Super-Resolution Stimulated Raman Scattering Microscopy with Graphical User Interface–Supported A-PoD
Hongje Jang, Hongje Jang, Yajuan Li, Yajuan Li, Shuang Wu, Shuang Wu, Lingyan Shi, Lingyan Shi
Abstract
Raman microscopy is a vibrational imaging technology that can detect molecular chemical bond vibrational signals. Since this signal is originated from almost every vibrational mode of molecules with different vibrational energy levels, it provides spatiotemporal distribution of various molecules in living organisms without the need for any labeling. The limitations of low signal strength in Raman microscopy have been effectively addressed by incorporating a stimulated emission process, leading to the development of stimulated Raman scattering (SRS) microscopy. Furthermore, the issue of low spatial resolution has been resolved through the application of computational techniques, specifically image deconvolution. In this article, we present a comprehensive guide to super-resolution SRS microscopy using an Adam-based pointillism deconvolution (A-PoD) algorithm, complemented by a user-friendly graphical user interface (GUI). We delve into the crucial parameters and conditions necessary for achieving super-resolved images through SRS imaging. Additionally, we provide a step-by-step walkthrough of the preprocessing steps and the use of GUI-supported A-PoD. This complete package offers a user-friendly platform for super-resolution SRS microscopy, enhancing the versatility and applicability of this advanced microscopy technique to reveal nanoscopic multimolecular nature. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Super-resolution stimulated Raman scattering microscopy with graphical user interface–supported A-PoD
Support Protocol : Deuterium labeling on cells with heavy water for metabolic imaging
INTRODUCTION
Vibrational microscopy, particularly Raman microscopy, holds significant promise in unveiling molecular structural details. Raman microscopy uses visible light sources, making it more versatile for configuring setups with various optical instruments. In theory, Raman scattering signals encompass structural information from every molecule within the sample. Nevertheless, when compared with fluorescence microscopy, this technique yields lower signal levels. To surmount this technical limitation, several approaches have been developed. By generating Raman scattering with one laser beam and stimulating the signal with another laser beam, the signal could be enhanced 108 times. This signal-enhancing process is called stimulated Raman scattering (SRS) and has been applied for microscopy (Freudiger et al., 2008). This signal enhancement has made practical imaging of biological samples feasible. However, the relatively modest resolution of SRS remains an obstacle to obtaining subcellular organelle-level information.
Numerous super-resolution microscopy methods have emerged, primarily relying on fluorescence signals (Betzig et al., 2006; Gustafsson, 2000; Hell & Wichmann, 1994; Rust et al., 2006). More recently, a handful of super-resolution techniques for Raman imaging have emerged. However, it is worth noting that the resolution limit of super-resolution SRS is still more than 10 times worse than that of super-resolution fluorescence microscopy. To address this limitation, we have adopted an image deconvolution method (Jang et al., 2023).
Image deconvolution is a computational technique used to recover details in distorted images (Sage et al., 2017; Sibarita, 2005), with blurring being a common type of distortion. Consequently, by solving the inverse problem of the relationship between the blurry images and the function to describe the pattern of blur (point spread function [PSF]), this method has been employed to eliminate blurriness and enhance spatial resolution. Nevertheless, it is important to note that due to potential artifacts that can arise during this process, image deconvolution does not qualify as a super-resolution microscopy technique.
Sparse deconvolution was initially used to address artifacts and reconstruct images in single-molecule localization microscopy (Hugelier et al., 2016; Min et al., 2014; Zhu et al., 2012). However, creating super-resolution images from a single frame of low-resolution images with a high emitter density proved to be a challenging task. To tackle this issue, we developed a more advanced sparse deconvolution algorithm known as Adam-based pointillism deconvolution (A-PoD; Jang et al., 2023). This approach mimics the concept of pointillism painting and describes images with multiple discontinuous spots (virtual emitters). The virtual emitters have the same unit intensity, and the total number of emitters is fixed. This characteristic allows for suppressing artifacts and describes real emitters’ distribution. Therefore, from low-resolution images of high-density emitters, super-resolution images can be restored. Accordingly, when applied to SRS microscopy, this method allowed us to achieve a spatial resolution below 60 nm.
Basic Protocol: SUPER-RESOLUTION STIMULATED RAMAN SCATTERING MICROSCOPY WITH GRAPHICAL USER INTERFACE–SUPPORTED A-PoD
The Basic Protocol provides information about how to measure SRS signals from samples and how to convert the images to super-resolution images using A-PoD. To ensure user-friendliness, we have created a graphical user interface (GUI) to support A-PoD. We guide the reader through the critical parameters and conditions necessary to obtain improved super-resolution images using this innovative concept, from the measurement process to the analysis stage.
