SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup
Disha Bhavsar, Disha Bhavsar, Florian Krammer, Daniel Stadlbauer, Daniel Stadlbauer, Fatima Amanat, Fatima Amanat, Veronika Chromikova, Veronika Chromikova, Kaijun Jiang, Kaijun Jiang, Shirin Strohmeier, Shirin Strohmeier, Guha Asthagiri Arunkumar, Guha Asthagiri Arunkumar, Jessica Tan, Jessica Tan, Christina Capuano, Christina Capuano, Ericka Kirkpatrick, Philip Meade, Ruhi Nichalle Brito, Catherine Teo, Meagan McMahon, Viviana Simon
Abstract
In late 2019, cases of atypical pneumonia were detected in China. The etiological agent was quickly identified as a betacoronavirus (named SARS-CoV-2), which has since caused a pandemic. Several methods allowing for the specific detection of viral nucleic acids have been established, but these only allow detection of the virus during a short period of time, generally during acute infection. Serological assays are urgently needed to conduct serosurveys, to understand the antibody responses mounted in response to the virus, and to identify individuals who are potentially immune to re-infection. Here we describe a detailed protocol for expression of antigens derived from the spike protein of SARS-CoV-2 that can serve as a substrate for immunological assays, as well as a two-stage serological enzyme-linked immunosorbent assay (ELISA). These assays can be used for research studies and for testing in clinical laboratories. © 2020 The Authors. Current Protocols in Microbiology published by Wiley Periodicals LLC.
Basic Protocol 1 : Mammalian cell transfection and protein purification
Basic Protocol 2 : A two-stage ELISA for high-throughput screening of human serum samples for antibodies binding to the spike protein of SARS-CoV-2
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COronaVIrus Disease 2019 (COVID19; often written COVID-19), emerged in late 2019 in Wuhan, China (Wu et al., 2020; Zhu et al., 2020). Rapid, global spread of the virus is presently causing a pandemic. Currently, no drugs or antivirals are available and countermeasures are limited to non-pharmaceutical interventions (NPIs). Nucleic acid−based tests for detection of the virus during acute disease are in use worldwide (Chu et al., 2020; Corman et al., 2020). However, the development of serological assays is lagging due to lack of suitable reagents. Serological assays are needed to perform serosurveys aimed at determining the real infection rate and infection fatality rate in a given population. Furthermore, they are useful to characterize the immune response to the virus in a detailed qualitative and quantitative manner. Serological assays are also of immediate practical use. They can be used to identify individuals who were infected (including severe, mild, and asymptomatic cases) and who are now potentially immune. A recent study in non-human primates showed that re-infection, at least in the small number of animals used in the study, does not occur (Bao et al., 2020) once antibody responses have been mounted. Infection with coronaviruses circulating in human populations, such as HKU, NL63, etc., also leads to immunity that protects from re-infection for months to years (Callow, Parry, Sergeant, & Tyrrell, 1990). Therefore, individuals who have mounted an immune response to SARS-CoV-2 are likely immune, which means that they are unlikely to transmit the virus to others. As an example, healthcare workers who are immune could potentially care for COVID19 patients with minimal risk to themselves, their colleagues, and other patients. In addition, the use of convalescent plasma may serve as a valuable treatment option for patients with severe COVID19, especially in the absence of other options. A serological assay is critical for identifying potential plasma donors.
The surface glycoprotein of the virus, termed the spike (S) protein, mediates attachment of the virus to human cells via its receptor-binding domain (RBD; Wrapp et al., 2020) and mediates fusion of viral and cellular membranes. Antibodies binding to the spike protein, and especially to the RBD domain, can neutralize SARS-CoV-2. Therefore, we used different recombinant spike protein preparations as the antigens for our ELISA. We reported in our earlier work that individuals not exposed to SARS-CoV-2 are completely naïve to the spike protein, and their serum samples show little or no reactivity in an ELISA (Amanat et al., 2020). It is, therefore, easy to distinguish between exposed/immune and naïve individuals.
In this report, we provide detailed protocols for expressing the required antigen(s) (Basic Protocol 1) as well as setting up the ELISA that we have developed (Basic Protocol 2). An overview of these protocols is shown in Figure 1. We believe that these protocols will be useful not only for research laboratories around the globe, but also for testing in diagnostic/clinical laboratories. The described protocol setup works well for us, but it can easily be modified, adapted to local needs, and improved by the research community in the future. Not every aspect of these protocols has been optimized in detail, and we provide notes and comments whenever further optimizations and testing are recommended. Mammalian expression plasmids for the generation of the recombinant proteins are available from the corresponding author and from BEI Resources.
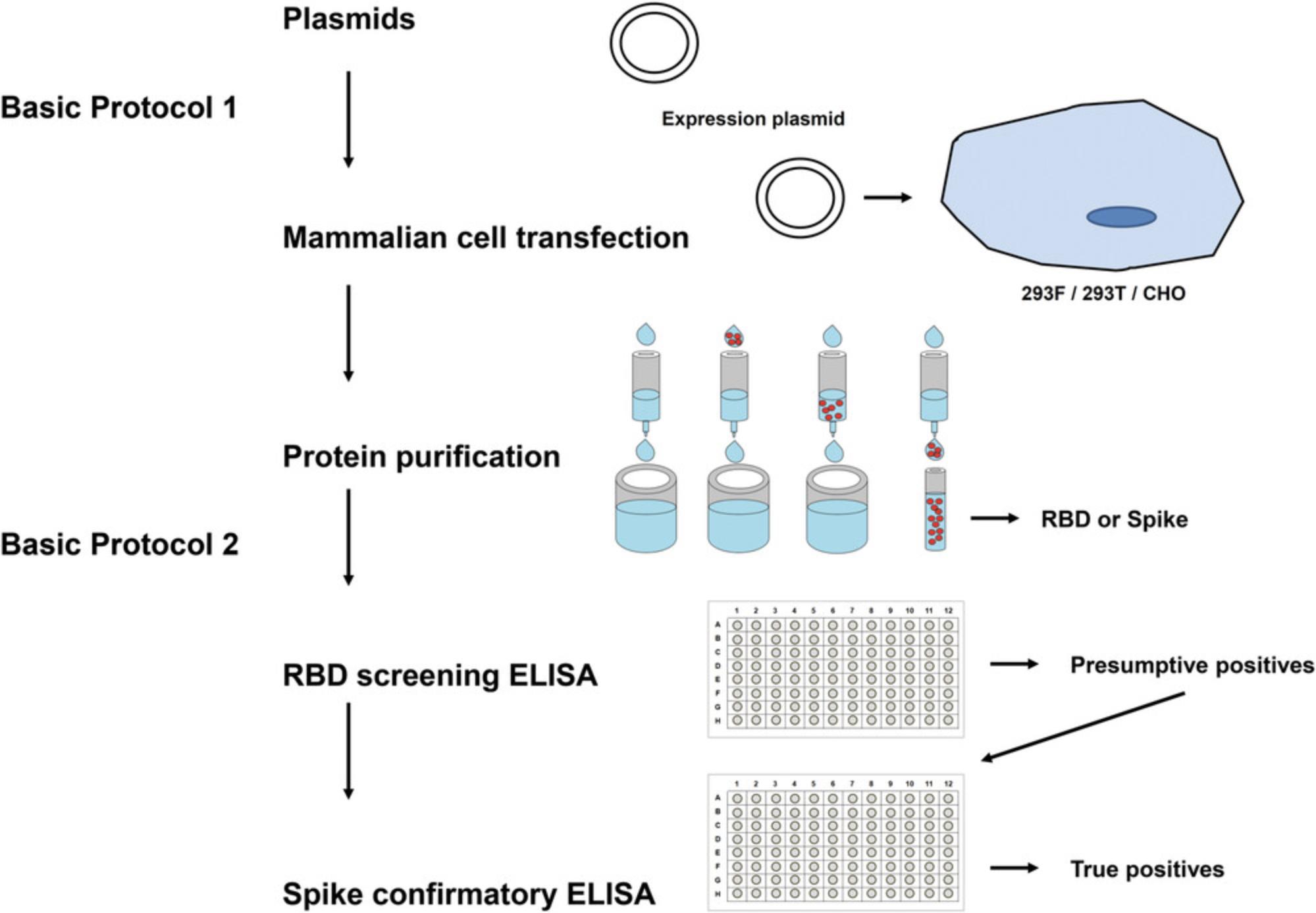
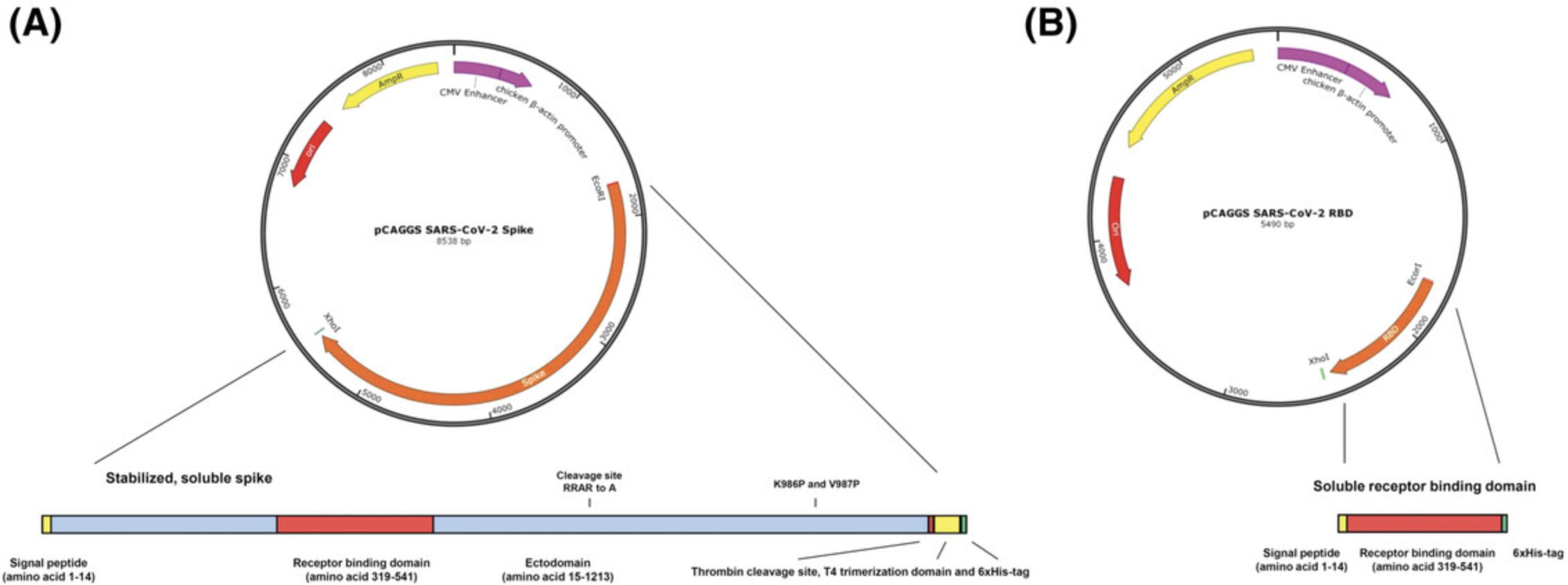
Basic Protocol 1: MAMMALIAN CELL TRANSFECTION AND PROTEIN PURIFICATION
This protocol can be used for both expression vectors: the one expressing secreted RBD as well as the one expressing a soluble, trimeric version of the SARS-CoV-2 spike protein. Expression levels of the RBD are very high in our hands (>20 mg/L culture), while expression levels for the full-length spike are lower (approximately 4 to 5 mg/L). Therefore, we use the recombinant RBD for initial screening ELISAs and the full-length spike for confirmatory ELISAs (as described in Basic Protocol 2). The expression vector constructs were described previously (Amanat et al., 2020). In brief, the sequences used for both proteins are based on the genomic sequence of the first isolate, Wuhan-Hu-1, which was released on January 10, 2020 (GenBank: MN908947.3). Sequences were codon-optimized for mammalian cell expression. The full-length spike protein sequence was modified to remove the polybasic cleavage site, which is recognized by furin, and to add a pair of stabilizing mutations (Figure 2). These two modifications were included to enhance the stability of the protein based on published literature (Amanat et al., 2020). The plasmids are grown in E. coli at 37°C (or 30°C) at 225 rpm in Luria-Bertani (LB) broth with ampicillin (LB-amp) in shaker flasks overnight. High-quality plasmid DNA can be obtained using commercially available maxiprep kits (ideally with an endotoxin-removal step). Importantly, other cell lines (293T, CHO, etc.), other media, transfection reagents, and more sophisticated protein purification methods might be used as alternatives if available.
