Hepatitis E Virus: Isolation, Propagation, and Quantification
Shaoli Lin, Shaoli Lin, Liping Yang, Liping Yang, Yan-Jin Zhang, Yan-Jin Zhang
Abstract
Hepatitis E virus (HEV) predominantly causes acute liver disease in humans and is transmitted via the fecal-oral route. HEV infection in pregnant women can result in grave consequences, with up to 30% fatality. The HEV strains infecting humans mainly belong to four genotypes. Genotypes 1 and 2 are restricted to human infection, while genotypes 3 and 4 are zoonotic. HEV genotype 3 (HEV-3) can cause both acute and chronic liver disease. Several cell lines (mainly hepatocytes) have been developed for HEV propagation and biological study. However, HEV production in these cell lines is suboptimal and inefficient. Here, we present methods for the isolation, propagation, and quantification of HEV. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Isolation and propagation of hepatitis E virus in cultured cells from clinical HEV specimens
Support Protocol 1 : Quantification of HEV RNA by RT-qPCR
Basic Protocol 2 : Recovery of HEV from infectious cDNA clones and purification of the virus
Support Protocol 2 : Quantification of HEV live particles by infectivity assay
INTRODUCTION
Hepatitis E is a liver disease caused by the hepatitis E virus (HEV; Nan & Zhang, 2016). According to the World Health Organization, hepatitis E caused ∼44,000 deaths in 2015, accounting for 3.3% of the mortality caused by viral hepatitis (World Health Organization, 2022). Among pregnant women with HEV infection, the case fatality rate can reach up to 30%. The virus is transmitted by the fecal-oral route, principally through contaminated water. HEV is a single-stranded, positive-sense RNA virus with a genome of about 7.2 kb in length. HEV belongs to the Hepeviridae , which contains two subfamilies: Orthohepevirinae and Parahepevirinae ( Purdy et al., 2022 ). The Orthohepevirinae consist of four genera: Avihepevirus , Chirohepevirus , Paslahepevirus , and Rocahepevirus. The Paslahepevirus genus includes two species: Paslahepevirus alci and Paslahepevirus balayani. There are eight genotypes in Paslahepevirus balayani , including the conventional genotypes 1, 2, 3, and 4 (Smith et al., 2014; Sridhar, Teng, Chiu, Lau, & Woo, 2017). Genotypes 1 and 2 have only been reported in humans, while genotypes 3 and 4 can infect both humans and animals. Genotypes 5 and 6 are isolated from wild boars, and genotypes 7 and 8 are found in camels (Sridhar et al., 2017). A genotype 7 HEV strain was recently found in a chronically infected liver transplant recipient who regularly consumed camel meat (Lee et al., 2016). HEV can infect a series of cells experimentally, including hepatocytes, lung cells, and colon epithelial cells (Emerson et al., 2004; Tanaka, Takahashi, Kusano, & Okamoto, 2007). However, it is hard to grow the virus robustly in cultured cells. For example, HEV Sar55 strain (genotype 1) can replicate in several cell lines post-transfection with full-length RNA of this virus (Table 1), but cell-to-cell spread cannot be achieved, which restricts effective virus proliferation. Although infectious cDNA clones have been established for the in vitro study of HEV (Emerson et al., 2001; Shukla et al., 2012), efficient production and propagation of HEV genotype 1 strains in cultured cells is still difficult. HEV strains of genotypes 3 and 4 appear to be able to infect cells in culture and grow robustly (Montpellier et al., 2017; Okamoto, 2011, 2013; Shukla et al., 2011). Using the HEV Kernow-C1 strain (genotype 3) as an example, the protocols presented here describe the isolation of HEV from clinical samples, recovery of virus from the cDNA clone, and propagation in HepG2/C3A cells, as well as purification and quantification of the virus.