Materials
-
Tissue or cell samples (see Support Protocol)
-
Immersion oil (e.g., Olympus, cat. no. IMMOIL-F30CC)
-
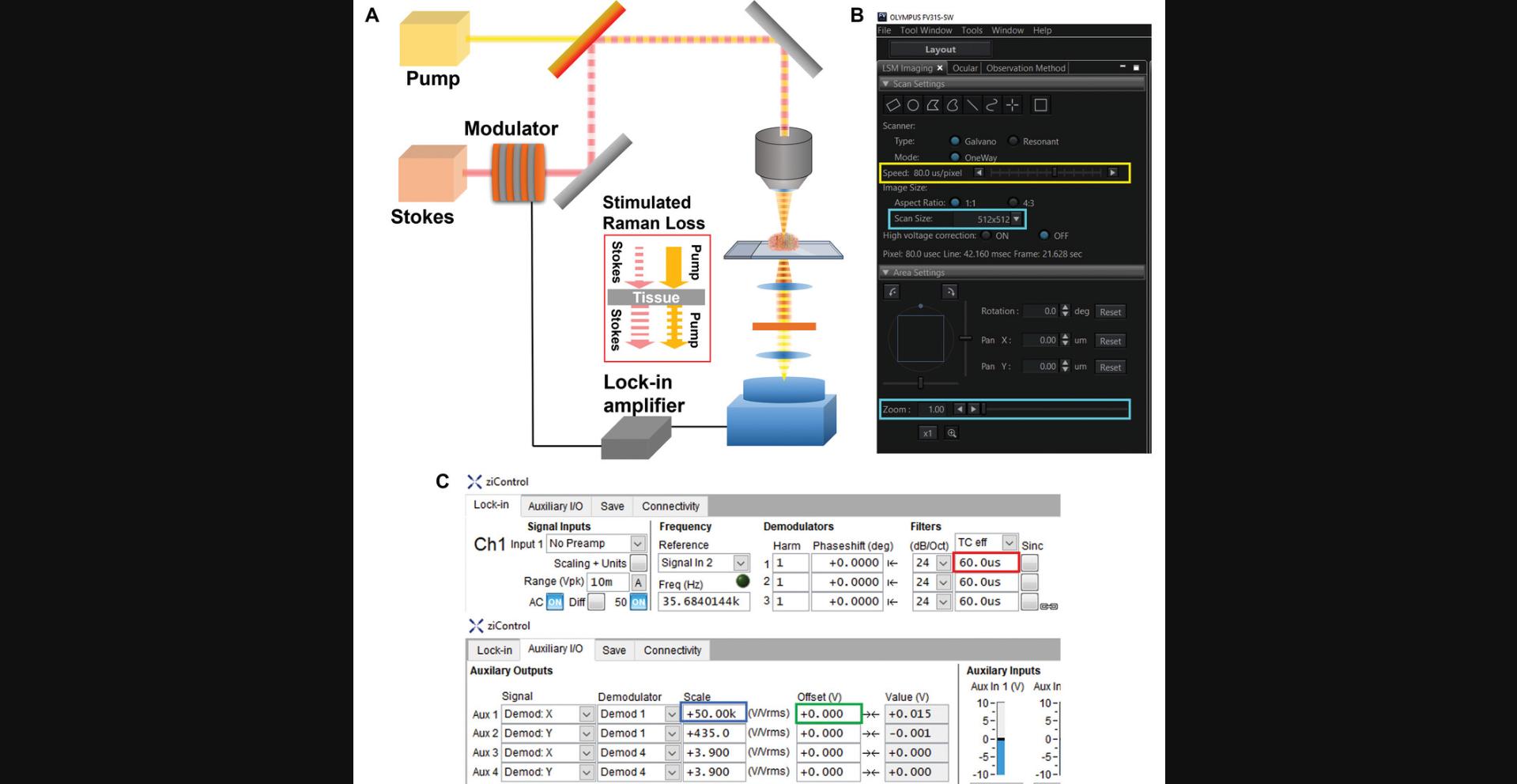
Custom-built upright laser-scanning microscope (e.g., Olympus FV1200MPE) for SRS setup (see Fig. 1A):
- Objective (e.g., Olympus XLPLN, WMP2, 1.05 NA)
- High-numerical aperture (NA) oil condenser (e.g., Olympus 6-U130)
- Photodiode detector (e.g., Hamamatsu S16008-1010)
-
picoEmerald Laser system (e.g., Applied Physics & Electronics)
-
Lock-in amplifier (e.g., Zurich Instruments HF2LI)
-
Computer with minimum requirements (tested with Intel Xeon Gold 5317 CPU, 128 GB RAM, and NVIDIA RTX A4500):
- 16 GB RAM
- Intel Core i5 10th Gen
- NVIDIA GTX 1060
-
ImageJ
-
Python 3.8.7 (with Tensorflow-gpu 2.6, Numpy 1.20.3, Pillow, Sci-kit image, opencv, pandas)
Measurement
1.Warm up laser and turn on microscope, Si photodiode detector, and photodiode detector.
2.Set measurement parameters of microscope and lock-in amplifier.
3.Set laser power (illumination intensity), wavelength, pulse width, and repetition rate (duration of laser pulse).
