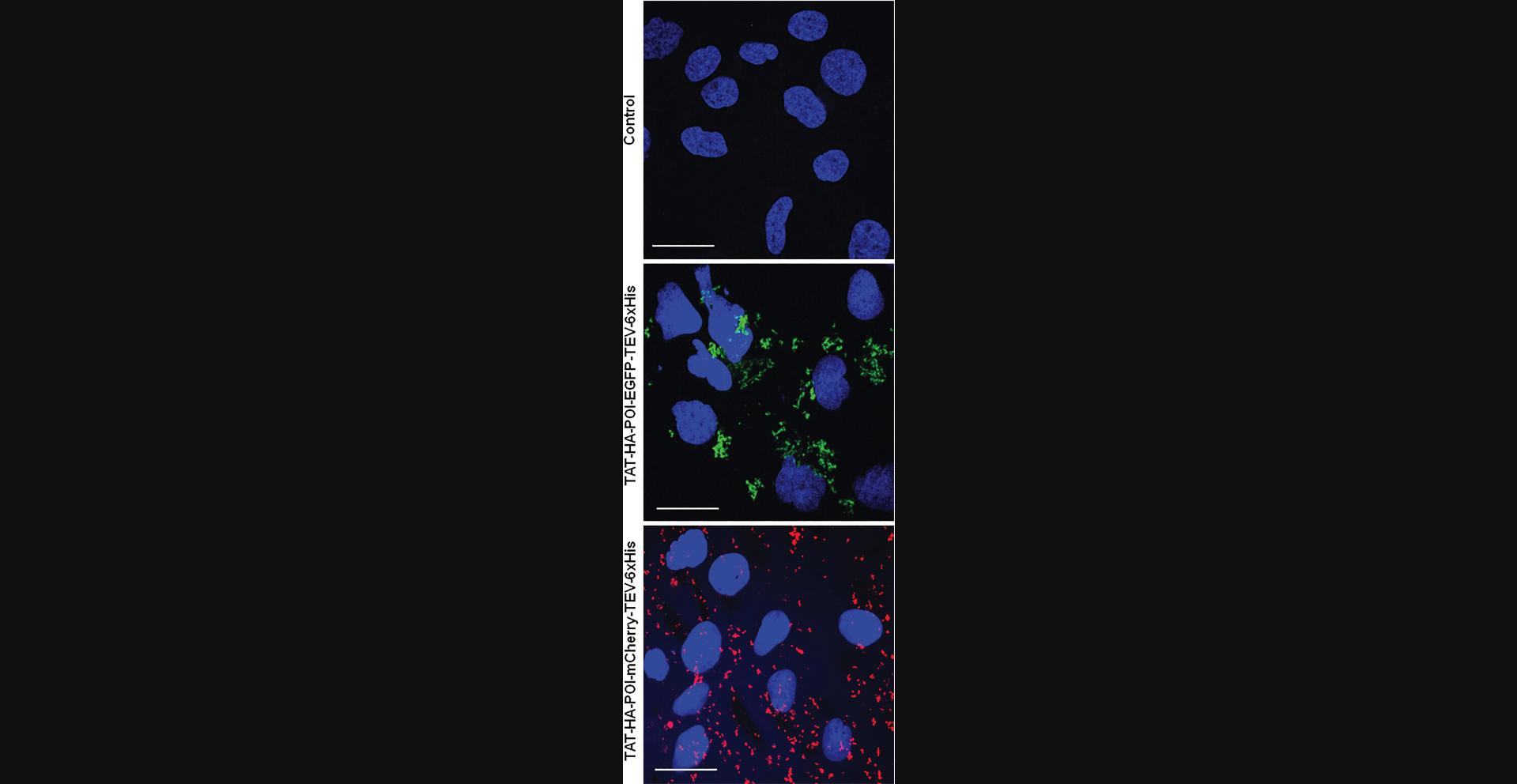
In this protocol, we describe the procedure of protein transduction within Hep3B mammalian cells. The investigation of protein entry into mammalian cells is a pivotal aspect of cellular and molecular biology, shedding light on essential physiological processes and enabling the development of targeted therapeutic strategies. This protocol delineates a fundamental approach for the transduction of proteins into mammalian cells, a process to efficiently deliver exogenous recombinant proteins. Furthermore, this protocol provides a detailed guide for monitoring the intracellular fate of transduced proteins through immunofluorescence microscopy, offering a powerful tool for visualizing and analyzing their subcellular localization and dynamics through fluorescent tags, such as EGFP and mCherry.