Definitions
- RBD = receptor-binding domain of SARS-CoV-2 (NR-52306)
- PBS = phosphate-buffered saline
- RT = room temperature (18° to 25°C)
- MEM = Minimum Essential Medium
- DNA = deoxyribonucleic acid
- Ni-NTA = nickel-nitrilotriacetic acid
Basic Protocol 2: A TWO-STAGE ELISA FOR HIGH-THROUGHPUT SCREENING OF HUMAN SERUM SAMPLES FOR ANTIBODIES BINDING TO THE SPIKE PROTEIN OF SARS-CoV-2
The purpose of this protocol is to describe the procedure for measuring human antibody responses to the recombinant receptor-binding domain (RBD) of the spike protein or full-length spike protein of SARS-CoV-2 and to ensure the reproducibility and consistency of the obtained results.
We developed this as a two-stage ELISA in which the first stage (‘a’ steps below) includes relatively high-throughput screening of samples in a single serum dilution against the RBD (which expresses very well and therefore can be produced in greater quantities). This is followed by a second stage (‘b’ steps below) in which positive samples from the first stage undergo a confirmatory ELISA against the full-length spike protein (which is harder to express; therefore there is usually less available). For the second stage, a dilution curve is performed. Typically, if only one operator is available, screening ELISAs can be run in the morning (760 samples/10 plates per run) and confirmatory ELISAs can be run in the afternoon (140 samples/10 plates per run). Of note, we describe the assay here as it is set up in our laboratory. We use a plate washer and a plate reader, but no automated system. The protocol can be adapted to use with an automated liquid handler. In addition, one of the difficulties in setting up the assay is the availability of appropriate negative and positive controls. Negative controls are easier to come by, and can be serum pools taken before 2020. Positive controls can be convalescent samples from COVID19 patients or monoclonal antibodies (mAbs) like CR3022 (ter Meulen et al., 2006; Tian et al., 2020). If no human sera or mAbs are available, mouse mAbs, mouse sera against SARS-CoV-2, other animal sera against SARS-CoV-2, or anti−His tag antibodies (the proteins are His-tagged) can be used. However, in this case, a different secondary antibody for the species from which the primary antibody is derived is needed for the positive control. Also, we recommend generating large batches of positive controls, which can be used for many runs. The positive control should be selected to result in a strong signal (recommend OD490 of about 2.0), and should be clearly distinguishable from the negative controls. ELISAs can be run with either serum or plasma.
CAUTION : Before starting to work with COVID19 samples, please consult with your local biosafety officer regarding which precautions, personal protective equipment and protective measures are required.
NOTE:
Definitions
- ELISA = enzyme-linked immunosorbent assay
- PBS = phosphate-buffered saline
- RT = room temperature (18° to 25°C)
- HRP = horseradish peroxidase
- HCl = hydrochloric acid
- OPD = O -phenylenediamine dihydrochloride
NOTE : RBD or full-length spike might be used for both ELISA stages if only one antigen is available. In addition, only the “a” steps (not recommended) or only the “b” steps might be performed, if fewer resources are available.
REAGENTS AND SOLUTIONS
Elution buffer (4 L)
- 31.74 g NaH2PO4.·H2O
- 70.16 g NaCl
- 64.0 g imidazole (Sigma-Aldrich # I5513 or equivalent; final concentration is 235 mM)
- 4 L distilled water
- Store at room temperature up to 4 months
Use distilled water filtered using a 0.22-µm Stericup vacuum filtration system.
Phosphate-buffered saline with 0.1% Tween 20 (PBS-T; 50 L)
- 45 L distilled water
- 5 L 10× PBS (Corning™ #46013CM or equivalent))
- 50 ml Tween 20 (Fisher Bioreagents #BP337-500 or equivalent)
- Store at room temperature for to 4 months
Wash buffer (4 L)
- 31.74 g NaH2PO4·H2O
- 70.16 g NaCl
- 5.44 g imidazole (Sigma-Aldrich # I5513 or equivalent; final concentration is 20 mM)
- 4 L distilled water
- Store at room temperature up to 4 months
Use distilled water filtered using a 0.22-µm Stericup vacuum filtration system.
COMMENTARY
Background Information
The protein expression and purification methods (Basic Protocol 1) described in this article are based on well-established techniques. The expression plasmids and protein sequences have been optimized to increase protein stability and yield (Amanat et al., 2020). Plasmids can be requested from the Krammer laboratory or can be found on BEI Resources. The ELISA protocol (Basic Protocol 2) has been designed to allow for high-throughput screening of many samples per day, followed by a confirmatory step to verify presumptive positive results. The ELISA assay itself is based on well-established protocols and has been optimized for the use of SARS-CoV-2 antigens.
Critical Parameters and Troubleshooting
The most common problem for the transfection (Basic Protocol 1) is low cell viability before performing the transfection. The cells need to be 90% to 95% viable. The absence of antibiotics/antifungals requires good sterile technique to prevent contamination. Sterile plasmid preparations are also recommended, and it is important to add the enhancer to the shaking flasks 16 hr post-transfection.
For the protein purification, we recommend always using fresh Ni-NTA resin to prevent cross-contamination with other proteins. Harvested supernatant should be ideally processed immediately to ensure protein integrity. To make filtering of the supernatant easier, an additional centrifugation step (after pelleting the cells) is recommended to pellet residual cells and other particles. When performing buffer exchange using Amicon Ultra Centrifugal Filter Units, make sure to use the right-size cut-off (use smaller cut-off for RBD). It is recommended that purified protein be diluted to a concentration of about 2 mg/ml. Storage at higher concentrations may result in aggregation of protein.
For the ELISA (Basic Protocol 2), performing all of the washing steps and adhering to the incubation times are important to achieve low background reactivity. Most critical are the incubation times for the secondary antibody and the substrate (OPD and HCl for stopping the reaction). In addition, touching wells with tips when transferring secondary antibody and substrate can result in higher background and possibly false positive wells, and needs to be avoided. In preparing the OPD, it is also important to dissolve the gold tablet fully and only add the silver tablet right before the substrate is added to the ELISA plate.
Understanding Results
We expect expression levels of the RBD to be above 20 mg per L of culture cells and expression of the full-length spike protein to be approximately 4 mg per L of 293Fs, using a gravity-flow protein-purification strategy. When running the SDS-PAGE to confirm protein integrity, clear single bands are expected for the RBD and full-length spike at around 25 to 30 kDa and ∼190 kDa, respectively. Additionally, ELISAs with positive and negative controls (e.g., monoclonal antibodies) are performed to confirm correct protein folding. We expect a good binding profile for the positive control and low-to-no background reactivity for the negative control.
Time Considerations
Basic Protocols 1 and 2 can be completed in about 6 days. Basic Protocol 1 takes about 4 days. Growing up a cryostock of 293F cells, bringing them to passage 4 (recommended before transfection), and obtaining a sufficient cell number would take another few days; this is not taken into account in the protocol. Basic Protocol 2 takes at least 2 days (antigen coating on day 1 and running the ELISA on day 2). The screening ELISA could be performed in the morning and the confirmatory ELISA in the afternoon, or the assays can be done on consecutive days.
Acknowledgements
We thank Dr. Raffael Nachbagauer (Icahn School for Medicine at Mount Sinai) and Dr. Aubree Gordon (University of Michigan) for critical reading and constructive comments. Development of this protocol was partially supported by the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C.
Philanthropic donations in support of our work are much appreciated, since the reagents are shared free of charge with the scientific community. Please contact Vanesa Saric (vanesa.saric@mountsinai.org) for further information.
Literature Cited
- Amanat, F., Nguyen, T., Chromikova, V., Strohmeier, S., Stadlbauer, D., Javier, A., … Krammer, F. (2020). A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv , 2020.2003.2017.20037713. doi: 10.1101/2020.03.17.20037713.
- Bao, L., Deng, W., Gao, H., Xiao, C., Liu, J., Xue, J., … Qin, C. (2020). Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv , 2020.2003.2013.990226. doi:10.1101/2020.03.13.990226.
- Callow, K. A., Parry, H. F., Sergeant, M., & Tyrrell, D. A. (1990). The time course of the immune response to experimental coronavirus infection of man. Epidemiology and Infection , 105(2), 435−446. doi: 10.1017/s0950268800048019.
- Chu, D. K. W., Pan, Y., Cheng, S. M. S., Hui, K. P. Y., Krishnan, P., Liu, Y., … Poon, L. L. M. (2020). Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clinical Chemistry , 66(4), 549−555. doi: 10.1093/clinchem/hvaa029.
- Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K. W., … Drosten, C. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance , 25(3). doi: 10.2807/1560-7917.ES.2020.25.3.2000045.
- Lovrien, R., & Matulis, M. (2005). Assays for total protein. Current Protocols in Microbiology , 00, A.3A.1–A.3A.14. doi: 10.1002/9780471729259.mca03as00.
- Manns, J. M. (2011). SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of proteins. Current Protocols in Microbiology , 22, A.3M.1–A.3M.13. doi: 10.1002/9780471729259.mca03ms22.
- Phelan, K., & May, K. M. (2015). Basic techniques in mammalian cell culture. Current Protocols in Cell Biology , 66, 1.1.1–1.1.22. doi: 10.1002/0471143030.cb0101s66.
- ter Meulen, J., van den Brink, E. N., Poon, L. L., Marissen, W. E., Leung, C. S., Cox, F., … Goudsmit, J. (2006). Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Medicine , 3(7), e237. doi: 10.1371/journal.pmed.0030237.
- Tian, X., Li, C., Huang, A., Xia, S., Lu, S., Shi, Z., … Ying, T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging Microbes & Infections, 9(1), 382–385. doi: 10.1080/22221751.2020.1729069.
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., … McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science , 367(6483), 1260–1263. doi: 10.1126/science.abb2507.
- Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., … Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature. 579(7798), 265−269. doi: 10.1038/s41586-020-2008-3.
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., … Team, C. N. C. I. a. R. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine , 382(8), 727–733. doi: 10.1056/NEJMoa2001017.
Citing Literature
Number of times cited according to CrossRef: 474
- Yoshihiro Takayama, Yusuke Shimakawa, Yoshiaki Aizawa, Christian Butcher, Naomi Chibana, Mary Collins, Kohei Kamegai, Tae Gyun Kim, Satoshi Koyama, Ryota Matsuyama, Melissa M. Matthews, Tomoari Mori, Tetsuharu Nagamoto, Masashi Narita, Ryosuke Omori, Noriko Shibata, Satoshi Shibata, Souichi Shiiki, Shunichi Takakura, Naoki Toyozato, Hiroyuki Tsuchiya, Matthias Wolf, Taro Yamamoto, Shuhei Yokoyama, Sho Yonaha, Kenji Mizumoto, SARS-CoV-2 IgG seroprevalence in the Okinawa Main Island and remote islands in Okinawa, Japan, 2020-2021., Japanese Journal of Infectious Diseases, 10.7883/yoken.JJID.2023.255, (2024).