| Genotype | Cells supporting HEV replication | Supplier |
|---|---|---|
| 1 | Caco-2 | ATCC #HTB-37, RRID: CVCL_0025 |
| Huh7.5 | RRID: CVCL_7927 | |
| A549 | NCI-DTP #A549, RRID: CVCL_0023 | |
| PLC/PRF/5 | ATCC #CRL-8024, RRID: CVCL_0485 | |
| Vero | ATCC #CCL-81, RRID: CVCL_0059 | |
| HepG2/C3A (Emerson et al., 2004) | ATCC #CRL-10741, RRID: CVCL_1098 | |
| 2 | Not reported | - |
| 3 | Caco-2 | ATCC #HTB-37, RRID: CVCL_0025 |
| PLC/PRF/5 and A549 (Tanaka et al., 2007) | ATCC #CRL-8024, RRID: CVCL_0485 and NCI-DTP #A549, RRID: CVCL_0023 | |
| LLP-CK1 | ATCC #CL-101, RRID: CVCL_0391 | |
| HepG2/C3A (Shukla et al., 2012) | ATCC #CRL-10741, RRID: CVCL_1098 | |
| HepaRG (Xu et al., 2017) | Millipore, cat. no. C103485, RRID: CVCL_9720 | |
| MRC5 (Xu et al., 2016) | ATCC #CCL-171, RRID: CVCL_0440 | |
| 4 | A549 | NCI-DTP #A549, RRID: CVCL_0023 |
| PLC/PRF/5 (Okamoto, 2011) | ATCC #CRL-8024, RRID: CVCL_0485 |
- a HEV, hepatitis E virus.
CAUTION : Hepatitis E virus is a Biosafety Level 2 (BSL-2) organism and must be handled under BSL-2 conditions, including use of a class II biological safety cabinet. See Burnett, Lunn, & Coico (2009) and other resources (Current Protocols, 2005) for more information.
Basic Protocol 1: ISOLATION AND PROPAGATION OF HEPATITIS E VIRUS IN CULTURED CELLS FROM CLINICAL HEV SPECIMENS
HepG2/C3A is the most frequently used cell line for persistent propagation of the HEV Kernow-C1 strain, and the virus can persist in the cells for up to six passages without significant titer drop (Table 1). Clinical fecal specimens to be tested should be kept at 4°C if processing within 48 hr is possible. For long-term storage, the specimens should be kept at –80°C for up to 1 year. Primary HEV specimens require the addition of antibiotics prior to inoculation of the cultured cells (Tanaka et al., 2007).
Materials
-
Complete growth medium (see recipe)
-
HepG2/C3A cells (ATCC #CRL-10741)
-
75% (v/v) ethanol
-
0.05% trypsin-EDTA (e.g., Corning, cat. no. 25-053-CI)
-
Clinical HEV fecal specimen
-
10 mM Tris·Cl, pH 7.5 (see Current Protocols article: Moore, 1996)
-
37°C water bath
-
15-ml centrifuge tubes
-
Centrifuge
-
25-cm2 and 75-cm2 tissue culture flasks
-
37°C, 5% CO2 humidified incubator
-
Inverted phase contrast microscope
-
Vortex
-
0.22-μm microfilter (e.g., Millipore Millex-GV)
-
Additional reagents and equipment for reverse-transcription quantitative PCR (RT-qPCR; see Support Protocol 1)
Prepare cell culture
1.Prewarm complete growth medium in a 37°C water bath.
2.Add 5 ml growth medium to a 15-ml centrifuge tube.
3.Remove a vial of HepG2/C3A cells from liquid nitrogen storage Dewar, and immediately thaw in a 37°C water bath. Remove the vial from the water bath when there is tiny piece of ice left in the vial.
4.Wipe the vial with 75% ethanol. Transfer the cell suspension from the vial to the warm growth medium (5 ml) in the 15-ml centrifuge tube, one drop at a time.
5.Centrifuge the tube 5 min at 200 × g , room temperature. Decant supernatant.
6.Resuspend the cell pellet with 5 ml warm complete growth medium, and transfer the cells to a 25-cm2 flask. Incubate the flask in a humidified incubator at 37°C with 5% CO2 for 24 to 48 hr.
7.Observe cell confluence daily through an inverted phase-contrast microscope. When cell monolayer reaches near complete confluence, detach the cells using trypsin incubation at 37°C for 3 to 5 min, and transfer the cells to a 75-cm2 flask for expansion of culture.