4.Mount sample on sample stage on an oil drop, and place a large water droplet on top of the sample slide.
5.Set size of images considering the wavelength and resolution limit.
6.Repeat steps to measure multiple channels with different wavenumbers.
Preprocessing for A-PoD (interpolation, denoising, PSF model generation)
7.Load image files in ImageJ.
8.Denoise images if signal-to-noise ratio is not enough.
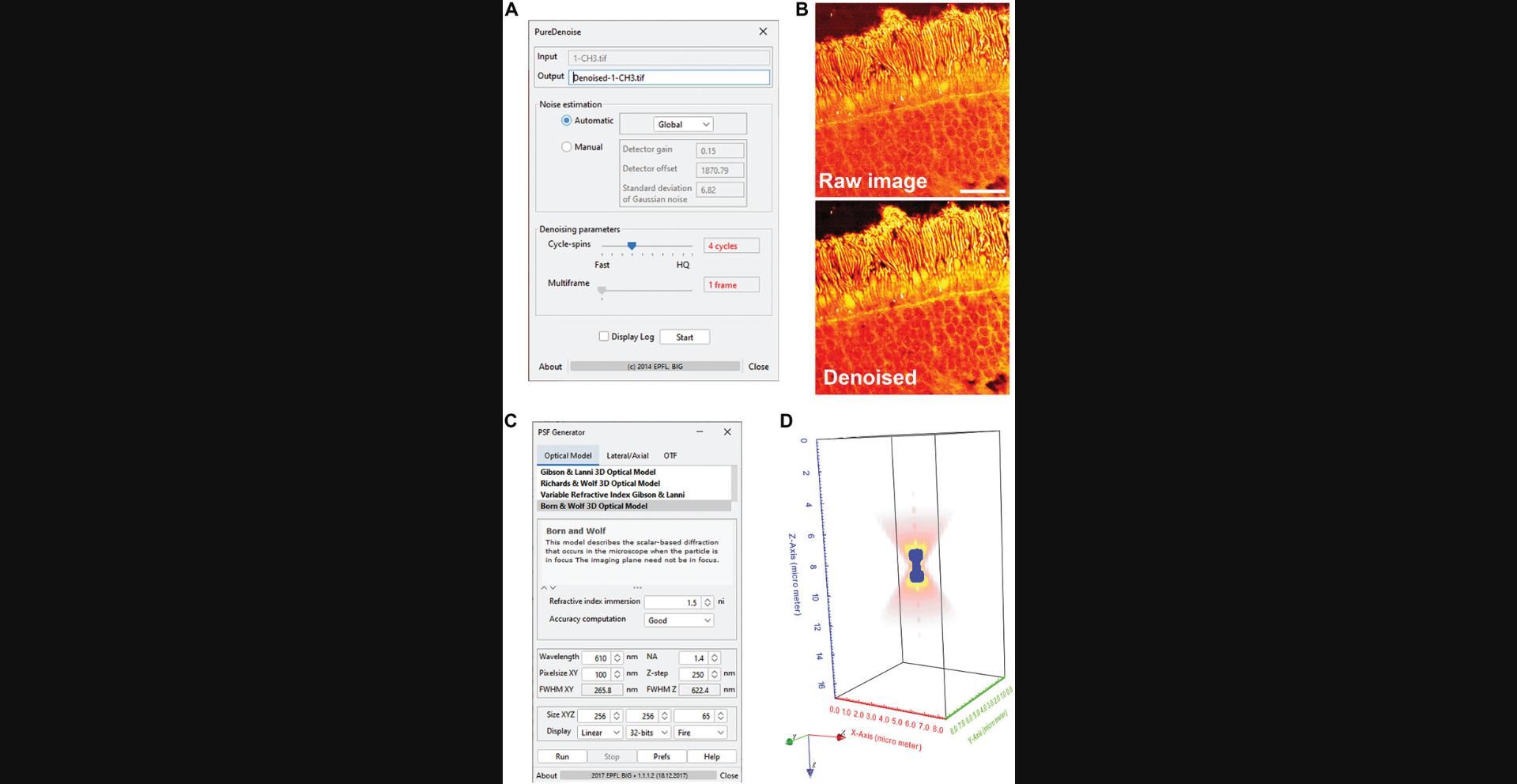
9.Resize images to make pixel size <100 nm, and save image as .tif format.
10.Install PSF generator plugin.
11.Run PSF generator, and enter parameters to generate PSF of Stokes beam.
12.Repeat step 11 to generate PSF of the Pump beam, with a different parameter of wavelength.
Installation of A-PoD
13.Install python 3.8.7.
14.Install plugins (Tensorflow-gpu, Numpy, Pillow, Scikit-image, OpenCV, and pandas).
15.Install GPU driver, CUDA, and CUDNN.
16.Download A-PoD code from Github (https://github.com/lingyanshi2020/A-PoD).