- Philip C. Mack, Chih-Yuan Hsu, Ananda M. Rodilla, Jorge E. Gomez, Jazz Cagan, Yuanhui Huang, Sooyun Tavolacci, Rajesh M. Valanparambil, Nicholas Rohs, Rachel Brody, Brittney Nichols, Juan Manuel Carreño, Sheena Bhalla, Christian Rolfo, David E. Gerber, Amy Moore, Jennifer C. King, Rafi Ahmed, John D. Minna, Paul A. Bunn, Adolfo García-Sastre, Florian Krammer, Fred R. Hirsch, Yu Shyr, Time-Dependent Effects of Clinical Interventions on SARS-CoV-2 Immunity in Patients with Lung Cancer, Vaccines, 10.3390/vaccines12070713, 12 , 7, (713), (2024).
- Mariângela de Oliveira Silva, Maria Fernanda Castro-Amarante, Alexia Adrianne Venceslau-Carvalho, Bianca da Silva Almeida, Isabela Pazotti Daher, Guilherme Antonio de Souza-Silva, Marcio Massao Yamamoto, Gabriela Koike, Edmarcia Elisa de Souza, Carsten Wrenger, Luís Carlos de Souza Ferreira, Silvia Beatriz Boscardin, Enhanced Immunogenicity and Protective Effects against SARS-CoV-2 Following Immunization with a Recombinant RBD-IgG Chimeric Protein, Vaccines, 10.3390/vaccines12040356, 12 , 4, (356), (2024).
- Christopher Franco, Alejandro Cornejo, Mariajosé Rodríguez, Alexis García, Inirida Belisario, Soriuska Mayora, Domingo José Garzaro, Rossana Celeste Jaspe, Mariana Hidalgo, Nereida Parra, Ferdinando Liprandi, José Luis Zambrano, Héctor Rafael Rangel, Flor Helene Pujol, Sputnik V-Induced Antibodies against SARS-CoV-2 Variants during the Dissemination of the Gamma Variant in Venezuela, Viruses, 10.3390/v16091480, 16 , 9, (1480), (2024).
- Jason S. Chwa, Minjun Kim, Yesun Lee, Wesley A. Cheng, Yunho Shin, Jaycee Jumarang, Jeffrey M. Bender, Pia S. Pannaraj, Detection of SARS-CoV-2-Specific Secretory IgA and Neutralizing Antibodies in the Nasal Secretions of Exposed Seronegative Individuals, Viruses, 10.3390/v16060852, 16 , 6, (852), (2024).
- Min Jung Kim, Izzati Haizan, Min Ju Ahn, Dong-Hyeok Park, Jin-Ha Choi, Recent Advances in Lateral Flow Assays for Viral Protein Detection with Nanomaterial-Based Optical Sensors, Biosensors, 10.3390/bios14040197, 14 , 4, (197), (2024).
- Flávia Lopes Adami, Mateus Vidigal de Castro, Bianca da Silva Almeida, Isabela Pazotti Daher, Márcio Massao Yamamoto, Keity Souza Santos, Mayana Zatz, Michel Satya Naslavsky, Daniela Santoro Rosa, Edecio Cunha-Neto, Vivian Leite de Oliveira, Jorge Kalil, Silvia Beatriz Boscardin, Anti-RBD IgG antibodies from endemic coronaviruses do not protect against the acquisition of SARS-CoV-2 infection among exposed uninfected individuals, Frontiers in Immunology, 10.3389/fimmu.2024.1396603, 15 , (2024).
- Jéromine Klingler, Shreyas Kowdle, Juan C. Bandres, Rozita Emami-Gorizi, Raymond A. Alvarez, Priyanka G. Rao, Fatima Amanat, Charles Gleason, Giulio Kleiner, Viviana Simon, Alexis Edelstein, Claudia Perandones, Chitra Upadhyay, Benhur Lee, Catarina E. Hioe, Heterologous Ad26/Ad5 adenovirus-vectored vaccines elicited SARS-CoV-2-specific antibody responses with potent Fc activities, Frontiers in Immunology, 10.3389/fimmu.2024.1382619, 15 , (2024).
- Ulises Zendejas-Hernandez, Nemi Alcántara-Martínez, Diana Tovar Vivar, Fermín Valenzuela, Alejandro Sosa Espinoza, Eduardo Emir Cervera Ceballos, Nebulized glycyrrhizin/enoxolone drug modulates IL-17A in COVID-19 patients: a randomized clinical trial, Frontiers in Immunology, 10.3389/fimmu.2023.1282280, 14 , (2024).
- TAYNÁ E. LIMA, MATHEUS V.F. FERRAZ, CARLOS A.A. BRITO, PAMELLA B. XIMENES, CAROLLINE A. MARIZ, CYNTHIA BRAGA, GABRIEL L. WALLAU, ISABELLE F.T. VIANA, ROBERTO D. LINS, Determination of prognostic markers for COVID-19 disease severity using routine blood tests and machine learning, Anais da Academia Brasileira de Ciências, 10.1590/0001-376520242023089, 96 , 2, (2024).
- Felipe Echeverri Tribin, Erin Williams, Valeska Testamarck, Juan Manuel Carreño, Dominika Bielak, Temima Yellin, Florian Krammer, Michael Hoffer, Suresh Pallikkuth, Savita Pahwa, Determinants of health as predictors for differential antibody responses following SARS-CoV-2 primary and booster vaccination in an at-risk, longitudinal cohort, PLOS ONE, 10.1371/journal.pone.0292566, 19 , 4, (e0292566), (2024).
- Gagandeep Singh, Anass Abbad, Giulio Kleiner, Komal Srivastava, Charles Gleason, Juan Manuel Carreño, Viviana Simon, Florian Krammer, Dalles Andre, Maria C Bermúdez-González, Dominika Bielak, Gianna Cai, Christian Cognigni, Yuexing Chen, Miriam Fried, Hyun Min Kang, Neko Lyttle, Jacob Mauldin, Brian Monahan, Sara Morris, Jessica Nardulli, Annika Oostenink, Ashley-Beathrese Salimbangon, Leeba Sullivan, Morgan Van Kesteren, Temima Yellin, The post-COVID-19 population has a high prevalence of cross-reactive antibodies to spikes from all Orthocoronavirinae genera , mBio, 10.1128/mbio.02250-23, 15 , 1, (2024).
- Allaura S. Cone, Yijun Zhou, Ryan P. McNamara, Anthony. B. Eason, Gabriel F. Arias, Justin T. Landis, Kyle W. Shifflett, Meredith G. Chambers, Runjie Yuan, Smaranda Willcox, Jack D. Griffith, Dirk P. Dittmer, CD81 fusion alters SARS-CoV-2 Spike trafficking, mBio, 10.1128/mbio.01922-24, 15 , 9, (2024).
- Baoling Ying, Chieh-Yu Liang, Pritesh Desai, Suzanne M. Scheaffer, Sayda M. Elbashir, Darin K. Edwards, Larissa B. Thackray, Michael S. Diamond, Ipsilateral or contralateral boosting of mice with mRNA vaccines confers equivalent immunity and protection against a SARS-CoV-2 Omicron strain, Journal of Virology, 10.1128/jvi.00574-24, 98 , 9, (2024).
- Benjamin A. Krishna, Eleanor Y. Lim, Marina Metaxaki, Sarah Jackson, Lenette Mactavous, Paul A. Lyons, Rainer Doffinger, John R. Bradley, Kenneth G. C. Smith, John Sinclair, Nicholas J. Matheson, Paul J. Lehner, Nyaradzai Sithole, Mark R. Wills, Spontaneous, persistent, T cell–dependent IFN-γ release in patients who progress to Long Covid, Science Advances, 10.1126/sciadv.adi9379, 10 , 8, (2024).
- Stephanie K. Lathrop, Jordan J. Clark, Karthik Siram, Robert Andreata-Santos, Jeremy Yong, Rebekah D. Tee, Clara J. Davison, Gagandeep Singh, David Burkhart, Florian Krammer, Jay T. Evans, Vaccination with ancestral SARS-CoV-2 spike adjuvanted with TLR agonists provides cross-protection against XBB.1, npj Viruses, 10.1038/s44298-024-00038-0, 2 , 1, (2024).
- Amanda Izeli Portilho, Valéria Oliveira Silva, Hernan Hermes Monteiro Da Costa, Rosemeire Yamashiro, Isabela Penteriche de Oliveira, Ivana Barros de Campos, Carlos Roberto Prudencio, Elaine Monteiro Matsuda, Luís Fernando de Macedo Brígido, Elizabeth De Gaspari, An unexpected IgE anti-receptor binding domain response following natural infection and different types of SARS-CoV-2 vaccines, Scientific Reports, 10.1038/s41598-024-71047-5, 14 , 1, (2024).
- Jae Kyu Ryu, Zhaoqi Yan, Mauricio Montano, Elif G. Sozmen, Karuna Dixit, Rahul K. Suryawanshi, Yusuke Matsui, Ekram Helmy, Prashant Kaushal, Sara K. Makanani, Thomas J. Deerinck, Anke Meyer-Franke, Pamela E. Rios Coronado, Troy N. Trevino, Min-Gyoung Shin, Reshmi Tognatta, Yixin Liu, Renaud Schuck, Lucas Le, Hisao Miyajima, Andrew S. Mendiola, Nikhita Arun, Brandon Guo, Taha Y. Taha, Ayushi Agrawal, Eilidh MacDonald, Oliver Aries, Aaron Yan, Olivia Weaver, Mark A. Petersen, Rosa Meza Acevedo, Maria del Pilar S. Alzamora, Reuben Thomas, Michela Traglia, Valentina L. Kouznetsova, Igor F. Tsigelny, Alexander R. Pico, Kristy Red-Horse, Mark H. Ellisman, Nevan J. Krogan, Mehdi Bouhaddou, Melanie Ott, Warner C. Greene, Katerina Akassoglou, Fibrin drives thromboinflammation and neuropathology in COVID-19, Nature, 10.1038/s41586-024-07873-4, 633 , 8031, (905-913), (2024).
- Chieh-Yu Liang, Saravanan Raju, Zhuoming Liu, Yuhao Li, Guha Asthagiri Arunkumar, James Brett Case, Suzanne M. Scheaffer, Seth J. Zost, Cory M. Acreman, Matthew Gagne, Shayne F. Andrew, Deborah Carolina Carvalho dos Anjos, Kathryn E. Foulds, Jason S. McLellan, James E. Crowe, Daniel C. Douek, Sean P. J. Whelan, Sayda M. Elbashir, Darin K. Edwards, Michael S. Diamond, Imprinting of serum neutralizing antibodies by Wuhan-1 mRNA vaccines, Nature, 10.1038/s41586-024-07539-1, 630 , 8018, (950-960), (2024).