Materials
-
Hep3B cells (ATCC, cat. no. HB-8064)
-
Gibco minimum essential medium (MEM) (Dutscher, cat. no. L0430-50), or other cell culture medium adapted to the cell line used
-
Fetal calf serum (FCS) (Corning, cat. no. 35-079-CV)
-
2 mM L-glutamine (Gibco by Life Technologies, cat. no. 25030-024)
-
1 mM sodium pyruvate (Gibco by Life Technologies, cat. no. 11360070)
-
50 mg/L antibiotics (streptomycin, penicillin) (Gibco by Life Technologies, cat. no. 15070-063)
-
DPBS w/o Mg, w/o Ca (Dutsher, cat. no. L0615-500)
-
Trypsin-EDTA, 1× in PBS w/o Ca, w/o Mg w/o phenol red (Dutsher, cat. no. L0940-100)
-
Trypan blue solution (Sigma-Aldrich, cat. no. T8154)
-
Purified POI in DPBS, 1×
-
DAPI (4´,6-diamidino-2-phenylindole dihydrochloride) (Sigma-Aldrich, cat. no. D9542)
-
70% ethanol
-
Moviol mounting medium (see recipe)
-
Water bath
-
T25 flasks (Corning, cat. no. 430720U)
-
Humidified cell culture incubator enriched with 5% (v/v) CO2
-
Leica DM IRB optical microscope
-
T75 flasks (Corning, cat. no. 430641U)
-
Suction pump
-
15-ml sterile conical tubes (Falcon, cat. no. 352097)
-
Malassez slide
-
Cell counter
-
24-well plates (Corning, cat. no. 3526)
-
120-mm coverslips
-
4% (w/v) paraformaldehyde (PAF) (Sigma-Aldrich, cat. no. P6148)
-
Humid chamber
-
Whatman paper
-
Parafilm
-
Zeiss LSM 780 confocal Microscope
-
Fiji ImageJ
CAUTION : Work consistently under the hood (with ventilation) and always wear gloves when handling PAF due to its toxic properties as a CMR substance.
NOTE : In cell culture, it is essential to maintain a sterile working environment. Clean the biological safety cabinet (BSC) thoroughly with 70% ethanol and disinfectant solution before each manipulation or experiment. Thoroughly clean all bottles used in cell culture with 70% ethanol before handling, and always manipulate under the BSC to prevent potential contamination.
Cell culture
1.Prepare the cell culture medium by mixing MEM to 10% (v/v) FCS, 2 mM L-glutamine, 1 mM sodium pyruvate, and 50 mg/L antibiotics. Place the cell culture medium, 1× DPBS, and trypsin in a water bath at 37°C for ∼10 to 20 min to achieve the optimal temperature for cell culture maintenance.
2.Initiate the culture of Hep3B cells in a T25 flask within an incubator set at 37°C with 5% (v/v) CO2.
3.Monitor cell proliferation using an optical (visible light) microscope to ensure viability; prevent contamination before the experiment and maintain confluency while adjusting the medium if necessary.
4.Once Hep3B cells reach 70% confluence in a T75 flask, discard the cell medium.
5.Wash the cells with DPBS and discard DPBS using a suction pump.
6.Add 2 ml trypsin and place the flask in the cell incubator at 37°C until the cells detach from the flask (∼5 to 10 min). Confirm cell detachment under the optical microscope.
7.Add 4 ml cell culture medium, twice the volume of trypsin on the cell monolayer. Discard the total 6 ml and transfer it to a sterile 15-ml sterile conical tube.
8.Take a 10 µl cell sample and mix with 10 µl trypan blue (1:1 ratio).
9.Use a Malassez slide for cell counting, placing 10 µl in each chamber and scan with a cell counting machine. Calculate the volume of cells to add to the culture medium for a final volume of 1 ml per well.
Protein transduction within living cells
10.In a 24-well plate, place a 120-mm coverslip in each well. Seed the cells for 24 hr by depositing ∼37,500 cells with cell culture medium to achieve a final volume of 1 ml per well.
11.After 24 hr, the cells should be well adherent and have reached ∼ 70% confluence.
12.Discard 1 ml medium, then add 1 to 5 µM of purified TAT-HA-POI-XFP (or POI-XFP if the TEV sequence has been cleaved) onto cells with the required culture medium volume to reach a final volume of 1 ml per well. Incubate the cells at 37°C for 2 hr.
13.Discard the medium, perform three quick washes with cold 1× DPBS, and fix the cells on the coverslips by adding 500 µl of 4% (w/v) PAF per well for 30 min at room temperature.
14.Wash the cells three times with DBPS for 5 min each.
15.Meanwhile, to avoid a drying out of the coverslips, prepare a humid chamber using a support, place a Whatman paper on it, wet it with ultrapure water, and cover it with parafilm.
16.Place the coverslip in the humid chamber and add 100 µl of 1× DPBS to cover the cells and prevent drying.
17.Discard DBPS and add DAPI to stain nuclei directly to each coverslip for 10 min at room temperature in three washes of 5 min each. Clean the coverslip with 70% ethanol and let it dry.
18.Add 5 µl Moviol (mounting liquid) to the coverslip and place it directly on the Moviol so that the side containing the fixed cells faces the liquid.
19.Let the coverslip dry at room temperature overnight in the dark. The next day, the coverslip is ready for observation under a confocal microscope.
20.Capture the images by using a confocal microscope:
-
Channel 1 (DAPI), λex405 nm, λem410 to 470 nm.
-
Channel 2 (EGFP), λex488 nm, λem500 to 543 nm.
-
Channel 3 (mCherry), λex561 nm, λem570-615 nm.
21.Images can be analyzed by using Fiji ImageJ software (Fig. 8).