- A-PoD_GUI.py: A-PoD for 2D nonlinear microscopy images (SRS and two-photon fluorescence)
- A-PoD_GUI_Single_PSF.py: A-PoD for 2D fluorescence images
- A-PoD_GUI_3D.py: A-PoD for 3D nonlinear microscopy images (SRS and two-photon fluorescence).
Deconvolution using A-PoD (2D SRS images)
17.Run A-PoD_GUI.py.
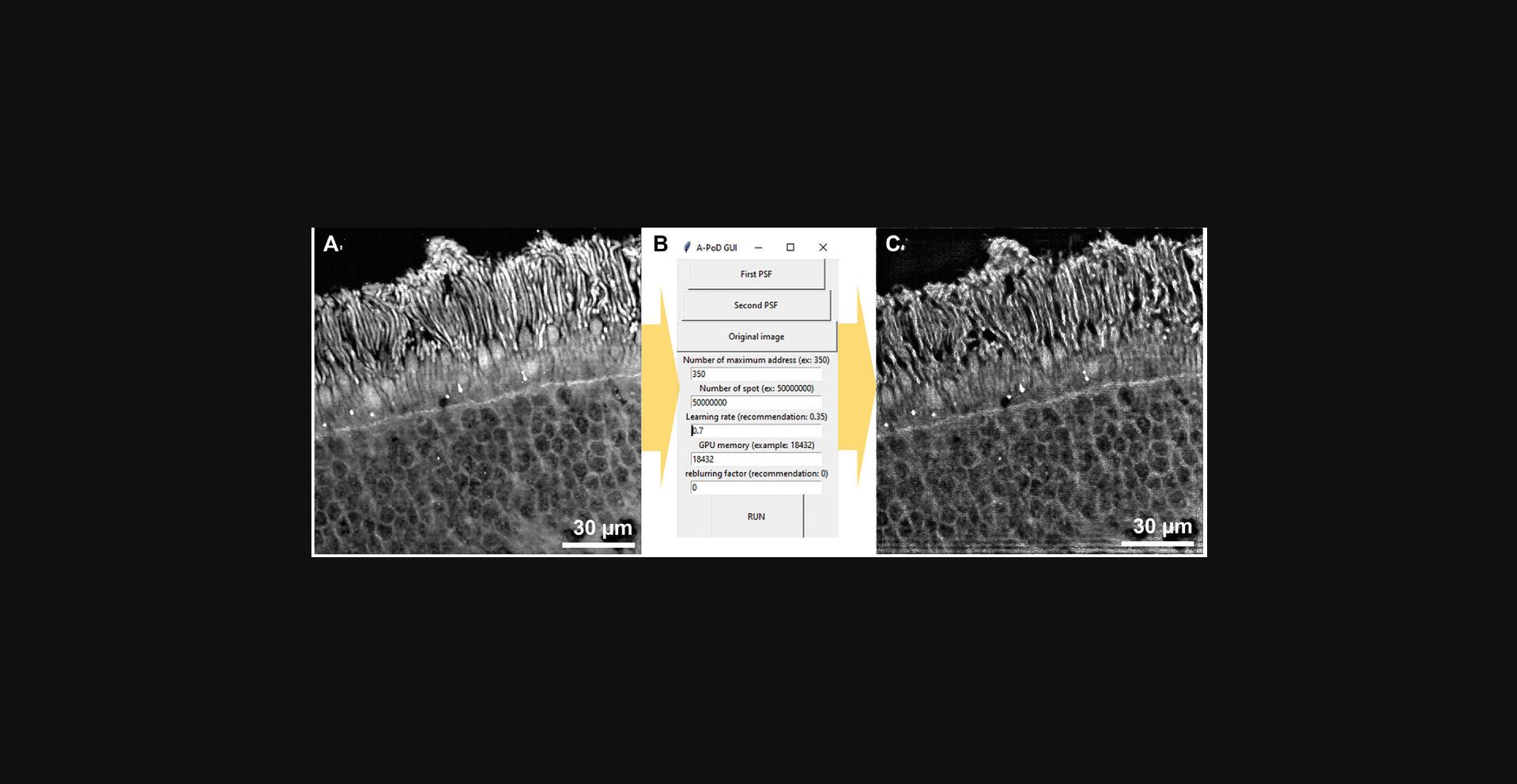
18.Set parameters:
- Number of maximum address: proportional constant of virtual emitter number
- Number of spot: maximum number of virtual emitters, which can be localized in a single process
- Learning rate: learning rate of Adam solver, which can be controlled from 0.01 to 1
- GPU memory: size of GPU memory in Megabyte
- Reblurring factor: sigma value of Gaussian function to reblur the resulting image to reduce overfitted results.
19.Click “First PSF,” and choose one of the PSF files.
20.Click “Second PSF,” and choose another PSF file.
21.Click “Original Image,” and choose image file that will be deconvolved.
22.Press “RUN” button.
23.Open deconvolved file in the folder.