- Juan Manuel Carreño, Abram L. Wagner, Brian Monahan, Gagandeep Singh, Daniel Floda, Ana S. Gonzalez-Reiche, Johnstone Tcheou, Ariel Raskin, Dominika Bielak, Sara Morris, Miriam Fried, Temima Yellin, Leeba Sullivan, Fatima Amanat, Guha Asthagiri Arunkumar, Christina Capuano, Jordan Ehrenhaus, Shelcie Fabre, Matthew M. Hernandez, Kaijun Jiang, Brian Lerman, Meagan McMahon, Daniel Stadlbauer, Jessica Tan, Catherine Teo, Kathryn Twyman, Emilia Mia Sordillo, Aubree Gordon, Harm van Bakel, Viviana Simon, Florian Krammer, SARS-CoV-2 serosurvey across multiple waves of the COVID-19 pandemic in New York City between 2020–2023, Nature Communications, 10.1038/s41467-024-50052-2, 15 , 1, (2024).
- Merve Eryilmaz, Artem Goncharov, Gyeo-Re Han, Hyou-Arm Joung, Zachary S. Ballard, Rajesh Ghosh, Yijie Zhang, Dino Di Carlo, Aydogan Ozcan, A Paper-Based Multiplexed Serological Test to Monitor Immunity against SARS-COV-2 Using Machine Learning, ACS Nano, 10.1021/acsnano.4c02434, 18 , 26, (16819-16831), (2024).
- Benjamin J. Des Soye, John P. McGee, Michael A. R. Hollas, Eleonora Forte, Ryan T. Fellers, Rafael D. Melani, John T. Wilkins, Philip D. Compton, Jared O. Kafader, Neil L. Kelleher, Automated Immunoprecipitation, Sample Preparation, and Individual Ion Mass Spectrometry Platform for Proteoforms, Analytical Chemistry, 10.1021/acs.analchem.4c01962, (2024).
- Anass Abbad, Temima Yellin, Gagandeep Singh, Miriam Fried, Ariel Raskin, Johnstone Tcheou, Brian Monahan, Charles Gleason, Dalles Andre, Maria C. Bermúdez-González, Dominika Bielak, Gianna Cai, Christian Cognigni, Yuexing Chen, Hyun Min Kang, Giulio Kleiner, Neko Lyttle, Jacob Mauldin, Sara Morris, Jessica Nardulli, Annika Oostenink, Ashley-Beathrese Salimbangon, Komal Srivastava, Leeba Sullivan, Morgan Van Kesteren, Viviana Simon, Juan Manuel Carreño, Florian Krammer, SARS-CoV-2 BA.1 and BA.2 breakthrough infections boost antibody responses to early Omicron subvariants but not BQ.1.1 or XBB.1.5, Cell Reports Medicine, 10.1016/j.xcrm.2024.101474, 5 , 3, (101474), (2024).
- Alla Kachko, Prabhuanand Selvaraj, Shufeng Liu, Jaekwan Kim, David Rotstein, Charles B. Stauft, Sylvie Chabot, Naveen Rajasagi, Yangqing Zhao, Tony Wang, Marian Major, Vaccine-associated respiratory pathology correlates with viral clearance and protective immunity after immunization with self-amplifying RNA expressing the spike (S) protein of SARS-CoV-2 in mouse models, Vaccine, 10.1016/j.vaccine.2023.12.052, 42 , 3, (608-619), (2024).
- Chien-Hsin Chiu, Chih-Hung Wang, Ying-Jun Lin, Chi-Chung Tang, Wei-Jei Peng, Wen-Yen Huang, Yan-Shen Shan, Huey-Pin Tsai, Gwo-Bin Lee, Simultaneous detection of SARS-CoV-2 and influenza A/B viruses on an electromagnetically-driven, integrated microfluidic system, Sensors and Actuators B: Chemical, 10.1016/j.snb.2024.135647, 410 , (135647), (2024).
- Frederieke A.J. Gigase, Rebecca H. Jessel, Elianna Kaplowitz, Natalie Boychuk, Sophie Ohrn, Erona Ibroci, Juliana Castro, Jezelle Lynch, Rushna Tubassum, Amy Balbierz, Nina M. Molenaar, Mara Graziani, Roy Missall, Tammy Flores, Toni Stern, Juan Manuel Carreno, Florian Krammer, Alan Adler, Rachel I. Brody, Corina Lesseur, Jia Chen, Sascha Ellington, Romeo R. Galang, Margaret C. Snead, Elizabeth Howell, Joanne Stone, Veerle Bergink, Siobhan Dolan, Whitney Lieb, Anna-Sophie Rommel, Lotje D. de Witte, Teresa Janevic, SARS-CoV-2 infection, inflammation and birth outcomes in a prospective NYC pregnancy cohort, Journal of Reproductive Immunology, 10.1016/j.jri.2024.104243, 163 , (104243), (2024).
- Juliana Castro, Frederieke A.J. Gigase, Nina M. Molenaar, Erona Ibroçi, M. Mercedes Perez-Rodriguez, Whitney Lieb, Teresa Janevic, Lot D. de Witte, Veerle Bergink, Anna-Sophie Rommel, Increased postpartum anxiety symptoms after perinatal SARS-CoV-2 infection in a large, prospective pregnancy cohort in New York City, Journal of Psychiatric Research, 10.1016/j.jpsychires.2023.12.020, 170 , (130-137), (2024).
- Hernan Hermes Monteiro da Costa, Valeria Oliveira Silva, Gustavo Carvalho Amorim, Marcia Grando Guereschi, Luciana Marciano Sergio, Carlos Henrique Rodrigues Gomes, Marisa Ailin Hong, Elaine Lopes de Oliveira, Luis Fernando de Macedo Brígido, Jose Angelo Lauletta Lindoso, Carlos Roberto Prudencio, Assessment of an in-house IgG ELISA targeting SARS-CoV-2 RBD: Applications in infected and vaccinated individuals, Journal of Immunological Methods, 10.1016/j.jim.2024.113683, 530 , (113683), (2024).
- Vincent J. Venditto, Brooke Hudspeth, Patricia R. Freeman, Lien Qasrawi, R. Kiplin Guy, Victoria H. Farley, Royce A. Johnson, Edward Freeman, David Henson, Ryan Marion, Sheridan B. Wagner, Brianna M. Doratt, Ilhem Messaoudi-Powers, Feasibility of pharmacy-based research opportunity to enhance community testing and surveillance, Journal of the American Pharmacists Association, 10.1016/j.japh.2024.102151, (102151), (2024).
- Komal Srivastava, Juan Manuel Carreño, Charles Gleason, Brian Monahan, Gagandeep Singh, Anass Abbad, Johnstone Tcheou, Ariel Raskin, Giulio Kleiner, Harm van Bakel, Emilia Mia Sordillo, Florian Krammer, Viviana Simon, Hala Alshammary, Angela A. Amoako, Dalles Andre, Mahmoud Awawda, Maria C. Bermúdez-González, Katherine F. Beach, Dominika Bielak, Gianna Y. Cai, Rachel L. Chernet, Christian Cognigni, Yuexing Chen, Lily Q. Eaker, Emily D. Ferreri, Daniel L. Floda, Miriam Fried, Joshua Z. Hamburger, Denise Jurczyszak, Hyun Min Kang, Neko Lyttle, Julia C. Matthews, Jacob Mauldin, Wanni A. Mendez, Jacob Mischka, Sara Morris, Lubbertus C.F. Mulder, Ismail Nabeel, Jessica R. Nardulli, Jose Polanco, Annika Oostenink, Aria Rooker, Kayla T. Russo, Ashley-Beathrese Salimbangon, Miti S. Saksena, Amber A. Shin, Levy A. Sominsky, Daniel Stadlbauer, Leeba Sullivan, Morgan van Kesteren, Temima Yellin, Ania Wajnberg, SARS-CoV-2-infection- and vaccine-induced antibody responses are long lasting with an initial waning phase followed by a stabilization phase, Immunity, 10.1016/j.immuni.2024.01.017, 57 , 3, (587-599.e4), (2024).
- Nuha Almulla, Raya Soltane, Ahlam Alasiri, Abdou Kamal Allayeh, Taha Alqadi, Fatma Alshehri, Ahlam Hamad Alrokban, Sameh S. Zaghlool, Abdallah Z. Zayan, Karam F. Abdalla, Ahmed M. Sayed, Advancements in SARS-CoV-2 detection: Navigating the molecular landscape and diagnostic technologies, Heliyon, 10.1016/j.heliyon.2024.e29909, 10 , 9, (e29909), (2024).
- Aled O’Neill, Chinmay Kumar Mantri, Chee Wah Tan, Wilfried A.A. Saron, Santhosh Kambaiah Nagaraj, Monica Palanichamy Kala, Christy Margarat Joy, Abhay P.S. Rathore, Shashank Tripathi, Lin-Fa Wang, Ashley L. St. John, Mucosal SARS-CoV-2 vaccination of rodents elicits superior systemic T central memory function and cross-neutralising antibodies against variants of concern, eBioMedicine, 10.1016/j.ebiom.2023.104924, 99 , (104924), (2024).
- Umut Karakus, Ignacio Mena, Jithesh Kottur, Sara S. El Zahed, Rocío Seoane, Soner Yildiz, Leanne Chen, Magdalena Plancarte, LeAnn Lindsay, Rebecca Halpin, Timothy B. Stockwell, David E. Wentworth, Geert-Jan Boons, Florian Krammer, Silke Stertz, Walter Boyce, Robert P. de Vries, Aneel K. Aggarwal, Adolfo García-Sastre, H19 influenza A virus exhibits species-specific MHC class II receptor usage, Cell Host & Microbe, 10.1016/j.chom.2024.05.018, 32 , 7, (1089-1102.e10), (2024).
- Disha Bhavsar, Kaori Sano, Gagandeep Singh, Florian Krammer, An ELISA‐Based Method to Measure Mucosal Antibody Responses Against SARS‐CoV‐2 in Human Saliva, Current Protocols, 10.1002/cpz1.1024, 4 , 4, (2024).
- Edit Ábrahám, Csaba Bajusz, Annamária Marton, Attila Borics, Thandiswa Mdluli, Norbert Pardi, Zoltán Lipinszki, Expression and purification of the receptor‐binding domain of SARS‐CoV‐2 spike protein in mammalian cells for immunological assays, FEBS Open Bio, 10.1002/2211-5463.13754, 14 , 3, (380-389), (2024).
- Shital Patil, Sham Toshniwal, Uttareshvar Dhumal, Ganesh Narwade, Dengue-COVID-19 overlap: A single-center prospective observational study in a tertiary care setting in India, Journal of Association of Pulmonologist of Tamil Nadu, 10.4103/japt.japt_37_22, 6 , 2, (45), (2023).
- Andrillene Laure Deutou Wondeu, Beatrice Metchum Talom, Giulia Linardos, Barnes Tanetsop Ngoumo, Aïchatou Bello, Aurele Marc Ndassi Soufo, Aimé Cesaire Momo, Christian Doll, Alaric Talom Tamuedjoun, Jules-Roger Kiuate, Giulia Cappelli, Cristina Russo, Carlo Federico Perno, Hyppolite K. Tchidjou, Lucia Scaramella, Andrea Galgani, The COVID-19 wave was already here: High seroprevalence of SARS-CoV-2 antibodies among staff and students in a Cameroon University, Journal of Public Health in Africa, 10.4081/jphia.2023.2242, 14 , 1, (2023).
- Michael Quinn, Luis Parra-Rodriguez, Wafaa B. Alsoussi, Chapelle Ayres, Michael K. Klebert, Chang Liu, Teresa Suessen, Suzanne M. Scheaffer, William D. Middleton, Sharlene A. Teefey, William G. Powderly, Michael S. Diamond, Rachel M. Presti, Ali H. Ellebedy, Jackson S. Turner, Jane A. O’Halloran, Philip A. Mudd, Persons with HIV Develop Spike-Specific Lymph Node Germinal Center Responses following SARS-CoV-2 Vaccination, The Journal of Immunology, 10.4049/jimmunol.2200920, 210 , 7, (947-958), (2023).