REAGENTS AND SOLUTIONS
Blocking buffer
- 5% (w/v) non-fatty-acid milk (for food use) or BSA (bovine serum albumin) (Sigma-Aldrich, cat. no. A9647) in TBS-Tween buffer (see recipe)
- Mix well
- Prepare fresh
Buffer A
- 3.15 g Tris (20 mM final; Euromedex, cat. no. EU001B)
- 17.53 g NaCl (300 mM final; Carlo Erba, cat. no. 479685)
- 1.36 g imidazole (20 mM final; Sigma-Aldrich, cat. no. I202)
- Adjust to 1 L with ultrapure H2O
- Adjust to pH 8.0 with HCl (37% w/v) (Carlo Erba, cat. no. 403871)
- Mix well and filter using a 0.22-µm filter
- Store up to 1 month at 4°C
Buffer B
- 3.15 g Tris (20 mM final; Euromedex, cat. no. EU001B)
- 17.53 g NaCl (300 mM final; Carlo Erba, cat. no. 479685)
- 34 g imidazole (500 mM final; Sigma-Aldrich, cat. no. I202)
- Adjust to 1 L with ultrapure H2O
- Adjust to pH 8.0 with HCl (37% w/v) (Carlo Erba, cat. no. 403871)
- Mix well and filter using a 0.22-µm filter
- Store up to 1 month at 4°C
Laemmli buffer, 5×
- 12.11 g Tris (250 mM final; Euromedex, cat. no. EU001B)
- 5% (w/v) SDS (Sigma-Aldrich, cat. no. L3771)
- 5% (v/v) 2-mercaptoethanol (Sigma-Aldrich, cat. no. M3148)
- 40% (v/v) glycerol (Sigma-Aldrich, cat. no. G9012)
- 0.02% (w/v) bromophenol blue (Sigma-Aldrich, cat. no. B0126)
- Bring up to 1 L with ultrapure H2O
- Adjust to pH 8.0 with HCl (37% w/v) (Carlo Erba, cat. no. 403871)
- Mix well
- Store at up to 1 month at 4°C
Polyacrylamide separating gel, 8%
- 3.75 ml of 1.5 M Tris·HCl pH 8.8 (Current Protocols, 2006)
- 50 µl of 20% (w/v) SDS (Sigma-Aldrich, cat. no. L3771)
- 2 ml of 40% (w/v) acrylamide/bis-acrylamide (ratio 29:1) (Sigma-Aldrich, cat. no. A2792)
- 30 µl Temed (N ,N ,N’ ,N’ -tetramethylethylenediamine) (Sigma-Aldrich, cat. no. T9281)
- 30 µl of 10% (w/v) ammonium persulfate (Sigma-Aldrich, cat. no. A7460)
- Bring up to 10 ml with ultrapure H2O
- Mix well
- Prepare fresh
Polyacrylamide stacking gel, 4%
- 1.5 ml of 0.5 M Tris·HCl pH 6.8 (Current Protocols, 2006)
- 50 µl of 20% (w/v) SDS (Sigma-Aldrich, cat. no. L3771)
- 1 ml of 40% (w/v) acrylamide/bis-acrylamide (ratio 29:1) (Sigma-Aldrich, cat. no. A2792)
- 60 µl Temed (N ,N ,N’ ,N’ -tetramethylethylenediamine) (Sigma-Aldrich, cat. no. T9281)
- 50 µl of 10% (w/v) ammonium persulfate (Sigma-Aldrich, cat. no. A7460)
- Bring up to 10 ml with ultrapure H2O
- Mix well
- Prepare fresh
Mowiol mounting medium
- 6 g glycerol
- 2.4 g mowiol 4-88 (biovalley, cat. no. 17951-500)
- 6 ml distilled H2O
- 12 ml 0.2 M Tris·HCl pH 8.5 (Current Protocols, 2006)
- Mix well and centrifuge 10 min at 1500 × g , room temperature, to clarify the solution
- Store up to 1 month at 4°C or 1 year at −20°C
TBS-Tween buffer
- 4.2 g Tris (15 mM final; Euromedex, cat. no. EU001B)
- 8.1 g NaCl (300 mM final; Carlo Erba, cat. no. 479685)
- 0.05% (v/v) Tween 20 (Fisher Scientific, cat. no. M12247)
- Adjust to 1 L with distilled H2O
- Adjust to pH 8.0 with HCl (37% w/v) (Carlo Erba, cat. no. 403871)
- Mix well
- Store up to 1 week at room temperature
COMMENTARY
Critical Parameters
Several steps in this experimental strategy are critical and therefore can greatly influence its success. It is important to correctly design the primers so that the different PCRs that precede the DNA molecular assembly are as specific as possible to generate a single band visualized on the agarose gel (Basic Protocol 1). This step is fundamental to getting off to a good start. Another crucial point is the bacteria subculturing stage. It is important to pick well-isolated bacterial colonies back into liquid culture so as to not mix different clones (Basic Protocol 1). The last step concerning the transduction of cells is relatively simple because the use of penetration peptides makes it possible to avoid many of the steps inherent in the transfection of plasmid vectors (Basic Protocol 3). Nevertheless, it is important here to test variable quantities of proteins to be transduced to optimize their subsequent study. If the POI is not used quickly after its purification, it is extremely important to store it in suitable conditions. Nevertheless, we encourage users to extemporaneously prepare the protein to transduce it immediately after purification (see Table 8). Other potential critical parameters are listed in Table 8.