Isolate and propagate HEV from clinical specimen
8.For isolation of HEV, suspend 1.5 g stool sample in 2 to 3 ml of 10 mM Tris·Cl in a 15-ml tube, and vortex for 30 s (also see Tanaka et al., 2007).
9.Centrifuge the specimen 10 min at 6200 × g , 4°C. Aliquot the supernatant as virus stocks, and store at –80°C.
10.Determine the HEV RNA copy numbers of the virus stock using RT-qPCR as described in Support Protocol 1.Culture the HepG2/C3A cells at 37°C with complete growth medium until the cell confluence reaches 60% to 70%.
11.Purify the virus stock by passaging through 0.22-μm microfilters. Assess the concentration of the filtered HEV stock by RT-qPCR as described in Support Protocol 1. Inoculate the cells with the specimen at 10,000 HEV genomes/cell, and incubate the flask for 5 hr at 37°C. Decant the inoculum and add fresh medium. Allow the virus to proliferate for 5 days.
12.Expand the cells to a 75-cm2 flask and monitor the virus release into the medium by RT-qPCR as described in Support Protocol 1.
Support Protocol 1: QUANTIFICATION OF HEV RNA BY RT-qPCR
Because HEV infection does not produce cytopathic effects in cultured cells, a plaque assay for virus quantitation is not feasible. Due to the slow growth rate and low production of HEV, indirect immunofluorescence assays for virus quantification may need a relatively long period. In contrast, RT-qPCR is considered a convenient and quick tool frequently used for virus quantification (Nan et al., 2015; Zhou et al., 2014).
Materials
-
HepG2/C3A cells infected with HEV (see Basic Protocol 1)
-
TRIzol Reagent (e.g., Thermo Fisher Scientific, cat. no. 15596018)
-
Chloroform
-
Isopropanol
-
Nuclease-free 75% (v/v) ethanol
-
Nuclease-free water
-
RevertAid First Strand cDNA Synthesis Kit (e.g., Thermo Fisher Scientific, cat. no. K1691)
-
Random hexamer primers
-
HEV forward and reverse primers (HEV-F and HEV-R)
-
SYBR Green I master mix (e.g., Thermo Fisher Scientific, cat. no. 4309155)
-
Centrifuge
-
1.5-ml microcentrifuge tubes, nuclease-free
-
Low profile 8-strip PCR tubes
-
Thermal cycler
-
Real-time PCR detection system
RNA isolation
1.For detection of virus copies in infected HepG2/C3A cells, remove the growth medium and add 0.3 to 0.4 ml TRIzol Reagent per 1 × 105 to 1 × 106 cells directly to the culture dish to lyse the cells.
2.Add 200 μl chloroform per 1 ml sample harvested by TRIzol and shake vigorously for 15 s. Keep the mixture standing for 5 min at room temperature, and then centrifuge 15 min at 14,000 × g , 4°C.
3.Transfer the top layer supernatant to a new 1.5-ml microcentrifuge tube. Add the same volume of cold isopropanol, and then mix thoroughly. Centrifuge 15 min at 14,000 × g , 4°C. Carefully decant the supernatant.
4.Add 1 ml cold 75% ethanol, and then centrifuge 5 min at 14,000 × g , 4°C. Decant the supernatant carefully, and aspirate remaining liquid in the tube. Let the RNA pellet dry for 1 min.
5.Add 20 μl nuclease-free water to dissolve the RNA.
Reverse transcription and real-time PCR
6.Using 5 μl RNA as the template, perform reverse transcription using RevertAid First Strand cDNA Synthesis Kit (according to kit instructions) with a random hexamer primer or a specific primer (i.e., HEV-R; Table 2).
| Primer | Sequence |
|---|---|
| HEV-Fa | 5′-CGTGTCGCTATCTCCACCTA-3′ |
| HEV-R | 5′-TGTCCTCAAGAACAGCAAGG-3′ |
- a
F, forward; HEV, hepatitis E virus; R, reverse.