- Karthik Siram, Stephanie K. Lathrop, Walid M. Abdelwahab, Rebekah Tee, Clara J. Davison, Haley A. Partlow, Jay T. Evans, David J. Burkhart, Co-Delivery of Novel Synthetic TLR4 and TLR7/8 Ligands Adsorbed to Aluminum Salts Promotes Th1-Mediated Immunity against Poorly Immunogenic SARS-CoV-2 RBD, Vaccines, 10.3390/vaccines12010021, 12 , 1, (21), (2023).
- María M. Lorenzo, Alejandro Marín-López, Kevin Chiem, Luis Jimenez-Cabello, Irfan Ullah, Sergio Utrilla-Trigo, Eva Calvo-Pinilla, Gema Lorenzo, Sandra Moreno, Chengjin Ye, Jun-Gyu Park, Alejandro Matía, Alejandro Brun, Juana M. Sánchez-Puig, Aitor Nogales, Walther Mothes, Pradeep D. Uchil, Priti Kumar, Javier Ortego, Erol Fikrig, Luis Martinez-Sobrido, Rafael Blasco, Vaccinia Virus Strain MVA Expressing a Prefusion-Stabilized SARS-CoV-2 Spike Glycoprotein Induces Robust Protection and Prevents Brain Infection in Mouse and Hamster Models, Vaccines, 10.3390/vaccines11051006, 11 , 5, (1006), (2023).
- Mariana Ulinici, Alen Suljič, Monica Poggianella, Rafaela Milan Bonotto, Katarina Resman Rus, Angela Paraschiv, Amedeo Marco Bonetti, Mihail Todiras, Alexandru Corlateanu, Stanislav Groppa, Emil Ceban, Miroslav Petrovec, Alessandro Marcello, Characterisation of the Antibody Response in Sinopharm (BBIBP-CorV) Recipients and COVID-19 Convalescent Sera from the Republic of Moldova, Vaccines, 10.3390/vaccines11030637, 11 , 3, (637), (2023).
- Shubhada K. Chothe, Padmaja Jakka, Veda Sheersh Boorla, Santhamani Ramasamy, Abhinay Gontu, Ruth H. Nissly, Justin Brown, Gregory Turner, Brent J. Sewall, DeeAnn M. Reeder, Kenneth A. Field, Julie B. Engiles, Saranya Amirthalingam, Abirami Ravichandran, Lindsey LaBella, Meera Surendran Nair, Costas D. Maranas, Suresh V. Kuchipudi, Little Brown Bats (Myotis lucifugus) Support the Binding of SARS-CoV-2 Spike and Are Likely Susceptible to SARS-CoV-2 Infection, Viruses, 10.3390/v15051103, 15 , 5, (1103), (2023).
- Hernan H. M. da Costa, Diego J. B. Orts, Andrew D. Moura, Amaro N. Duarte-Neto, Cinthya S. Cirqueira, Rodrigo A. Réssio, Cristina T. Kanamura, Karen Miguita, Jerenice E. Ferreira, Raimunda T. M. Santos, Patricia P. Adriani, Jair P. Cunha-Junior, Renato M. Astray, Regina M. Catarino, Marcelo Lancelotti, Carlos R. Prudencio, RBD and Spike DNA-Based Immunization in Rabbits Elicited IgG Avidity Maturation and High Neutralizing Antibody Responses against SARS-CoV-2, Viruses, 10.3390/v15020555, 15 , 2, (555), (2023).
- Ha Nui Kim, Jung Yoon, Woong Sik Jang, Chae Seung Lim, Performance Evaluation of RapiSure (EDGC) COVID-19 S1 RBD IgG/Neutralizing Ab Test for the Rapid Detection of SARS-CoV-2 Antibodies, Diagnostics, 10.3390/diagnostics13040643, 13 , 4, (643), (2023).
- Erin Williams, Devin J. Kennedy, Michael Hoffer, Juan Manuel Carreño, Florian Krammer, Suresh Pallikkuth, Savita Pahwa, Chronic False Positive Rapid Plasma Reagin (RPR) Tests Induced by COVID-19 Vaccination, COVID, 10.3390/covid3090090, 3 , 9, (1304-1309), (2023).
- Kritika Srinivasan Rajsri, Michael P. McRae, Nicolaos J. Christodoulides, Isaac Dapkins, Glennon W. Simmons, Hanover Matz, Helen Dooley, David Fenyö, John T. McDevitt, Simultaneous Quantitative SARS-CoV-2 Antigen and Host Antibody Detection and Pre-Screening Strategy at the Point of Care, Bioengineering, 10.3390/bioengineering10060670, 10 , 6, (670), (2023).
- Yoichiro Natori, Eric Martin, Adela Mattiazzi, Leopoldo Arosemena, Mariella Ortigosa-Goggins, Sivan Shobana, David Roth, Warren Lee Kupin, George William Burke, Gaetano Ciancio, Mahmoud Morsi, Anita Phancao, Mrudula R. Munagala, Hoda Butrous, Suresh Manickavel, Neeraj Sinha, Katherine Sota, Suresh Pallikkuth, Julia Bini, Jacques Simkins, Shweta Anjan, Rodrigo M. Vianna, Giselle Guerra, A Pilot Single-Blinded, Randomized, Controlled Trial Comparing BNT162b2 vs. JNJ-78436735 Vaccine as the Third Dose After Two Doses of BNT162b2 Vaccine in Solid Organ Transplant Recipients, Transplant International, 10.3389/ti.2023.10938, 36 , (2023).
- Erin C. Williams, Alexander Kizhner, Valerie S. Stark, Aria Nawab, Daniel D. Muniz, Felipe Echeverri Tribin, Juan Manuel Carreño, Dominika Bielak, Gagandeep Singh, Michael E. Hoffer, Florian Krammer, Suresh Pallikkuth, Savita Pahwa, Predictors for reactogenicity and humoral immunity to SARS-CoV-2 following infection and mRNA vaccination: A regularized, mixed-effects modelling approach, Frontiers in Immunology, 10.3389/fimmu.2023.971277, 14 , (2023).
- Jenny M. Lee, Alexis Figueroa, Jaiprasath Sachithanandham, Maggie Li, Caoilfhionn M. Connolly, Janna R. Shapiro, Yiqun Chen, Michelle Jones, Venkata Gayatri Dhara, Marilyn Towns, John S. Lee, Stephanie R. Peralta, Aaron M. Milstone, Michael Betenbaugh, Amanda K. Debes, Joel Blankson, Ioannis Sitaras, Steve Yoon, Elizabeth A. Thompson, Clifton O. Bingham, Sabra L. Klein, Andrew Pekosz, Justin R. Bailey, Three doses of COVID-19 mRNA vaccine induce class-switched antibody responses in inflammatory arthritis patients on immunomodulatory therapies, Frontiers in Immunology, 10.3389/fimmu.2023.1266370, 14 , (2023).
- Kelsey E. Lesteberg, Paula Araya, Katherine A. Waugh, Lakshmi Chauhan, Joaquin M. Espinosa, J. David Beckham, Severely ill and high-risk COVID-19 patients exhibit increased peripheral circulation of CD62L+ and perforin+ T cells, Frontiers in Immunology, 10.3389/fimmu.2023.1113932, 14 , (2023).
- Abdelhadi Djaïleb, Étienne Lavallée, Megan-Faye Parker, Marie-Pierre Cayer, Florence Desautels, Marie Joëlle de Grandmont, Matthew Stuible, Christian Gervais, Yves Durocher, Sylvie Trottier, Denis Boudreau, Jean-Francois Masson, Danny Brouard, Joelle N. Pelletier, Assessment of the longitudinal humoral response in non-hospitalized SARS-CoV-2-positive individuals at decentralized sites: Outcomes and concordance, Frontiers in Immunology, 10.3389/fimmu.2022.1052424, 13 , (2023).
- Benjamin J. Krajacich, Djibril Samaké, Adama Dao, Moussa Diallo, Zana Lamissa Sanogo, Alpha Seydou Yaro, Amatigue Zeguime, Josué Poudiougo, Kadiatou Cissé, Mamadou Traoré, Alassane dit Assitoun, Roy Faiman, Irfan Zaidi, John Woodford, Patrick E. Duffy, Tovi Lehmann, Tracking SARS-CoV-2 seropositivity in rural communities using blood-fed mosquitoes: a proof-of-concept study, Frontiers in Epidemiology, 10.3389/fepid.2023.1243691, 3 , (2023).
- Horacio Pérez-Juárez, Angélica Serrano-Vázquez, Héctor Godínez-Alvarez, Enrique González, Liliana Rojas-Velázquez, Patricia Moran, Tobías Portillo-Bobadilla, Manuel Ramiro, Eric Hernández, Clara Lau, Marcela Martínez, Ma. de los Ángeles Padilla, Martha E. Zaragoza, Blanca Taboada, Laura A. Palomares, Susana López, Alejandro Alagón, Carlos F. Arias, Cecilia Ximénez, Longitudinal anti-SARS-CoV-2 antibody immune response in acute and convalescent patients, Frontiers in Cellular and Infection Microbiology, 10.3389/fcimb.2023.1239700, 13 , (2023).
- Janikua Nelson-Mora, Diana Rubio, Amairani Ventura-Martínez, Luis A. González, Diana Del-Rio, Yuli Aranda-López, Edgar Jiménez-Díaz, Diego Zamarrón-Hernández, Diana G. Ríos-López, Stephanie Aguirre, Yasab Ruiz-Hernandez, Aarón Cruz-Ramírez, Jonás S. Barjau, Miguel A. Jáurez, Jehú Lopez-Aparicio, Andrea Campa-Higareda, Tatiana Fiordelisio, New detection method of SARS-CoV-2 antibodies toward a point-of-care biosensor, Frontiers in Bioengineering and Biotechnology, 10.3389/fbioe.2023.1202126, 11 , (2023).
- Lipeng Liu, Kangping Zhou, Yan Xing, Wei Luo, Bing Pei, Junqiang Xu, Lei Yi, Honglei Wang, Juan Wang, Wei Zhang, Fei Yu, Kun Cai, Peng Liu, Clinical Application of SARS-CoV-2 IgM and IgG Antibody Detection Using the Colloidal Gold Immunochromatography Assay, Zoonoses, 10.15212/ZOONOSES-2023-0020, 3 , 1, (2023).
- Darryl P. Leong, Ali Zhang, Jessica A. Breznik, Rumi Clare, Angela Huynh, Maha Mushtaha, Sumathy Rangarajan, Hannah Stacey, Paul Y. Kim, Mark Loeb, Judah A. Denburg, Dominik Mertz, Zain Chagla, Ishac Nazy, Matthew S. Miller, Dawn M. E. Bowdish, MyLinh Duong, Comparison of three dosing intervals for the primary vaccination of the SARS-CoV-2 mRNA Vaccine (BNT162b2) on magnitude, neutralization capacity and durability of the humoral immune response in health care workers: A prospective cohort study, PLOS ONE, 10.1371/journal.pone.0281673, 18 , 2, (e0281673), (2023).
- Muhammad Hakimin Shafie, Marie Antony Dass, Hazlam Shamin Ahmad Shaberi, Zainuddin Zafarina, Screening and confirmation tests for SARS-CoV-2: benefits and drawbacks, Beni-Suef University Journal of Basic and Applied Sciences, 10.1186/s43088-023-00342-3, 12 , 1, (2023).
- Charles T. Semelka, Michael E. DeWitt, Maria W. Blevins, Beth C. Holbrook, John W. Sanders, Martha A. Alexander-Miller, Frailty impacts immune responses to Moderna COVID-19 mRNA vaccine in older adults, Immunity & Ageing, 10.1186/s12979-023-00327-x, 20 , 1, (2023).