| Problem | Possible Cause | Solution |
|---|---|---|
| Poor protein stability | Inappropriate storage conditions or buffer | Store protein at recommended temperature; optimize buffer composition; add proteases inhibitors to buffers |
| Low protein yield | Inefficient cell lysis or purification method | Optimize lysis conditions; consider alternative purification methods |
| Contaminants in elution | Carryover from previous steps or column issues | Ensure proper column equilibration; optimize washing steps |
| Low/no signal in western blot results | Poor antibody binding | Ensure primary and secondary antibodies are compatible and correctly diluted |
| High background or low ratio expected signal/background in western blot | Antibodies too much interact with the membrane or with the blocking reagent | Increase the time and the number of washings; change the blocking reagent; increase the dilution of the primary or/and the secondary antibodies |
| Weak or no fluorescence signal | Too low protein concentration in contact with the cell | Increase the concentration of the protein |
| Cell autofluorescence | Presence of residual medium or contaminants | Ensure thorough washing after treatment |
| Protein aggregation | Improper storage conditions or handling | Store protein appropriately; handle with care |
Troubleshooting Table
Table 8 lists the most common problems that may be encountered during protein production detailed in this article (Table 8).
Statistical Analysis
GraphPad Prism 9 or any other statistics tool can be used for calculating the standard deviations of the growth curves of bacteria, as those shown in Figure 4.
Understanding Results
Basic Protocols 1 and 2
The data obtained by Basic Protocols 1 and 2 generate agarose gel electrophoretic profiles to assess the quantity and quality of the PCR products, and polyacrylamide gel profiles that can be visualized either by treatment of the gels with a solution of Coomassie blue or by electro-blotting of the resolved proteins by SDS-PAGE onto nitrocellulose. These experiments require carefully supporting the samples to be analyzed with size markers for agarose gels and molecular mass markers for polyacrylamide gels; be careful not to confuse the types of markers, the first being expressed in base pairs (bp), the second in kilodaltons (kDa).
The presentation of the data requires labeling the figures by clearly identifying at the top of the gels the samples loaded on the left or the right lateral side, and the values corresponding to each mark of size (bp) or molecular mass (kDa). By comparing the migration distance of the band corresponding to the PCR product or to the protein produced, it is possible to prove not only that the product is indeed present, but also to estimate its abundance and purity. The western blot provides an advantage since the use of antibodies guarantees an additional argument simply because the apparent molecular mass is indeed what is expected. Make sure to check the supplier's information sheet whether these antibodies can be used for western blotting. Some antibodies are more suitable for immunoprecipitation, ELISA, or immunofluorescence since they recognize proteins in their native form while others are better for western blot because they were generated against a short peptide sequence. This is the case for the latter in this series of protocols, we used antibodies directed against tag “labels” (HA and 6×His), which generally guarantees their use in western blotting, while anti-HA antibodies are better for western blotting than anti-6×His antibodies preferentially used for protein purification. The problem of non-recognition can, however, be encountered with anti-POI antibodies. In fact, in Figure 3 and Figure 7C-D, we can consider on the one hand the good purity of the different PCR products (this must be ensured before moving on to the molecular cloning stage, which will be more perilous if the PCRs are not sufficiently pure) and the protein thus produced. It is common to observe, as we do in Figure 7C-D, bands of molecular mass lower than the expected band; these are generally proteolytic degradation bands generated during production and purification. In our case it turns out that the mCherry form of our POI is less stable than the GFP form (it may be useful in this case to use larger quantities of protease inhibitors during the process).
Basic Protocol 3
Regarding images obtained by microscopy in confocal mode (Basic Protocol 3), they must be clearly labeled by indicating the corresponding condition. It is very useful to merge with the DAPI image, which allows to locate the POI at a glance. Our microscopy images show a cytoplasmic distribution of our POI in both the EGFP and mCherry versions. Finally, a scale bar (20 µm in our experience) makes it possible to judge the distances on the microscopy images. It is common and even advisable to show separate images and merge for immunofluorescence microscopy experiments.
Time Considerations
Total time required to complete the full experimental strategy: 3 to 4 weeks.
Basic Protocol 1 (7 days)
Amplification of the various PCR products
Samples preparation: 10 min PCR: 3 hr Agarose gel preparation and polymerization: 30 min Agarose gel electrophoresis: 30 min Ethidium bromide staining: 15 min Bands visualization by using UV transilluminator: 10 min Bands excision by using UV transilluminator: 15 min PCR gel purification: 1 hr Total: 1 day
DNA molecular assembly
Plasmid assembly procedure: 1 hr Transformation and selection procedure (E. coli 5-alpha): 1 days DNA extraction and purification: 1 hr DNA sequencing: 2 days Total: 4 days
Transformation and selection procedure in E. coli BL21(DE3)
Total: 1 day
Recombinant protein expression
Total: 1 day
Basic Protocol 2 (5 days)
Protein fractionation
Total: 1 day
Protein desalting, concentration, and quantification
Total: 3 days
Analysis of recombinant protein expression by SDS-PAGE
Coomassie blue staining Western blot Total: 1 day
Basic Protocol 3 (3 days)
Cell seeding
Total: 1 day
Protein transduction
Protein transduction: 2 hr Slide and coverslip preparation Total: 1 day
Data collection and analysis
Immunofluorescence microscopy: data collecting Data analysis: 1 to 2 hr Total: 1 day
Acknowledgments
The authors are grateful to the European Union's Horizon 2020 research and innovation program that funded this work (Marie Sklodowska-Curie grant agreement No 847568), the Centre National de la Recherche Scientifique (CNRS) and the University of Lille for their recurrent fundings, and the Plateformes Lilloises en Biologie et Santé (PLBS) for help in using fluorescent microscopy.