7.Mix the reaction of the reverse transcription in a PCR tube. Perform the reverse transcription reaction on a thermal cycler using the following protocol:
| 1 cycle | 5 min | 25°C |
| 60 min | 42°C | |
| 5 min | 75°C | |
| 5 min | 4°C |
8.Dilute the product of reverse transcription (step 7) at a ratio of 1:10, and use the diluted product as a template for real-time PCR with primer HEV-F and HEV-R (Table 2). Add the following components to a PCR tube:
- 7 μl SYBR Green I master mix
- 1 μl of 2.5 μM primer HEV-F
- 1 μl of 2.5 μM primer HEV-R
- 5 μl diluted reverse transcription product.
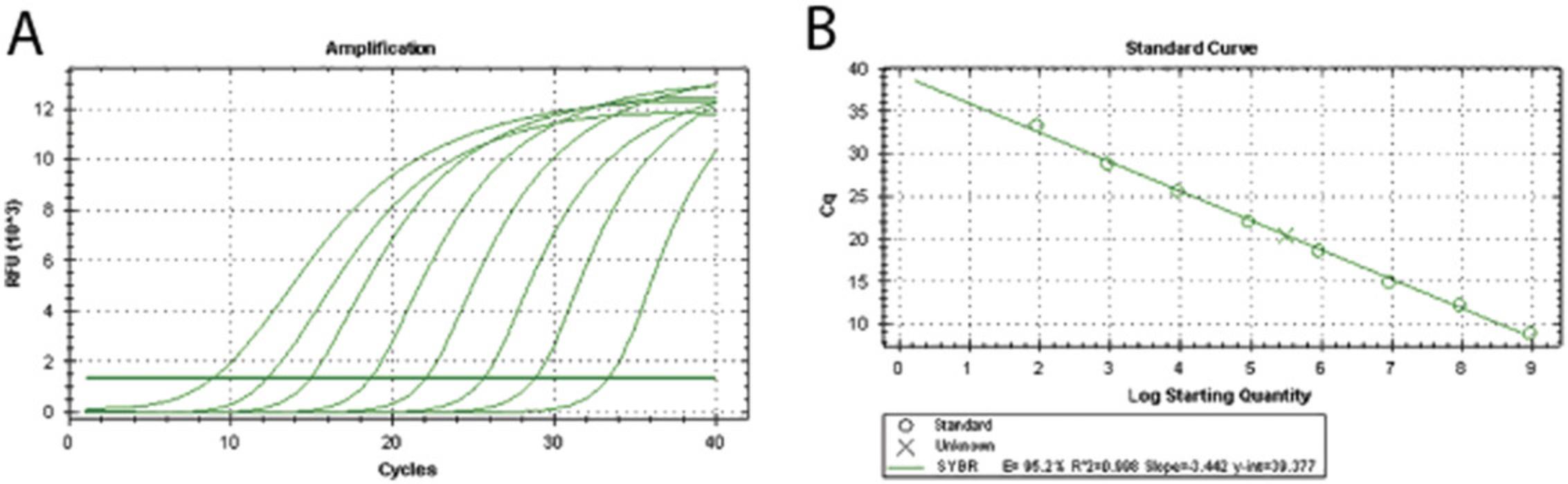
9.Dilute DNA of linearized HEV cDNA infectious clone in a 10-fold series from 108 to 1 copies/μl for a standard curve in the real-time PCR. Compose the reaction volume as described in step 8.
10.Perform the real-time PCR using the following protocol:
| Pre-denature | 5 min | 95°C |
| 40 cycles | 10 s | 95°C |
| 40 s | 60°C | |
| Melting curve | 5 s each | 0.5°C increments from 65°C to 95°C |
| Final step | 20°C |
Basic Protocol 2: RECOVERY OF HEV FROM INFECTIOUS cDNA CLONES AND PURIFICATION OF THE VIRUS
Infectious cDNA clones of HEV have been established for recovery of the virus and study of its biology (Emerson et al., 2001; Shukla et al., 2012). Here, the Kernow-C1 p6 clone containing the full-length cDNA of the HEV Kernow-C1 strain (GenBank accession number JQ679013) is used as an example in the following protocol (Shukla et al., 2011; Xu et al., 2017). For purification of the virus, density gradient centrifugation can be used (Emerson et al., 2010).