- Erica A. Green, Nathaniel K. Hamaker, Kelvin H. Lee, Comparison of vector elements and process conditions in transient and stable suspension HEK293 platforms using SARS-CoV-2 receptor binding domain as a model protein, BMC Biotechnology, 10.1186/s12896-023-00777-7, 23 , 1, (2023).
- Kiran Iqbal Masood, Shama Qaiser, Syed Hani Abidi, Erum Khan, Syed Faisal Mahmood, Areeba Hussain, Zara Ghous, Khekahsan Imtiaz, Natasha Ali, Muhammad Hasan, Haris Ali Memon, Maliha Yameen, Shiza Ali, Sadaf Baloch, Gulzar Lakhani, Paula M. Alves, Najeeha Talat Iqbal, Kumail Ahmed, Junaid Iqbal, Zulfiqar A. Bhutta, Rabia Hussain, Martin Rottenberg, J. Pedro Simas, Marc Veldhoen, Kulsoom Ghias, Zahra Hasan, Humoral and T cell responses to SARS-CoV-2 reveal insights into immunity during the early pandemic period in Pakistan, BMC Infectious Diseases, 10.1186/s12879-023-08829-1, 23 , 1, (2023).
- Lina M. Marin, George S. Katselis, Paulos Chumala, Stephen Sanche, Lucas Julseth, Erika Penz, Robert Skomro, Walter L. Siqueira, Identification of SARS-CoV-2 biomarkers in saliva by transcriptomic and proteomics analysis, Clinical Proteomics, 10.1186/s12014-023-09417-w, 20 , 1, (2023).
- Shang-Chuen Wu, Connie M. Arthur, Hau-Ming Jan, Wilfredo F. Garcia-Beltran, Kashyap R. Patel, Matthew F. Rathgeber, Hans P. Verkerke, Narayanaiah Cheedarla, Ryan Philip Jajosky, Anu Paul, Andrew S. Neish, John D. Roback, Cassandra D. Josephson, Duane R. Wesemann, Daniel Kalman, Seth Rakoff-Nahoum, Richard D. Cummings, Sean R. Stowell, Blood group A enhances SARS-CoV-2 infection, Blood, 10.1182/blood.2022018903, 142 , 8, (742-747), (2023).
- Juliana Gonçalves, Magda Melro, Marta Alenquer, Catarina Araújo, Júlia Castro-Neves, Daniela Amaral-Silva, Filipe Ferreira, José S. Ramalho, Nádia Charepe, Fátima Serrano, Carlos Pontinha, Maria João Amorim, Helena Soares, Balance between maternal antiviral response and placental transfer of protection in gestational SARS-CoV-2 infection, JCI Insight, 10.1172/jci.insight.167140, 8 , 17, (2023).
- Siriruk Changrob, Peter J. Halfmann, Hejun Liu, Jonathan L. Torres, Joshua J.C. McGrath, Gabriel Ozorowski, Lei Li, G. Dewey Wilbanks, Makoto Kuroda, Tadashi Maemura, Min Huang, Nai-Ying Zheng, Hannah L. Turner, Steven A. Erickson, Yanbin Fu, Atsuhiro Yasuhara, Gagandeep Singh, Brian Monahan, Jacob Mauldin, Komal Srivastava, Viviana Simon, Florian Krammer, D. Noah Sather, Andrew B. Ward, Ian A. Wilson, Yoshihiro Kawaoka, Patrick C. Wilson, Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants, Journal of Clinical Investigation, 10.1172/JCI166844, 133 , 8, (2023).
- Masoud Norouzi, Thang Truong, Katariina Jaenes, Bryce M. Warner, Robert Vendramelli, Kevin Tierney, Darwyn Kobasa, Nikesh Tailor, Pamela Plant, Claudia dos Santos, Shawn Babiuk, Aruna Ambagala, Keith Pardee, Cell-Free Dot Blot: an Ultra-Low-Cost and Practical Immunoassay Platform for Detection of Anti-SARS-CoV-2 Antibodies in Human and Animal Sera, Microbiology Spectrum, 10.1128/spectrum.02457-22, 11 , 2, (2023).
- Fatima Amanat, Jordan Clark, Juan Manuel Carreño, Shirin Strohmeier, Temima Yellin, Philip S. Meade, Disha Bhavsar, Hiromi Muramatsu, Weina Sun, Lynda Coughlan, Norbert Pardi, Florian Krammer, Immunity to Seasonal Coronavirus Spike Proteins Does Not Protect from SARS-CoV-2 Challenge in a Mouse Model but Has No Detrimental Effect on Protection Mediated by COVID-19 mRNA Vaccination, Journal of Virology, 10.1128/jvi.01664-22, 97 , 3, (2023).
- Juan Manuel Carreño, Ariel Raskin, Gagandeep Singh, Johnstone Tcheou, Hisaaki Kawabata, Charles Gleason, Komal Srivastava, Vladimir Vigdorovich, Nicholas Dambrauskas, Sneh Lata Gupta, Irene González Domínguez, Jose Luis Martinez, Stefan Slamanig, D. Noah Sather, Rama Raghunandan, Ponthip Wirachwong, Sant Muangnoicharoen, Punnee Pitisuttithum, Jens Wrammert, Mehul S. Suthar, Weina Sun, Peter Palese, Adolfo García-Sastre, Viviana Simon, Florian Krammer, An inactivated NDV-HXP-S COVID-19 vaccine elicits a higher proportion of neutralizing antibodies in humans than mRNA vaccination, Science Translational Medicine, 10.1126/scitranslmed.abo2847, 15 , 683, (2023).
- Valerie S. Stark, Erin C. Williams, Felipe Echeverri Tribin, Jennifer Coto, Adam Carrico, Juan Manuel Carreño, Dominika Bielak, Parnavi Desai, Florian Krammer, Michael E. Hoffer, Suresh Pallikkuth, Savita Pahwa, Examining the Effect of SARS-CoV-2 Pandemic-Induced Stress and Anxiety on Humoral Immunity in Health Care Workers, Journal of Occupational & Environmental Medicine, 10.1097/JOM.0000000000003014, 66 , 2, (e48-e53), (2023).
- Sopisa Benjakul, Aina Karen Anthi, Anette Kolderup, Marina Vaysburd, Heidrun Elisabeth Lode, Donna Mallery, Even Fossum, Elisabeth Lea Vikse, Anna Albecka, Aleksandr Ianevski, Denis Kainov, Karine Flem Karlsen, Siri Aastedatter Sakya, Mari Nyquist-Andersen, Torleif Tollefsrud Gjølberg, Morten C Moe, Magnar Bjørås, Inger Sandlie, Leo C James, Jan Terje Andersen, A pan-SARS-CoV-2-specific soluble angiotensin-converting enzyme 2-albumin fusion engineered for enhanced plasma half-life and needle-free mucosal delivery, PNAS Nexus, 10.1093/pnasnexus/pgad403, 2 , 12, (2023).
- Minjun Kim, Wesley A Cheng, Zion Congrave-Wilson, Carolyn Jennifer Marentes Ruiz, Lauren Turner, Shirley Mendieta, Jaycee Jumarang, Jennifer Del Valle, Yesun Lee, Thomas Fabrizio, E Kaitlynn Allen, Paul G Thomas, Richard Webby, Aubree Gordon, Pia S Pannaraj, Comparisons of Pediatric and Adult SARS-CoV-2-Specific Antibodies up to 6 Months after Infection, Vaccination, or Hybrid Immunity, Journal of the Pediatric Infectious Diseases Society, 10.1093/jpids/piad107, 13 , 1, (91-99), (2023).
- Maria Rebelo, Cong Tang, Ana R Coelho, Carlos Labão-Almeida, Matthias M Schneider, Laurie Tatalick, Pedro Ruivo, Marta Pires de Miranda, Andreia Gomes, Tânia Carvalho, Matthew J Walker, Hannes Ausserwoeger, J Pedro Simas, Marc Veldhoen, Tuomas P J Knowles, Daniel-Adriano Silva, David Shoultz, Gonçalo J L Bernardes, De Novo Human Angiotensin-Converting Enzyme 2 Decoy NL-CVX1 Protects Mice From Severe Disease After Severe Acute Respiratory Syndrome Coronavirus 2 Infection, The Journal of Infectious Diseases, 10.1093/infdis/jiad135, 228 , 6, (723-733), (2023).
- Gagandeep Singh, Anass Abbad, Johnstone Tcheou, Demodara Rao Mendu, Adolfo Firpo-Betancourt, Charles Gleason, Komal Srivastava, Carlos Cordon-Cardo, Viviana Simon, Florian Krammer, Juan Manuel Carreño, Binding and Avidity Signatures of Polyclonal Sera From Individuals With Different Exposure Histories to Severe Acute Respiratory Syndrome Coronavirus 2 Infection, Vaccination, and Omicron Breakthrough Infections, The Journal of Infectious Diseases, 10.1093/infdis/jiad116, 228 , 5, (564-575), (2023).
- Aaron M Frutos, Guillermina Kuan, Roger Lopez, Sergio Ojeda, Abigail Shotwell, Nery Sanchez, Saira Saborio, Miguel Plazaola, Carlos Barilla, Eben Kenah, Angel Balmaseda, Aubree Gordon, Infection-Induced Immunity Is Associated With Protection Against Severe Acute Respiratory Syndrome Coronavirus 2 Infection and Decreased Infectivity, Clinical Infectious Diseases, 10.1093/cid/ciad074, 76 , 12, (2126-2133), (2023).
- Jaimi L. Allen, Benjamin C. Amick, Mark L. Williams, Joshua L. Kennedy, Karl W. Boehme, J. Craig Forrest, Brian Primack, Erica Ashley Sides, Wendy N. Nembhard, Stephanie F. Gardner, Jessica N. Snowden, Laura P. James, Ericka Olgaard, Jay Gandy, A longitudinal study of SARS-CoV-2 antibody seroprevalence and mitigation behaviors among college students at an Arkansas University, Journal of American College Health, 10.1080/07448481.2023.2217456, (1-10), (2023).
- Masato Hirota, Miho Tamai, Sachie Yukawa, Naoyuki Taira, Melissa M. Matthews, Takeshi Toma, Yu Seto, Makiko Yoshida, Sakura Toguchi, Mio Miyagi, Tomoari Mori, Hiroaki Tomori, Osamu Tamai, Mitsuo Kina, Eishin Sakihara, Chiaki Yamashiro, Masatake Miyagi, Kentaro Tamaki, Matthias Wolf, Mary K. Collins, Hiroaki Kitano, Hiroki Ishikawa, Human immune and gut microbial parameters associated with inter-individual variations in COVID-19 mRNA vaccine-induced immunity, Communications Biology, 10.1038/s42003-023-04755-9, 6 , 1, (2023).
- Marta Ribes, Júlia Montañà, Marta Vidal, Ruth Aguilar, Patricia Nicolás, Uxue Alfonso, Natalia Rodrigo, Carlo Carolis, Carlota Dobaño, Gemma Moncunill, Carlos Chaccour, Seroprevalence and socioeconomic impact of the first SARS-CoV-2 infection wave in a small town in Navarre, Spain, Scientific Reports, 10.1038/s41598-023-30542-x, 13 , 1, (2023).
- Marina S. Boukhvalova, Emma Mortensen, Jessica Caple, John Joseph, Fatoumata Sylla, Arash Kamali, Daniel Stylos, Diego Lopez, Thomas March, Kevin Matthew Byrd, Gregory A. Prince, Ariel Arndt, Adriana Kajon, Jorge C. G. Blanco, SARS-CoV-2 infection augments species- and age-specific predispositions in cotton rats, Scientific Reports, 10.1038/s41598-022-27328-y, 13 , 1, (2023).