Support Protocol: DEUTERIUM LABELING ON CELLS WITH HEAVY WATER FOR METABOLIC IMAGING
Metabolic imaging with SRS and deuterium labeling can be applied to various types of animals and cells (Bagheri et al., 2023; Li et al., 2023; Shi et al., 2018; Zhang et al., 2019). The Support Protocol outlines the process for deuterium labeling of cells. Given the diversity in methods for feeding animals and maintaining cells, the specific protocols may vary depending on the cell and animal types. Within this protocol, we focus on elucidating the most common cell culture procedure, which includes the deuterium labeling method.
Materials
-
Dulbecco's modified Eagle's medium (DMEM) powder
-
Double-distilled water
-
Heavy water (deuterium oxide [D20]; e.g., Cambridge Isotope Laboratories, cat. no. 7732-18-5)
-
Fetal bovine serum (FBS; e.g., Corning, cat. no. SH300880340)
-
Penicillin/streptomycin (e.g., Thermo Fisher Scientific, cat. no. 15140122)
-
HeLa cells (e.g., ATCC, cat. no. CCL-2)
-
Liquid DMEM (e.g., Corning, cat. no. 10027CV)
-
70% (v/v) ethanol
-
1× Dulbecco's phosphate-buffered saline (PBS), no calcium or magnesium (e.g., Thermo Fisher Scientific, cat. no. 14190136)
-
0.25% trypsin (e.g., Cytiva, cat. no. SH30042.02)
-
0.4% (w/v) trypan blue, 0.85% (w/v) NaCl (e.g., Lonza, cat. no. 17-942E)
-
1× PBS, with calcium and magnesium (e.g., Thermo Fisher Scientific, cat. no. 14040117)
-
16% (w/v) formaldehyde, methanol free (e.g., Thermo Fisher Scientific, cat. no. 28906)
-
Clear nail polish
-
15-ml centrifuge tubes (e.g., VWR, cat. no. 89039-664)
-
Vortex mixer
-
25-mm syringe filter, 2-μm polyethersulfone (PES) membrane (e.g., Foxx Life Sciences, cat. no. 381-2216-OEM)
-
Parafilm
-
10- or 25-cm2 flask
-
37°C, 5% CO2 cell culture incubator
-
Centrifuge
-
Hemocytometer (e.g., Millipore Sigma, cat. no. Z359629)
-
24-well plate (e.g., Fisher Scientific, cat. no. FB0112929)
-
Poly-D-lysine-coated 12-mm round coverslips
-
Light microscope
-
Microscope slides
-
9-mm diameter imaging spacers
-
Tweezers, sterile
Medium preparation
1.Weigh and mix 10 mg DMEM powder with 4.7 ml double-distilled water in a 15-ml conical tube. Gently vortex and invert tube to achieve thorough mixing of the solution. To prepare another solution containing heavy water (50% D2O), instead of 4.7 ml double-distilled water, use 2.35 ml double-distilled water and 2.35 ml heavy water.
2.Add 4.7 ml D2O, 0.5 ml FBS (5% [v/v] final), and 0.1 ml penicillin/streptomycin (1% [v/v] final). Gently vortex and invert to ensure complete homogenization of the solution.
3.Use a 25-mm syringe filter with 0.22-μm PES membrane to filter media.
4.Seal conical tubes containing media using parafilm, and store at 4°C for up to 3 weeks. Use prewarmed (37°C) media to treat cells in the next section.
Cell labeling
5.To maintain HeLa cells, culture in a 10- or 25-cm2 flask in standard DMEM supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin at 37°C with 5% CO2.
6.When HeLa cells reach ≥80% confluence, perform subculturing at a 1:10 split ratio.
7.To prepare cells for SRS imaging, when cells reach 80% confluency, rinse once with 1× PBS without magnesium or calcium.
8.Add 2 ml of 0.25% trypsin to detach adherent cells from the flask, and incubate at 37°C with 5% CO2 for 3 min.
9.Add 2 ml DMEM supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin. Gently pipette up and down, and collect cells by centrifuging 3 min at 300 × g , room temperature.
10.Resuspend cells in 1 ml DMEM supplemented with 0.5% (v/v) FBS and 1% (v/v) penicillin/streptomycin.
11.Count cells using trypan blue and a hemocytometer under a light microscope, and seed at a density of 2 × 105 cells per well in a 24-well plate with poly-D-lysine-coated glass coverslips.
12.Incubate cells at 37°C with 5% CO2, and gently shake plate to distribute the cells evenly.
13.After 8 hr, gently aspirate original medium using a serological pipet, and add 50% D2O as prepared in step 1 along the wall of culture well. Return cells to the 37°C, 5% CO2 incubator for 36 hr.
14.Optional : After 36 hr, fix cells on microscope slide:
-
Prepare microscope slides with 9-mm diameter imaging spacers, and place 15 μl of 1× PBS with calcium and magnesium on spacers.
-
Rinse cells with 1× PBS with calcium and magnesium.