Materials
-
cDNA clone of Kernow-C1 p6 (see Shukla et al., 2011)
-
20 U/μl Mlu I with 10× reaction buffer (e.g., New England BioLabs, cat. no. R0198S)
-
Nuclease-free water
-
6× DNA loading dye (e.g., Thermo Fisher Scientific, cat. no. R1151)
-
1% (w/v) agarose Tris-acetate-EDTA (agarose-TAE) gel (see recipe)
-
1× TAE buffer (see recipe)
-
GeneJET PCR Purification Kit (e.g., Thermo Fisher Scientific, cat. no. K0701)
-
mMESSAGE mMACHINE T7 Transcription Kit (e.g., Thermo Fisher Scientific, cat. no. AM1344)
-
TRIzol Reagent (e.g., Thermo Fisher Scientific, cat. no. 15596018)
-
Chloroform
-
Isopropanol
-
Nuclease-free 70% (v/v) ethanol
-
2× RNA loading dye
-
HepG2/C3A cells infected with HEV (see Basic Protocol 1)
-
Complete growth medium (see recipe)
-
0.05% trypsin-EDTA (e.g., Corning, cat. no. 25-053-CI)
-
Opti-MEM Reduced Serum Medium (e.g., Thermo Fisher Scientific, cat. no. 51985091)
-
TNE buffer (see recipe)
-
15%, 20%, 30%, 40%, and 50% sucrose buffer solutions (see recipe)
-
37°C, 5% CO2 incubator
-
75°C incubator
-
Centrifuge
-
1.5-ml microcentrifuge tubes, sterile
-
Spectrophotometer
-
25-cm2 flask
-
4-mm electroporation cuvette
-
Gene Pulser Xcell Electroporation System (e.g., Bio-Rad)
-
37°C, 5% CO2 humidified incubator
-
Ultracentrifuge with swinging bucket rotor (e.g., SW41 rotor)
-
Centrifuge tubes suitable for ultracentrifugation
-
Syringe with long needle
-
RNeasy Micro Kit (Qiagen, cat. no. 74004)
-
Additional reagents and equipment for gel electrophoresis (see Current Protocols article: Voytas, 2001)
Plasmid preparation
1.Linearize the Kernow-C1 p6 plasmid with Mlu I restriction enzyme by adding the following components to a 1.5-ml tube:
- 10 μg Kernow-C1 p6 plasmid
- 20 U Mlu I enzyme
- 2 μl 10× reaction buffer
- Nuclease-free water to a final volume of 20 μl.
2.Incubate the reaction at 37°C for 4 to 8 hr.
3.After the incubation, take 2 μl of the reaction mixture and add 1/6 vol of 6× DNA loading dye. Load the mixture onto a 1% agarose-TAE gel, and then run the gel in TAE buffer. Include the same amount of undigested DNA plasmid as a control.
4.Optional : If the plasmid is not fully linearized, add an additional 10 U Mlu I, and continue the digestion.
5.Purify the linearized plasmid DNA with GeneJET Purification Kit following the manufacturer's instructions.
In vitro transcription and purification
6.Use the purified linear plasmid as the template to conduct in vitro transcription using the mMESSAGE mMACHINE in vitro RNA transcription kit according to the manufacturer's instructions to generate capped HEV RNA.
7.After RNA generation, digest the DNA in the reaction mixture with 2 μl DNase provided in the kit for 15 min.
8.For purification of the RNA, add 900 μl TRIzol Reagent to the RNA product, and invert the tube four times. Next, add 200 μl chloroform, and shake vigorously for 15 s. Keep the mixture standing for 5 min at room temperature, and then centrifuge 15 min at 14,000 × g , 4°C.
9.Transfer the top liquid layer to a new 1.5-ml microcentrifuge tube. Add the same volume of cold isopropanol, and mix thoroughly.
10.Centrifuge the mixture 15 min at 14,000 × g , 4°C. Carefully decant the supernatant.
11.Add 1 ml cold 70% ethanol, and then centrifuge 5 min at 14,000 × g , 4°C.
12.Decant the supernatant carefully, and aspirate any liquid residue. Let the RNA pellet dry for 1 min.
13.Add 50 μl nuclease-free water to dissolve the RNA.