Author Contributions
Christer Abou Anny : Conceptualization; data curation; formal analysis; methodology; writing original draft. Sébastien Nouaille : Conceptualization; investigation; methodology; supervision; validation; writing review and editing. Régis Fauré : Conceptualization; investigation; methodology; supervision; validation; writing review and editing. Céline Schulz : Conceptualization; formal analysis; methodology; writing review and editing. Corentin Spriet : Conceptualization; methodology; writing review and editing. Isabelle Huvent : Conceptualization; investigation; methodology; supervision; writing review and editing. Christophe Biot : Conceptualization; formal analysis; funding acquisition; investigation; resources; supervision; validation; writing review and editing. Tony Lefebvre : Conceptualization; formal analysis; funding acquisition; investigation; project administration; resources; supervision; writing original draft.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created in this study.
Literature Cited
- Ai, H., Baird, M. A., Shen, Y., Davidson, M. W., & Campbell, R. E. (2014). Engineering and characterizing monomeric fluorescent proteins for live-cell imaging applications. Nature Protocols , 9(4), 910–928. https://doi.org/10.1038/nprot.2014.054
- Andersen, D. C., & Krummen, L. (2002). Recombinant protein expression for therapeutic applications. Current Opinion in Biotechnology , 13(2), 117–123. https://doi.org/10.1016/S0958-1669(02)00300-2
- MacKay, A. J., & Szoka, F. C. Jr. (2003). HIV TAT protein transduction domain mediated cell binding and intracellular delivery of nanoparticles. Journal of Dispersion Science and Technology , 24(3–4), 465–473. https://doi.org/10.1081/DIS-120021802
- Anné, J., Maldonado, B., Van Impe, J., Van Mellaert, L., & Bernaerts, K. (2012). Recombinant protein production and streptomycetes. Journal of Biotechnology , 158(4), 159–167. https://doi.org/10.1016/j.jbiotec.2011.06.028
- Assenberg, R., Wan, P. T., Geisse, S., & Mayr, L. M. (2013). Advances in recombinant protein expression for use in pharmaceutical research. Current Opinion in Structural Biology , 23(3), 393–402. https://doi.org/10.1016/j.sbi.2013.03.008
- Baneyx, F. (1999). Recombinant protein expression in Escherichia coli. Current Opinion in Biotechnology , 10(5), 411–421. https://doi.org/10.1016/S0958-1669(99)00003-8
- Baneyx, F., & Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnology , 22(11), 1399–1408. https://doi.org/10.1038/nbt1029
- Barnes, L. M., Bentley, C. M., & Dickson, A. J. (2003). Stability of protein production from recombinant mammalian cells. Biotechnology and Bioengineering , 81(6), 631–639. https://doi.org/10.1002/bit.10517
- Becker-Hapak, M., McAllister, S. S., & Dowdy, S. F. (2001). TAT-mediated protein transduction into mammalian cells. Methods , 24(3), 247–256. https://doi.org/10.1006/meth.2001.1186
- Block, H., Maertens, B., Spriestersbach, A., Brinker, N., Kubicek, J., Fabis, R., Labahn, J., & Schäfer, F. (2009). Immobilized-metal affinity chromatography (IMAC): A review. Methods in Enzymology , 463, 439–473. https://doi.org/10.1016/S0076-6879(09)63027-5
- Bordat, A., Houvenaghel, M.-C., & German-Retana, S. (2015). Gibson assembly: An easy way to clone potyviral full-length infectious cDNA clones expressing an ectopic VPg. Virology Journal , 12(1), 89. https://doi.org/10.1186/s12985-015-0315-3
- Brondyk, W. H. (2009). Selecting an appropriate method for expressing a recombinant protein. Methods in Enzymology , 463, 131–147. https://doi.org/10.1016/S0076-6879(09)63011-1
- Burnett, M. J. B., & Burnett, A. C. (2020). Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants, People, Planet , 2(2), 121–132. https://doi.org/10.1002/ppp3.10073
- Casini, A., Storch, M., Baldwin, G. S., & Ellis, T. (2015). Bricks and blueprints: Methods and standards for DNA assembly. Nature Reviews Molecular Cell Biology , 16(9), 568–576. https://doi.org/10.1038/nrm4014
- Chen, R. (2012). Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnology Advances , 30(5), 1102–1107. https://doi.org/10.1016/j.biotechadv.2011.09.013