- Wafaa B. Alsoussi, Sameer Kumar Malladi, Julian Q. Zhou, Zhuoming Liu, Baoling Ying, Wooseob Kim, Aaron J. Schmitz, Tingting Lei, Stephen C. Horvath, Alexandria J. Sturtz, Katherine M. McIntire, Birk Evavold, Fangjie Han, Suzanne M. Scheaffer, Isabella F. Fox, Senaa F. Mirza, Luis Parra-Rodriguez, Raffael Nachbagauer, Biliana Nestorova, Spyros Chalkias, Christopher W. Farnsworth, Michael K. Klebert, Iskra Pusic, Benjamin S. Strnad, William D. Middleton, Sharlene A. Teefey, Sean P. J. Whelan, Michael S. Diamond, Robert Paris, Jane A. O’Halloran, Rachel M. Presti, Jackson S. Turner, Ali H. Ellebedy, SARS-CoV-2 Omicron boosting induces de novo B cell response in humans, Nature, 10.1038/s41586-023-06025-4, 617 , 7961, (592-598), (2023).
- Juan Carlos Yam-Puc, Zhaleh Hosseini, Emily C. Horner, Pehuén Pereyra Gerber, Nonantzin Beristain-Covarrubias, Robert Hughes, Aleksei Lulla, Maria Rust, Rebecca Boston, Magda Ali, Katrin Fischer, Edward Simmons-Rosello, Martin O’Reilly, Harry Robson, Lucy H. Booth, Lakmini Kahanawita, Andrea Correa-Noguera, David Favara, Lourdes Ceron-Gutierrez, Baerbel Keller, Andrew Craxton, Georgina S. F. Anderson, Xiao-Ming Sun, Anne Elmer, Caroline Saunders, Areti Bermperi, Sherly Jose, Nathalie Kingston, Thomas E. Mulroney, Lucia P. G. Piñon, Michael A. Chapman, Sofia Grigoriadou, Marion MacFarlane, Anne E. Willis, Kiran R. Patil, Sarah Spencer, Emily Staples, Klaus Warnatz, Matthew S. Buckland, Florian Hollfelder, Marko Hyvönen, Rainer Döffinger, Christine Parkinson, Sara Lear, Nicholas J. Matheson, James E. D. Thaventhiran, Age-associated B cells predict impaired humoral immunity after COVID-19 vaccination in patients receiving immune checkpoint blockade, Nature Communications, 10.1038/s41467-023-38810-0, 14 , 1, (2023).
- Lian C. T. Shoute, Gaser N. Abdelrasoul, Yuhao Ma, Pedro A. Duarte, Cole Edwards, Ran Zhuo, Jie Zeng, Yiwei Feng, Carmen L. Charlton, Jamil N. Kanji, Shawn Babiuk, Jie Chen, Label-free impedimetric immunosensor for point-of-care detection of COVID-19 antibodies, Microsystems & Nanoengineering, 10.1038/s41378-022-00460-5, 9 , 1, (2023).
- Joann Diray-Arce, Slim Fourati, Naresh Doni Jayavelu, Ravi Patel, Cole Maguire, Ana C. Chang, Ravi Dandekar, Jingjing Qi, Brian H. Lee, Patrick van Zalm, Andrew Schroeder, Ernie Chen, Anna Konstorum, Anderson Brito, Jeremy P. Gygi, Alvin Kho, Jing Chen, Shrikant Pawar, Ana Silvia Gonzalez-Reiche, Annmarie Hoch, Carly E. Milliren, James A. Overton, Kerstin Westendorf, Charles B. Cairns, Nadine Rouphael, Steven E. Bosinger, Seunghee Kim-Schulze, Florian Krammer, Lindsey Rosen, Nathan D. Grubaugh, Harm van Bakel, Michael Wilson, Jayant Rajan, Hanno Steen, Walter Eckalbar, Chris Cotsapas, Charles R. Langelier, Ofer Levy, Matthew C. Altman, Holden Maecker, Ruth R. Montgomery, Elias K. Haddad, Rafick P. Sekaly, Denise Esserman, Al Ozonoff, Patrice M. Becker, Alison D. Augustine, Leying Guan, Bjoern Peters, Steven H. Kleinstein, James Abraham, Michael Adkisson, Marisa Albert, Luz Torres Altamirano, Bonny Alvarenga, Matthew L. Anderson, Evan J. Anderson, Azlann Arnett, Hiromitsu Asashima, Mark A. Atkinson, Lindsey R. Baden, Brenda Barton, Katherine Beach, Elizabeth Beagle, Patrice M. Becker, Matthew R. Bell, Mariana Bernui, Christian Bime, Arun Kumar Boddapati, J. Leland Booth, Brittney Borresen, Scott C. Brakenridge, Laurel Bristow, Robert Bryant, Carolyn S. Calfee, Juan Manuel Carreño, Sidney Carrillo, Suzanna Chak, Iris Chang, Jennifer Connors, Michelle Conway, David B. Corry, David Cowan, Brett Croen, Charles S. Dela Cruz, Gina Cusimano, Lily Eaker, Carolyn Edwards, Lauren I.R. Ehrlich, David Elashoff, Heidi Erickson, David J. Erle, Shelli Farhadian, Keith Farrugia, Benoit Fatou, Andrea Fernandes, Ana Fernandez-Sesma, Gabriela K. Fragiadakis, Sara Furukawa, Janelle N. Geltman, Rajani Ghale, Maria Carolina Bermúdez González, I. Michael Goonewardene, Estella Sanchez Guerrero, Faheem W. Guirgis, David A. Hafler, Sydney Hamilton, Paul Harris, Arash Nemati Hayati, Carolyn M. Hendrickson, Nelson I. Agudelo Higuita, Thomas Hodder, Steven M. Holland, Catherine L. Hough, Christopher Huerta, Kerin C. Hurley, Scott R. Hutton, Akiko Iwasaki, Alejandra Jauregui, Meenakshi Jha, Brandi Johnson, David Joyner, Kirsten N. Kangelaris, Geoffrey Kelly, Zain Khalil, Zenab Khan, Farrah Kheradmand, James N. Kim, Hiroki Kimura, Albert I. Ko, Bernard Kohr, Monica Kraft, Matthew Krummel, Michele A. Kutzler, Jessica Lasky-Su, Serena Lee, Deanna Lee, Michael Leipold, Claudia Lentucci, Carolyn Leroux, Edward Lin, Shanshan Liu, Christina Love, Zhengchun Lu, Lenka Maliskova, Brittany Roth Manning, Monali Manohar, Mark Martens, Grace A. McComsey, Kerry McEnaney, Renee McLin, Esther Melamed, Nataliya Melnyk, Kevin Mendez, William B. Messer, Jordan P. Metcalf, Gregory Michelotti, Eran Mick, Subhasis Mohanty, Jarrod Mosier, Lubbertus C.F. Mulder, Maimouna Murphy, Kari R.C. Nadeau, Ebony Nelson, Allison Nelson, Viet Nguyen, Jordan Oberhaus, Bernadine Panganiban, Kathryn L. Pellegrini, Harry C. Pickering, Debra L. Powell, Scott Presnell, Bali Pulendran, Adeeb H. Rahman, Ahmad Sadeed Rashid, Ariel Raskin, Elaine F. Reed, Susan Pereira Ribeiro, Adreanne M. Rivera, Jacob E. Rogers, Angela Rogers, Brandon Rogowski, Rebecca Rooks, Yael Rosenberg-Hasson, Jessica Rothman, Justin F. Rousseau, Ramin Salehi-Rad, Mehmet Saluvan, Hady Samaha, Joanna Schaenman, Ron Schunk, Nicholas C. Semenza, Subha Sen, Jonathan Sevransky, Vicki Seyfert-Margolis, Tanzia Shaheen, Albert C. Shaw, Scott Sieg, Sarah A.R. Siegel, Natalia Sigal, Nadia Siles, Brent Simmons, Viviana Simon, Gagandeep Singh, Lauren Sinko, Cecilia M. Smith, Kinga K. Smolen, Li-Zhen Song, Komal Srivastava, Peter Sullivan, Caitlin Syphurs, Johnstone Tcheou, George P. Tegos, Greg K. Tharp, Alexandra Tong, Alexandra Tsitsiklis, Ricardo F. Ungaro, Tatyana Vaysman, Arthur Viode, Randi Vita, Xiaomei Wang, Alyssa Ward, Dawn C. Ward, Andrew Willmore, Kyra Woloszczuk, Kari Wong, Prescott G. Woodruff, Leqi Xu, Simon van Haren, Adriana van de Guchte, Yujiao Zhao, Multi-omic longitudinal study reveals immune correlates of clinical course among hospitalized COVID-19 patients, Cell Reports Medicine, 10.1016/j.xcrm.2023.101079, 4 , 6, (101079), (2023).
- Yen-Fang Huang, Fang-Chi Hsu, Jiunn-Jong Wu, Yi-Ling Lin, Ming-Tsan Liu, Chin-Hui Yang, Hsu-Sung Kuo, Yen-Ju Chen, Chien-Yu Cheng, His-Hsun Lin, Chun-Che Liao, Chih-Shin Chang, Jian-Jong Liang, Wen-Yueh Cheng, Jason C. Huang, Cheng-Pin Chen, Shu-Hsing Cheng, Yi-Chun Lin, Shung-Haur Yang, Yiing-Jenq Chou, Longitudinal neutralizing antibody responses after SARS-CoV-2 infection: A convalescent cohort study in Taiwan, Journal of Microbiology, Immunology and Infection, 10.1016/j.jmii.2023.03.004, 56 , 3, (506-515), (2023).
- Christopher W Dukes, Renata AM Rossetti, Jonathan A Hensel, Sebastian Snedal, Christopher L Cubitt, Michael J Schell, Martha Abrahamsen, Kimberly Isaacs-Soriano, Kayoko Kennedy, Leslie N Mangual, Junmin Whiting, Veronica Martinez-Brockhus, Jessica Y Islam, Julie Rathwell, Matthew Beatty, Amy M Hall, Daniel Abate-Daga, Anna R Giuliano, Shari Pilon-Thomas, SARS-CoV-2 antibody response duration and neutralization following natural infection, Journal of Clinical Virology Plus, 10.1016/j.jcvp.2023.100158, 3 , 3, (100158), (2023).
- Kimberley Cousins, Kaori Sano, Brandon Lam, Katharina Röltgen, Disha Bhavsar, Gagandeep Singh, Oliver McRae, Stephanie Jeong, Nouran Aboelregal, Hsi-en Ho, Scott Boyd, Florian Krammer, Charlotte Cunningham-Rundles, Detection of SARS-CoV-2 Antibodies in Immunoglobulin Products, The Journal of Allergy and Clinical Immunology: In Practice, 10.1016/j.jaip.2023.05.005, 11 , 8, (2534-2541.e2), (2023).