14.To 3 μl RNA, add 3 μl of 2× RNA loading dye containing the denaturing agent. Incubate the mixture for 5 min at 75°C, and then allow it to cool for 2 min on ice. Perform electrophoresis of the RNA product using a 1% agarose-TAE gel to test the integrity of the RNA. Add an RNA ladder to allow for size measurement of the purified RNA. Assess the concentration and purity of RNA using a spectrophotometer at OD260/280.
Cell electroporation with the HEV RNA
15.Culture the HepG2/C3A cells with complete growth medium in a 25-cm2 flask until the cells reach confluence, and then detach the cells using trypsin and incubation at 37°C for 3 to 5 min.
16.Rinse the cells three times with Opti-MEM by centrifugation, and then resuspend the cell pellet in 200 μl Opti-MEM.
17.Mix the cells with 10 μg p6 full-length HEV RNA (from step 13), and then transfer the mixture to a 4-mm cuvette. Keep the mixture on ice before the electroporation.
18.Conduct electroporation of the HepG2/C3A cells in the 4-mm cuvette using the Bio-Rad Gene Pulser Xcell Electroporation Systems (Bio-Rad).
19.Add 5 ml complete growth medium to a 25-cm2 flask. Transfer the cells from the cuvette after electroporation to the flask. Incubate the flask at 37°C with 5% CO2 for up to 1 week. Check for HEV-infected cells by immunofluorescence assay as described in Support Protocol 2.
20.Expand the culture or inoculate fresh cells to propagate the virus as described in Basic Protocol 1.
Concentration of the virus
21.Collect the supernatant that contains HEV viral particles from HEV-infected cells, and then centrifuge 30 min at 8000 × g , 4°C, to remove cell debris.
22.Subject the supernatant to ultracentrifugation 2 hr at 100,000 × g , 4°C, to concentrate HEV virus particles.
23.Decant the supernatant carefully. Resuspend the pellet with 1 ml TNE buffer.
Preparation of a sucrose gradient
24.Prepare five sucrose solutions (15%, 20%, 30%, 40%, and 50%).
25.Divide the total volume of a centrifuge tube by 5.
26.Add 2.44 ml of the 15% sucrose buffer solution to every centrifuge tube.
27.Inject 2.44 ml of the 20% sucrose buffer solution underneath the 15% solution using a syringe with a long needle.
28.Repeat step 27 for the 30%, 40%, and 50% sucrose buffer solutions.
Purification of the virus
29.Add 1 ml of concentrated virus suspension (from step 23) carefully on top of the gradient. Balance the tubes by weight to within 1 mg using TNE buffer.
30.Insert the tubes carefully in the buckets and hang the buckets in the correct orientation in their appropriate place on the SW41 rotor.
31.Centrifuge 5 hr at 100,000 × g , 4°C.
32.After centrifugation, carefully remove the centrifuge tubes from each bucket.
33.Collect the band(s) in the tubes with a disposable syringe and needle. To do so, slide the needle along the tube wall to just below the band, and withdraw the band slowly into the syringe. Then, add the collected bands to separate tubes.
34.Dilute the virus with 35 ml TNE buffer, and centrifuge 2 hr at 100,000 × g , 4°C, to remove the sucrose from the virus suspension. Remove the supernatant and resuspend the pellet in 1 ml TNE buffer.
35.Analyze 10 μl for the presence of the virus using RT-qPCR as described in Support Protocol 1.
36.Dilute the virus with TNE buffer to 1 × 107 copies/ml. Aliquot and store at –80°C.