- Cohen, S. N. (2013). DNA cloning: A personal view after 40 years. Proceedings of the National Academy of Sciences , 110(39), 15521–15529. https://doi.org/10.1073/pnas.1313397110
- Cregg, J. M., Cereghino, J. L., Shi, J., & Higgins, D. R. (2000). Recombinant protein expression in Pichia pastoris. Molecular Biotechnology , 16(1), 23–52. https://doi.org/10.1385/MB:16:1:23
- Current Protocols. (2006). Commonly Used Reagents. Current Protocols in Microbiology , 00, A.2A.1–A.2A.15. https://doi.org/10.1002/9780471729259.mca02as00
- Eguchi, A., Akuta, T., Okuyama, H., Senda, T., Yokoi, H., Inokuchi, H., Fujita, S., Hayakawa, T., Takeda, K., Hasegawa, M., & Nakanishi, M. (2001). Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. Journal of Biological Chemistry , 276(28), 26204–26210. https://doi.org/10.1074/jbc.M010625200
- Farzaneh, M., Hassani, S.-N., Mozdziak, P., & Baharvand, H. (2017). Avian embryos and related cell lines: A convenient platform for recombinant proteins and vaccine production. Biotechnology Journal , 12(5), 1600598. https://doi.org/10.1002/biot.201600598
- Ferro, M. M. M., Ramos-Sobrinho, R., Xavier, C. A. D., Zerbini, F. M., Lima, G. S. A., Nagata, T., & Assunção, I. P. (2019). New approach for the construction of infectious clones of a circular DNA plant virus using Gibson assembly. Journal of Virological Methods , 263, 20–23. https://doi.org/10.1016/j.jviromet.2018.10.017
- Fittipaldi, A., & Giacca, M. (2005). Transcellular protein transduction using the Tat protein of HIV-1. Advanced Drug Delivery Reviews , 57(4), 597–608. https://doi.org/10.1016/j.addr.2004.10.011
- Ford, K. G., Souberbielle, B. E., Darling, D., & Farzaneh, F. (2001). Protein transduction: An alternative to genetic intervention? Gene Therapy , 8(1), 1–4. https://doi.org/10.1038/sj.gt.3301383
- Froger, A., & Hall, J. E. (2007). Transformation of plasmid DNA into E. coli using the heat shock method. Journal of Visualized Experiments , 6, e253. https://doi.org/10.3791/253
- Gaberc-Porekar, V., & Menart, V. (2001). Perspectives of immobilized-metal affinity chromatography. Journal of Biochemical and Biophysical Methods , 49(1), 335–360. https://doi.org/10.1016/S0165-022X(01)00207-X
- Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., & Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods , 6(5), 343–345. https://doi.org/10.1038/nmeth.1318
- Gopal, G. J., & Kumar, A. (2013). Strategies for the production of recombinant protein in Escherichia coli. The Protein Journal , 32(6), 419–425. https://doi.org/10.1007/s10930-013-9502-5
- Greenblatt, J., & Schleif, R. (1971). Arabinose C protein: Regulation of the arabinose operon in vitro. Nature New Biology , 233(40), 166–170. https://doi.org/10.1038/newbio233166a0
- Gump, J. M., & Dowdy, S. F. (2007). TAT transduction: The molecular mechanism and therapeutic prospects. Trends in Molecular Medicine , 13(10), 443–448. https://doi.org/10.1016/j.molmed.2007.08.002
- Gump, J. M., June, R. K., & Dowdy, S. F. (2010). Revised role of glycosaminoglycans in TAT protein transduction domain-mediated cellular transduction. Journal of Biological Chemistry , 285(2), 1500–1507. https://doi.org/10.1074/jbc.M109.021964
- Guo, Q., Zhao, G., Hao, F., & Guan, Y. (2012). Effects of the TAT peptide orientation and relative location on the protein transduction efficiency. Chemical Biology & Drug Design, 79(5), 683–690. https://doi.org/10.1111/j.1747-0285.2011.01315.x
- Helmuth, J. A., Burckhardt, C. J., Greber, U. F., & Sbalzarini, I. F. (2009). Shape reconstruction of subcellular structures from live cell fluorescence microscopy images. Journal of Structural Biology , 167(1), 1–10. https://doi.org/10.1016/j.jsb.2009.03.017
- Hopkins, R. F., Wall, V. E., & Esposito, D. (2012). Optimizing transient recombinant protein expression in mammalian cells. Methods in Molecular Biology , 801, 251–268. https://doi.org/10.1007/978-1-61779-352-3_16
- Jäger, V., Büssow, K., & Schirrmann, T. (2015). Transient recombinant protein expression in mammalian cells. In M. Al-Rubeai (Ed.), Animal Cell Culture. Cell Engineering (Vol. 9, pp. 27–64). Springer. https://doi.org/10.1007/978-3-319-10320-4_2