-
Add 0.5 ml of 4% (w/v) formaldehyde, methanol free, and leave plate in a biosafety hood for ∼15 min.
-
Aspirate formaldehyde and rinse twice with 1× PBS with calcium and magnesium.
-
Add 1.5 ml of 1× PBS with calcium and magnesium to each well.
15.Gently pick up coverslips with sterile tweezers, invert, and place cell-containing side onto the imaging spacers to contact the PBS.
16.Seal outer layer of the coverslip with the imaging spacer using clear nail polish.
COMMENTARY
Background Information
In the past, sparse deconvolution techniques managed data sparsity by incorporating L1 or L0 penalty parameters (Hugelier et al., 2016; Min et al., 2014; Zhu et al., 2012). These methods proved effective for deconvolving images with high emitter densities, particularly in single-molecule localization microscopy. However, the challenge arises in the context of SRS images, which typically exhibit exceptionally high emitter densities. Consequently, conventional sparse deconvolution methods are inadequate for reconstructing super-resolution SRS images. To address this limitation, a Pointillism deconvolution approach (A-PoD) was introduced (Jang et al., 2023). This innovative concept restricts the total number of emitters with the same unit intensity, ensuring a sufficient level of data sparsity required for reconstructing super-resolution SRS images.
One of the primary strengths of SRS imaging lies in its ability to capture multiple molecular signals without labeling. When it comes to measuring various molecular groups and their subtypes, traditional methods often rely on a repeated measurement scheme involving the attachment and detachment of fluorophores. In contrast, SRS microscopy directly captures vibrational signals from numerous functional groups, eliminating the necessity for repetitive labeling processes to detect multiple molecular signals. By acquiring a limited number of channels that represent vibrational modes specific to various functional groups and then separating the spectral overlaps through unmixing, it becomes possible to quantitatively determine the molecular ratios (Lu et al., 2015; Shi et al., 2018).
By combining deuterium labeling with SRS imaging, metabolic activity such as turnover of biomolecules can be measured (Shi et al., 2018; Zhang et al., 2019). When we supply heavy water or deuterated glucose into cells and animals, carbon-deuterium bonds are formed in newly synthesized biomolecules during metabolism, with characteristic Raman peaks in the cell-silent region. In a previous study, we successfully demonstrated the concept of super-resolution metabolic imaging using heavy water. This metabolic imaging approach is now being applied to investigate the effects of aging and diet on metabolism in multiple organs (Li et al., 2023; Li, Bagheri et al., 2022; Li, Chen et al., 2022; Li, Zhang et al., 2022). By combining SRS, deuterium labeling, and A-PoD, we can uncover nanoscopic changes in subcellular organelle-level metabolism induced by factors such as aging, diet, and diseases. Furthermore, to make the approach more broadly applicable, it is essential to create various PSF models tailored to different microscopy setups and imaging systems. After the PSF model preparations for various imaging setups, achieving A-PoD could serve as a significant milestone in advancing multiple generations of super-resolution imaging techniques.
Furthermore, the A-PoD algorithm holds great potential in a broader context. When the PSF is accurately defined either through theoretical calculations or experimental data, A-PoD becomes a valuable tool for enhancing the spatial resolution of various microscopy images. This improvement extends to diverse imaging modalities, including phase contrast microscopy, photoacoustic microscopy, medical imaging devices, and fluorescence microscopy. Additionally, A-PoD can even enhance the spatial resolution of telescope images. The GUI introduced in this article serves to streamline the A-PoD process, making it more accessible and widening its range of applications.
Critical Parameters
To obtain reliable and clear images using A-PoD, it is essential to carefully consider several key parameters. First, optimizing the number of virtual emitters is crucial. The A-PoD process mimics pointillism painting concepts. Therefore, resulting images of A-PoD comprise multiple discontinuous spots (virtual emitters). Generally, a greater number of virtual emitters is preferable, but it can also lead to longer processing times. To streamline this parameter's estimation, it is calculated based on the variance and mean value of intensity. Occasionally, manual adjustments may be required, particularly in cases involving strong noise or tilted background signals. For manual corrections, the GUI now includes the option to adjust the “Number of maximum address,” typically ranging between 100 and 1000, with increases recommended if noisy results are observed.
Second, because the algorithm employs an Adam solver (Kingma & Ba, 2014), which is a gradient descent algorithm, the learning rate plays a significant role in avoiding local minima and expediting calculations. In this context, the learning rate represents the step size used to move each virtual emitter. If the learning rate is too high, processing times can become longer due to difficulties in precise localization. Conversely, if the learning rate is too low, the results may become trapped in local minima, resulting in artifacts. These artifacts are often related to noise patterns or unique patterns generated by various instrumental conditions.