- Marc Emmenegger, Elena De Cecco, David Lamparter, Raphaël P.B. Jacquat, Julien Riou, Dominik Menges, Tala Ballouz, Daniel Ebner, Matthias M. Schneider, Itzel Condado Morales, Berre Doğançay, Jingjing Guo, Anne Wiedmer, Julie Domange, Marigona Imeri, Rita Moos, Chryssa Zografou, Leyla Batkitar, Lidia Madrigal, Dezirae Schneider, Chiara Trevisan, Andres Gonzalez-Guerra, Alessandra Carrella, Irina L. Dubach, Catherine K. Xu, Georg Meisl, Vasilis Kosmoliaptsis, Tomas Malinauskas, Nicola Burgess-Brown, Ray Owens, Stephanie Hatch, Juthathip Mongkolsapaya, Gavin R. Screaton, Katharina Schubert, John D. Huck, Feimei Liu, Florence Pojer, Kelvin Lau, David Hacker, Elsbeth Probst-Müller, Carlo Cervia, Jakob Nilsson, Onur Boyman, Lanja Saleh, Katharina Spanaus, Arnold von Eckardstein, Dominik J. Schaer, Nenad Ban, Ching-Ju Tsai, Jacopo Marino, Gebhard F.X. Schertler, Nadine Ebert, Volker Thiel, Jochen Gottschalk, Beat M. Frey, Regina R. Reimann, Simone Hornemann, Aaron M. Ring, Tuomas P.J. Knowles, Milo A. Puhan, Christian L. Althaus, Ioannis Xenarios, David I. Stuart, Adriano Aguzzi, Continuous population-level monitoring of SARS-CoV-2 seroprevalence in a large European metropolitan region, iScience, 10.1016/j.isci.2023.105928, 26 , 2, (105928), (2023).
- Aviraag Vijaya Prakash, R.Ross Welliver, Sanjiti Mirmire, Sarah Baron, Mark D Hicar, Presence of coronary aneurysms during Kawasaki Disease (KD) correlates with lower levels of autoantibodies to both full form and spliced variant of immune regulator Del-1, Immunology Letters, 10.1016/j.imlet.2023.03.007, 256-257 , (34-41), (2023).
- J. Denis, A. Garnier, D. Claverie, F. De Laval, S. Attoumani, B. Tenebray, G.A. Durand, B. Coutard, I. Leparc-Goffart, J.N. Tournier, S. Briolant, C. Badaut, The Wood equation allows consistent fitting of individual antibody-response profiles of Zika virus or SARS-CoV-2 infected patients, Heliyon, 10.1016/j.heliyon.2023.e21945, 9 , 11, (e21945), (2023).
- Ji Youn Yoo, Sai Lata De, Anujit Sarkar, John H. Adams, Maureen Groer, A pilot study: Validation of dried blood spots (DBS) to assess SARS-CoV2 IgG antibody immunoassays in underserved minority population, Heliyon, 10.1016/j.heliyon.2023.e14729, 9 , 4, (e14729), (2023).
- Kirtan Kaur, Corina Lesseur, Lixian Chen, Syam S. Andra, Srinivasan Narasimhan, Divya Pulivarthi, Vishal Midya, Yula Ma, Erona Ibroci, Frederieke Gigase, Molly Lieber, Whitney Lieb, Teresa Janevic, Lotje D. De Witte, Veerle Bergink, Anna-Sophie Rommel, Jia Chen, Cross-sectional associations of maternal PFAS exposure on SARS-CoV-2 IgG antibody levels during pregnancy, Environmental Research, 10.1016/j.envres.2022.115067, 219 , (115067), (2023).
- Adolfo Aleman, Morgan van Kesteren, Ariel Kogan Zajdman, Komal Srivastava, Christian Cognigni, Jacob Mischka, Lucia Y. Chen, Bhaskar Upadhyaya, Kseniya Serebryakova, Jessica R. Nardulli, Neko Lyttle, Katerina Kappes, Hayley Jackson, Charles R. Gleason, Annika Oostenink, Gianna Y. Cai, Oliver Van Oekelen, Harm van Bakel, Emilia Mia Sordillo, Carlos Cordon-Cardo, Miriam Merad, Sundar Jagannath, Ania Wajnberg, Viviana Simon, Samir Parekh, Hala Alshammary, Dalles Andre, Radhika Banu, Katherine Beach, María Carolina Bermúdez-González, Ajai Chari, Yuexing Chen, Hearn Cho, Adolfo Firpo, Ana Silvia Gonzalez-Reiche, Eun Hye Kim, Giulio Kleiner, Florian Krammer, Jacob Mauldin, Rao Mendu, Brian Monahan, Shambavi Richard, Joshua Richter, Cesar Rodriguez, Adrianna Rossi, Ashley Salimbangon, Laryssa Sanchez, Daniel Verina, Cellular mechanisms associated with sub-optimal immune responses to SARS-CoV-2 bivalent booster vaccination in patients with Multiple Myeloma, eBioMedicine, 10.1016/j.ebiom.2023.104886, 98 , (104886), (2023).
- Fabio Salvatore Macaluso, Mariabeatrice Principi, Federica Facciotti, Antonella Contaldo, Alessia Todeschini, Simone Saibeni, Cristina Bezzio, Fabiana Castiglione, Olga Maria Nardone, Rocco Spagnuolo, Massimo Claudio Fantini, Gaia Riguccio, Flavio Caprioli, Chiara Viganò, Carla Felice, Gionata Fiorino, Carmen Correale, Giorgia Bodini, Monica Milla, Giulia Scardino, Marta Vernero, Federico Desideri, Mariella Mannino, Giuseppe Rizzo, Ambrogio Orlando, Arnaldo Amato, Marta Ascolani, Giulio Calabrese, Angelo Casà, Michele Comberlato, Francesco Simone Conforti, Manuela De Bona, Maria Giulia Demarzo, Patrizia Doldo, Gabriele Dragoni, Federica Furfaro, Giacomo Mulinacci, Oriana Olmo, Nicole Piazza O'Sed, Salvatore Paba, Simona Radice, Sara Renna, Davide Giuseppe Ribaldone, Giulia Rizzuto, Reduced humoral response to two doses of COVID-19 vaccine in patients with inflammatory bowel disease: Data from ESCAPE-IBD, an IG-IBD study, Digestive and Liver Disease, 10.1016/j.dld.2022.08.027, 55 , 2, (154-159), (2023).
- Chaouki Benabdessalem, Wafa Ben Hamouda, Soumaya Marzouki, Rokhaya Faye, Adji Astou Mbow, Babacar Diouf, Oumar Ndiaye, Ndongo Dia, Ousmane Faye, Amadou A. Sall, Cheikh Tidiane Diagne, Houda Amellal, Sayeh Ezzikouri, Diary Juliannie Ny Mioramalala, Fanirisoa Randrianarisaona, Khaled Trabelsi, Mohamed Boumaiza, Sonia Ben Hamouda, Rym Ouni, Soumaya Bchiri, Amani Chaaban, Mariem Gdoura, Yousr Gorgi, Imen Sfar, Sadok Yalaoui, Jalila Ben Khelil, Agnes Hamzaoui, Meya Abdallah, Yosra Cherif, Stéphane Petres, Chris Ka Pun Mok, Nicolas Escriou, Sébastien Quesney, Koussay Dellagi, Matthieu Schoenhals, M'hammed Sarih, Inès Vigan-Womas, Jihene Bettaieb, Samia Rourou, Mohamed Ridha Barbouche, Melika Ben Ahmed, Development and comparative evaluation of SARS-CoV-2 S-RBD and N based ELISA tests in various African endemic settings, Diagnostic Microbiology and Infectious Disease, 10.1016/j.diagmicrobio.2023.115903, 105 , 4, (115903), (2023).
- Vilja Pietiäinen, Minttu Polso, Ede Migh, Christian Guckelsberger, Maria Harmati, Akos Diosdi, Laura Turunen, Antti Hassinen, Swapnil Potdar, Annika Koponen, Edina Gyukity Sebestyen, Ferenc Kovacs, Andras Kriston, Reka Hollandi, Katalin Burian, Gabriella Terhes, Adam Visnyovszki, Eszter Fodor, Zsombor Lacza, Anu Kantele, Pekka Kolehmainen, Laura Kakkola, Tomas Strandin, Lev Levanov, Olli Kallioniemi, Lajos Kemeny, Ilkka Julkunen, Olli Vapalahti, Krisztina Buzas, Lassi Paavolainen, Peter Horvath, Jussi Hepojoki, Image-based and machine learning-guided multiplexed serology test for SARS-CoV-2, Cell Reports Methods, 10.1016/j.crmeth.2023.100565, 3 , 8, (100565), (2023).
- Hung-Bin Wu, Chih-Hung Wang, Yi-Da Chung, Yan-Shen Shan, Ying-Jun Lin, Huey-Pin Tsai, Gwo-Bin Lee, Highly-specific aptamer targeting SARS-CoV-2 S1 protein screened on an automatic integrated microfluidic system for COVID-19 diagnosis, Analytica Chimica Acta, 10.1016/j.aca.2023.341531, 1274 , (341531), (2023).
- Gonzalo Rivas, Nuria Labiod, Joanna Luczkowiak, Fátima Lasala, Marta Rolo, Mikel Mancheño‐Losa, David Rial‐Crestelo, Jaime Lora‐Tamayo, Alfredo Pérez‐Rivilla, María Dolores Folgueira, Rafael Delgado, Superior neutralizing response after first versus second SARS‐CoV‐2 infection in fully vaccinated individuals, Journal of Medical Virology, 10.1002/jmv.29225, 95 , 11, (2023).
- Zahra Hasan, Kiran Iqbal Masood, Shama Qaiser, Erum Khan, Areeba Hussain, Zara Ghous, Unab Khan, Maliha Yameen, Imran Hassan, Muhammad Imran Nasir, Muhammad Farrukh Qazi, Haris Ali Memon, Shiza Ali, Sadaf Baloch, Zulfiqar A. Bhutta, Marc Veldhoen, J. Pedro Simas, Syed Faisal Mahmood, Kulsoom Ghias, Rabia Hussain, Investigating the impact of prior COVID‐19 on IgG antibody and interferon γ responses after BBIBP‐CorV vaccination in a disease endemic population: A prospective observational study, Health Science Reports, 10.1002/hsr2.1521, 6 , 9, (2023).
- Zijie Zhang, Jiuxing Li, Ryan Amini, Alexandria Mansfield, Jimmy Gu, Jianrun Xia, John D. Brennan, Yingfu Li, Comparative Characterization of Diverse DNA Aptamers for Recognition of Spike Proteins of Multiple SARS‐CoV‐2 Variants, Analysis & Sensing, 10.1002/anse.202300001, 3 , 5, (2023).
- Shital Patil, Shubhangi Khule, Gajanan Gondhali, Dengue Fever with Corona Virus Disease 2019: Is it a “Double Trouble” with Concurrent Both Diseases or Single Disease with “Polyhedron” Nature and Antigenic Cross Reactivity?, Journal of Translational Critical Care Medicine, 10.4103/JTCCM-D-22-00009, 4 , 1, (17), (2022).
- Corey Gallen, Christopher W. Dukes, Amy Aldrich, Lauren Macaisa, Qianxing Mo, Christopher L. Cubitt, Shari Pilon-Thomas, Anna R. Giuliano, Brian J. Czerniecki, Ricardo L. B. Costa, Long-Term CD4+ T-Cell and Immunoglobulin G Immune Responses in Oncology Workers following COVID-19 Vaccination: An Interim Analysis of a Prospective Cohort Study, Vaccines, 10.3390/vaccines10111931, 10 , 11, (1931), (2022).
- Atsuhiko Sakamoto, Michinobu Yoshimura, Ryota Itoh, Ryo Ozuru, Kazunari Ishii, Yusuke Sechi, Shigeki Nabeshima, Kenji Hiromatsu, Longitudinal Dynamics of SARS-CoV-2 IgG Antibody Responses after the Two-Dose Regimen of BNT162b2 Vaccination and the Effect of a Third Dose on Healthcare Workers in Japan, Vaccines, 10.3390/vaccines10060830, 10 , 6, (830), (2022).
- See more