- Jakobs, S., Subramaniam, V., Schönle, A., Jovin, T. M., & Hell, S. W. (2000). EGFP and DsRed expressing cultures of Escherichia coli imaged by confocal, two-photon and fluorescence lifetime microscopy. FEBS Letters , 479(3), 131–135. https://doi.org/10.1016/S0014-5793(00)01896-2
- Jang, C.-W., & Magnuson, T. (2013). A novel selection marker for efficient DNA cloning and recombineering in E. coli. PLoS ONE , 8(2), e57075. https://doi.org/10.1371/journal.pone.0057075
- Joung, Y. H., Park, S. H., Moon, K.-B., Jeon, J.-H., Cho, H.-S., & Kim, H.-S. (2016). The last ten years of advancements in plant-derived recombinant vaccines against Hepatitis B. International Journal of Molecular Sciences , 17(10), 1715. https://doi.org/10.3390/ijms17101715
- Kabouridis, P. S. (2003). Biological applications of protein transduction technology. Trends in Biotechnology , 21(11), 498–503. https://doi.org/10.1016/j.tibtech.2003.09.008
- Kaplan, I. M., Wadia, J. S., & Dowdy, S. F. (2005). Cationic TAT peptide transduction domain enters cells by macropinocytosis. Journal of Controlled Release , 102(1), 247–253. https://doi.org/10.1016/j.jconrel.2004.10.018
- Kirchhausen, T. (2009). Imaging endocytic clathrin structures in living cells. Trends in Cell Biology , 19(11), 596–605. https://doi.org/10.1016/j.tcb.2009.09.002
- Kostylev, M., Otwell, A. E., Richardson, R. E., & Suzuki, Y. (2015). Cloning should be simple: Escherichia coli DH5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS ONE , 10(9), e0137466. https://doi.org/10.1371/journal.pone.0137466
- Leifert, J. A., Harkins, S., & Whitton, J. L. (2002). Full-length proteins attached to the HIV tat protein transduction domain are neither transduced between cells, nor exhibit enhanced immunogenicity. Gene Therapy , 9(21), 1422–1428. https://doi.org/10.1038/sj.gt.3301819
- Li, C., Ji, C., Huguet-Tapia, J. C., White, F. F., Dong, H., & Yang, B. (2019). An efficient method to clone TAL effector genes from Xanthomonas oryzae using Gibson assembly. Molecular Plant Pathology , 20(10), 1453–1462. https://doi.org/10.1111/mpp.12820
- Li, L., Jiang, W., & Lu, Y. (2018). A modified Gibson assembly method for cloning large DNA fragments with high GC contents. Methods in Molecular Biology , 1671, 203–209. https://doi.org/10.1007/978-1-4939-7295-1_13
- Li, Z.-N., Jelkmann, W., Sun, P., & Zhang, L. (2020). Construction of full-length infectious cDNA clones of apple stem grooving virus using Gibson assembly method. Virus Research , 276, 197790. https://doi.org/10.1016/j.virusres.2019.197790
- Mattanovich, D., Branduardi, P., Dato, L., Gasser, B., Sauer, M., & Porro, D. (2012). Recombinant protein production in yeasts. Methods in Molecular Biology , 824, 329–358. https://doi.org/10.1007/978-1-61779-433-9_17
- McDonald, D., Vodicka, M. A., Lucero, G., Svitkina, T. M., Borisy, G. G., Emerman, M., & Hope, T. J. (2002). Visualization of the intracellular behavior of HIV in living cells. Journal of Cell Biology , 159(3), 441–452. https://doi.org/10.1083/jcb.200203150
- McRae, S. R., Brown, C. L., & Bushell, G. R. (2005). Rapid purification of EGFP, EYFP, and ECFP with high yield and purity. Protein Expression and Purification , 41(1), 121–127. https://doi.org/10.1016/j.pep.2004.12.030
- Newman, R. H., Fosbrink, M. D., & Zhang, J. (2011). Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chemical Reviews , 111(5), 3614–3666. https://doi.org/10.1021/cr100002u
- Nienhaus, K., & Nienhaus, G. U. (2014). Fluorescent proteins for live-cell imaging with super-resolution. Chemical Society Reviews , 43(4), 1088–1106. https://doi.org/10.1039/C3CS60171D
- Noguchi, H., & Matsumoto, S. (2006). Protein transduction technology: A novel therapeutic perspective. Acta Medica Okayama , 60(1), 1–11. http://doi.org/10.18926/AMO/30757
- Nolte, C., Matyash, M., Pivneva, T., Schipke, C. G., Ohlemeyer, C., Hanisch, U.-K., Kirchhoff, F., & Kettenmann, H. (2001). GFAP promoter-controlled EGFP-expressing transgenic mice: A tool to visualize astrocytes and astrogliosis in living brain tissue. Glia , 33(1), 72–86.