Lastly, pixel size, referred to as spatial frequency, is critical for accurately localizing virtual emitters. A-PoD lacks the ability to localize within subpixel ranges, and the fitting precision is determined by the pixel size. For instance, if the pixel size is 50 nm, the resolution limit cannot be smaller than 50 nm. Therefore, capturing images with high spatial frequency and a large number of pixels will yield better spatial resolution in the deconvolution results compared with images with lower spatial frequency. Ideally, every sample should be measured with a very high spatial frequency. However, practical constraints like sample integrity and stage stability limit the extent to which we can increase spatial frequency. To address this limitation, interpolation methods, as described in step 9 of the Basic Protocol, can be employed.
Troubleshooting
Most of the problems and challenges are related to the parameters of A-PoD. The representative problems are listed in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| Strong noise in super-resolution images | Failed optimization due to insufficient number of virtual emitters | Increase parameter “Number of maximum address” or increase reblurring factor |
| Horizontal or vertical lines on the boundary of images | Signal goes out from the images; partial loss of emitter signal on the boundaries | Add more pixels on the image boundaries, zero-padding |
| Calculation too slow | Failed optimization due to insufficient number of virtual emitters; too large of a learning ratea | Increase parameter “Number of maximum address”; set lower value of learning rate |
- A-PoD, Adam-based pointillism deconvolution.
- a Too large of a learning rate can slow down the optimization process.
Time Considerations
The time required for imaging can vary based on factors such as image size and pixel number. Typically, imaging can take anywhere from a few seconds to several minutes. The time needed for deconvolution with A-PoD also depends on the image quality. For instance, in Figure 3, the deconvolution process for the retina image took <10 min when using a computer with the same specifications mentioned in the materials section.
Acknowledgments
The authors acknowledge the University of California, San Diego startup funds, grants from the National Institutes of Health (U54CA132378, 5R01NS111039, R21NS125395, U54DK134301, U54 HL165443, R01GM149976, NIAID U01AI167892, NSF 2320437), and a Hellman Fellow Award.
Author Contributions
Hongje Jang : software, writing—original draft; Yajuan Li : resources; Shuang Wu : software; Lingyan Shi : supervision, writing—review and editing.
Conflict of Interest
A provisional patent application has been filed by the University of California, San Diego patent office for L.S. and H.J. under the title “Super-resolution stimulated Raman scattering microscopy with A-PoD,” U.S. provisional application serial no. 63/379,226, filed October 12, 2022. All other authors declare no competing interests.
Open Research
Data Availability Statement
All the data and source code of this protocol are available with explanation at https://github.com/lingyanshi2020/A-PoD/.
Literature Cited
- Bagheri, P., Hoang, K., Kuo, C. Y., Trivedi, H., Jang, H., & Shi, L. (2023). Bioorthogonal chemical imaging of cell metabolism regulated by aromatic amino acids. Journal of Visualized Experiments , 195, e65121. https://doi.org/10.3791/65121
- Betzig, E., Patterson, G. H., Sougrat, R., Lindwasser, O. W., Olenych, S., Bonifacino, J. S., Davidson, M. W., Lippincott-Schwartz, J., & Hess, H. F. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science , 313(5793), 1642–1645. https://doi.org/10.1126/science.1127344
- Freudiger, C. W., Min, W., Saar, B. G., Lu, S., Holtom, G. R., He, C., Tsai, J. C., Kang, J. X., & Xie, X. S. (2008). Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science , 322(5909), 1857–1861. https://doi.org/10.1126/science.1165758
- Gustafsson, M. G. (2000). Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Journal of Microscopy , 198(2), 82–87. https://doi.org/10.1046/j.1365-2818.2000.00710.x
- Hell, S. W., & Wichmann, J. (1994). Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Optics Letters , 19(11), 780–782. https://doi.org/10.1364/OL.19.000780
- Hugelier, S., de Rooi, J. J., Bernex, R., Duwé, S., Devos, O., Sliwa, M., Dedecker, P., Eilers, P. H., & Ruckebusch, C. (2016). Sparse deconvolution of high-density super-resolution images. Scientific Reports , 6, 21413. https://doi.org/10.1038/srep21413
- Jang, H., Li, Y., Fung, A. A., Bagheri, P., Hoang, K., Skowronska-Krawczyk, D., Chen, X., Wu, J. Y., Bintu, B., & Shi, L. (2023). Super-resolution SRS microscopy with A-PoD. Nature Methods , 20(3), 448–458. https://doi.org/10.1038/s41592-023-01779-1
- Kingma, D. P., & Ba, J. (2014). Adam: A method for stochastic optimization. arXiv , 1412.6980 [preprint]. https://arxiv.org/abs/1412.6980
- Li, Q., Chen, Y., Feng, W., Cai, J., Gao, J., Ge, F., Zhou, T., Wang, Z., Ding, F., Marshall, C., Sheng, C., Zhang, Y., Sun, M., Shi, J., & Xiao, M. (2022). Drainage of senescent astrocytes from brain via meningeal lymphatic routes. Brain, Behavior, and Immunity , 103, 85–96. https://doi.org/10.1016/j.bbi.2022.04.005
- Li, Y., Bagheri, P., Chang, P., Zeng, A., Hao, J., Fung, A., Wu, J. Y., & Shi, L. (2022). Direct imaging of lipid metabolic changes in Drosophila ovary during aging using DO-SRS microscopy. Frontiers in Aging , 2, 819903. https://doi.org/10.3389/fragi.2021.819903
- Li, Y., Bagheri, P., Chang, P., Zeng, A., Hao, J., Fung, A., Wu, J. Y., & Shi, L. (2023). Bioorthogonal stimulated Raman scattering imaging uncovers lipid metabolic dynamics in Drosophila brain during aging. GEN Biotechnology , 2(3), 247–261. https://doi.org/10.1089/genbio.2023.0017
- Li, Y., Zhang, W., Fung, A. A., & Shi, L. (2022). DO-SRS imaging of diet regulated metabolic activities in Drosophila during aging processes. Aging Cell , 21(4), e13586. https://doi.org/10.1111/acel.13586
- Lu, F. K., Basu, S., Igras, V., Hoang, M. P., Ji, M., Fu, D., Holtom, G. R., Neel, V. A., Freudiger, C. W., Fisher, D. E., & Xie, X. S. (2015). Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proceedings of the National Academy of Sciences of the United States of America , 112(37), 11624–11629. https://doi.org/10.1073/pnas.1515121112
- Min, J., Vonesch, C., Kirshner, H., Carlini, L., Olivier, N., Holden, S., Manley, S., Ye, J. C., & Unser, M. (2014). FALCON: Fast and unbiased reconstruction of high-density super-resolution microscopy data. Scientific Reports , 4, 4577. https://doi.org/10.1038/srep04577
- Rust, M. J., Bates, M., & Zhuang, X. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods , 3(10), 793–796. https://doi.org/10.1038/nmeth929
- Sage, D., Donati, L., Soulez, F., Fortun, D., Schmit, G., Seitz, A., Guiet, R., Vonesch, C., & Unser, M. (2017). DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods , 115, 28–41. https://doi.org/10.1016/j.ymeth.2016.12.015
- Shi, L., Zheng, C., Shen, Y., Chen, Z., Silveira, E. S., Zhang, L., Wei, M., Liu, C., de Sena-Tomas, C., Targoff, K., & Min, W. (2018). Optical imaging of metabolic dynamics in animals. Nature Communications , 9, 2995. https://doi.org/10.1038/s41467-018-05401-3
- Sibarita, J.-B. (2005). Deconvolution microscopy. Microscopy Techniques , 201–243. https://doi.org/10.1007/b102215
- Zhang, L., Shi, L., Shen, Y., Miao, Y., Wei, M., Qian, N., Liu, Y., & Min, W. (2019). Spectral tracing of deuterium for imaging glucose metabolism. Nature Biomedical Engineering , 3(5), 402–413. https://doi.org/10.1038/s41551-019-0393-4
- Zhu, L., Zhang, W., Elnatan, D., & Huang, B. (2012). Faster STORM using compressed sensing. Nature Methods , 9(7), 721–723. https://doi.org/10.1038/nmeth.1978
Internet Resources
- https://github.com/lingyanshi2020/A-PoD
- Available A-PoD code repository, including brief instructions on how to install the code.
- https://imagej.net/software/fiji/
- FIJI (ImageJ) image-processing package; the package can be downloaded and executed without any additional installation.
- https://www.python.org/downloads/release/python-387/
- Python 3.8.7: A-PoD was mainly written in this language; the package can be downloaded and installed following the instructions.
- https://www.tensorflow.org/versions/r2.6/api_docs/python/tf
- Tensorflow-gpu 2.6: Python plugin for neural network programming; following the instructions, the package can be installed.
- https://pypi.org/project/numpy/1.20.3/
- Numpy 1.20.3: Mathematical library for Python; because of the tensorflow-gpu version, use of this specific version is recommended.
- https://pypi.org/project/Pillow/
- Pillow: Python imaging library; following the instructions, the newest version can be installed on Python.
- https://scikit-image.org/
- Sci-kit image: Image-processing plugin for Python; this provides a wide range of image-processing capabilities, encompassing both fundamental filtering and sophisticated image clustering functions.
- https://pypi.org/project/opencv-python/
- OpenCV: Open computer vision library; this library supports multiple functions for advanced image processing.
- https://pypi.org/project/pandas/
- pandas: Python data analysis library; this library supports functions for data analysis and manipulation.
- https://bigwww.epfl.ch/algorithms/denoise/
- PURE-denoise filter: Image denoising filter developed by Florian Luisier at the Biomedical Imaging Group (École Polytechnique Fédérale de Lausanne, Switzerland).
Citing Literature
Number of times cited according to CrossRef: 1
- William J. Tipping, Karen Faulds, Duncan Graham, Advances in Super-resolution Stimulated Raman Scattering Microscopy, Chemical & Biomedical Imaging, 10.1021/cbmi.4c00057, (2024).