- Norris, A. D., Kim, H.-M., Colaiácovo, M. P., & Calarco, J. A. (2015). Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics , 201(2), 449–458. https://doi.org/10.1534/genetics.115.180679
- Overton, T. W. (2014). Recombinant protein production in bacterial hosts. Drug Discovery Today , 19(5), 590–601. https://doi.org/10.1016/j.drudis.2013.11.008
- Pawson, T., & Nash, P. (2000). Protein-protein interactions define specificity in signal transduction. Genes & Development, 14(9), 1027–1047. https://doi.org/10.1101/gad.14.9.1027
- Porath, J. (1992). Immobilized metal ion affinity chromatography. Protein Expression and Purification , 3(4), 263–281. https://doi.org/10.1016/1046-5928(92)90001-D
- Porro, D., Sauer, M., Branduardi, P., & Mattanovich, D. (2005). Recombinant protein production in yeasts. Molecular Biotechnology , 31(3), 245–259. https://doi.org/10.1385/MB:31:3:245
- Rizzuto, R., Brini, M., Pizzo, P., Murgia, M., & Pozzan, T. (1995). Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Current Biology , 5(6), 635–642. https://doi.org/10.1016/S0960-9822(95)00128-X
- Roberts, M. A. J. (2019). Recombinant DNA technology and DNA sequencing. Essays in Biochemistry , 63(4), 457–468. https://doi.org/10.1042/EBC20180039
- Rosano, G. L., & Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli : Advances and challenges. Frontiers in Microbiology , 5, 172. https://www.frontiersin.org/articles/10.3389/fmicb.2014.00172
- Rudenko, O., & Barnes, A. C. (2018). Gibson assembly facilitates bacterial allelic exchange mutagenesis. Journal of Microbiological Methods , 144, 157–163. https://doi.org/10.1016/j.mimet.2017.11.023
- Salomonnson, E., Mihalko, L. A., Luker, K. E., Verkhusha, V. V., & Luker, G. D. (2012). Cell-based and in vivo spectral analysis of fluorescent proteins for multiphoton microscopy. Journal of Biomedical Optics , 17(9), 096001. https://doi.org/10.1117/1.JBO.17.9.096001
- Sands, B., & Brent, R. (2016). Overview of post Cohen-Boyer methods for single segment cloning and for multisegment DNA assembly. Current Protocols in Molecular Biology , 113(1), 3.26.1–3.26.20. https://doi.org/10.1002/0471142727.mb0326s113
- Schleif, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics , 16(12), 559–565. https://doi.org/10.1016/S0168-9525(00)02153-3
- Schleif, R., Hess, W., Finkelstein, S., & Ellis, D. (1973). Induction kinetics of the L-arabinose operon of Escherichia coli. Journal of Bacteriology , 115(1), 9–14. https://doi.org/10.1128/jb.115.1.9-14.1973
- Schwarze, S. R., Hruska, K. A., Dowdy, S. F., Schwarze, S. R., Hruska, K. A., & Dowdy, S. F. (2000). Protein transduction: Unrestricted delivery into all cells? Trends in Cell Biology , 10(7), 290–295. https://doi.org/10.1016/S0962-8924(00)01771-2
- Shen, Y., Chen, Y., Wu, J., Shaner, N. C., & Campbell, R. E. (2017). Engineering of mCherry variants with long Stokes shift, red-shifted fluorescence, and low cytotoxicity. PLoS ONE , 12(2), e0171257. https://doi.org/10.1371/journal.pone.0171257
- Shokolenko, I. N., Alexeyev, M. F., LeDoux, S. P., & Wilson, G. L. (2005). TAT-mediated protein transduction and targeted delivery of fusion proteins into mitochondria of breast cancer cells. DNA Repair , 4(4), 511–518. https://doi.org/10.1016/j.dnarep.2004.11.009
- Sørensen, H. P., & Mortensen, K. K. (2005). Advanced genetic strategies for recombinant protein expression in Escherichia coli. Journal of Biotechnology , 115(2), 113–128. https://doi.org/10.1016/j.jbiotec.2004.08.004
- Subach, F. V., Patterson, G. H., Manley, S., Gillette, J. M., Lippincott-Schwartz, J., & Verkhusha, V. V. (2009). Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nature Methods , 6(2), 153–159. https://doi.org/10.1038/nmeth.1298
- Sun, X., Chiu, J.-F., & He, Q.-Y. (2005). Application of immobilized metal affinity chromatography in proteomics. Expert Review of Proteomics , 2(5), 649–657. https://doi.org/10.1586/14789450.2.5.649
- Sunley, K., & Butler, M. (2010). Strategies for the enhancement of recombinant protein production from mammalian cells by growth arrest. Biotechnology Advances , 28(3), 385–394. https://doi.org/10.1016/j.biotechadv.2010.02.003
- Thomas, S., Maynard, N. D., & Gill, J. (2015). DNA library construction using Gibson assembly®. Nature Methods , 12(11), i–ii. https://doi.org/10.1038/nmeth.f.384
- Wadia, J. S., & Dowdy, S. F. (2002). Protein transduction technology. Current Opinion in Biotechnology , 13(1), 52–56. https://doi.org/10.1016/S0958-1669(02)00284-7
- Wang, J. W., Wang, A., Li, K., Wang, B., Jin, S., Reiser, M., & Lockey, R. F. (2015). CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques , 58(4), 161–170. https://doi.org/10.2144/000114261
- Wang, Y., Shyy, J. Y. J., & Chien, S. (2008). Fluorescence proteins, live-cell imaging, and mechanobiology: Seeing is believing. Annual Review of Biomedical Engineering , 10(1), 1–38. https://doi.org/10.1146/annurev.bioeng.010308.161731
- Wiedenmann, J., Oswald, F., & Nienhaus, G. U. (2009). Fluorescent proteins for live cell imaging: Opportunities, limitations, and challenges. IUBMB Life , 61(11), 1029–1042. https://doi.org/10.1002/iub.256
- Wurm, F. M. (2004). Production of recombinant protein therapeutics in cultivated mammalian cells. Nature Biotechnology , 22(11), 1393–1398. https://doi.org/10.1038/nbt1026
- Yin, J., Zhu, D., Zhang, Z., Wang, W., Fan, J., Men, D., Deng, J., Wei, H., Zhang, X.-E., & Cui, Z. (2013). Imaging of mRNA–protein interactions in live cells using novel mCherry trimolecular fluorescence complementation systems. PLoS ONE , 8(11), e80851. https://doi.org/10.1371/journal.pone.0080851
- Zhang, H., Ma, Y., Gu, J., Liao, B., Li, J., Wong, J., & Jin, Y. (2012). Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials , 33(20), 5047–5055. https://doi.org/10.1016/j.biomaterials.2012.03.061