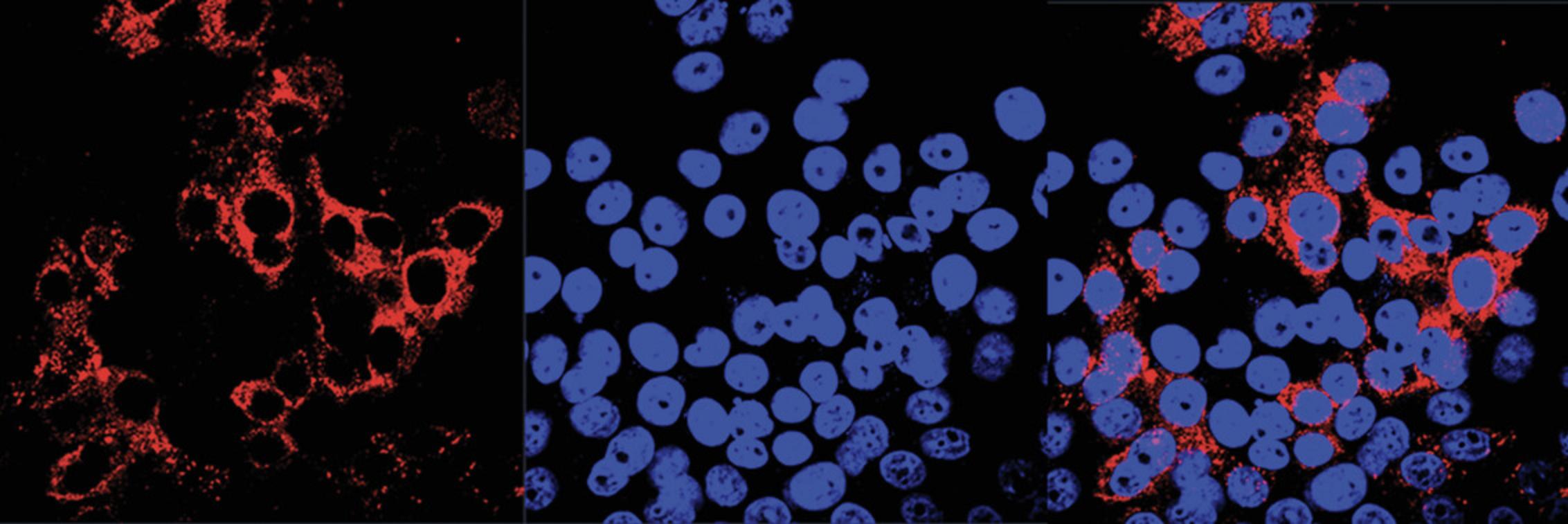
Support Protocol 2: QUANTIFICATION OF HEV LIVE PARTICLES BY INFECTIVITY ASSAY
It was previously reported that HepG2/C3A cells can be infected by the HEV Kernow-C1 strain at high efficiency (Shukla et al., 2012). Here, infected HepG2/C3A cells are used to illustrate determination of HEV infectivity using an immunofluorescence assay.
Materials
-
HepG2/C3A cells infected with HEV (see Basic Protocol 1)
-
0.05% trypsin-EDTA (e.g., Corning, cat. no. 25-053-CI)
-
Complete growth medium (see recipe)
-
1:1 methanol:acetone
-
Phosphate-buffered saline (PBS; see recipe)
-
Mouse monoclonal antibody against HEV open reading frame 2 (ORF2; e.g., Millipore, cat. no. MAB8002, RRID: AB_11212382)
-
Goat anti-mouse IgG (H&L) antibody, Dylight 549–conjugated (e.g., Rockland Immunologicals, cat. no. 610-142-002, RRID: AB_11182796)
-
SlowFade Gold antifade reagent containing DAPI (e.g., Thermo Fisher Scientific, cat. no. S36939)
-
12-well cell culture plate
-
Circular cover glass (12 mm diameter)
-
37°C, 5% CO2 humidified incubator
-
37°C microbiological incubator
-
Glass microscope slides
-
Fluorescence microscope with 63× oil immersion lens
Inoculate HepG2/C3A cells with HEV
1.Prepare HepG2/C3A cells as described in Basic Protocol 1.
2.Detach confluent cell monolayer using trypsin and incubation at 37°C for 3 to 5 min, and plate cells into 12-well culture plates with a cover glass. Incubate the plate at 37°C with 5% CO2 in a humidified incubator overnight.
3.Incubate cells with HEV samples to be tested at 37°C with 5% CO2 for 5 hr, and replace the medium with complete growth medium. Incubate the plate for 5 to 6 days at 37°C.
Indirect immunofluorescence assay
4.Fix cells with cold 1:1 methanol:acetone for 15 min.
5.Rinse the cover glass with PBS once and add anti-HEV ORF2 antibody diluted with PBS (at a working concentration recommended starting 1:200). Incubate the plate at 37°C for 35 min.
6.Rinse the cover glass three times with PBS, each time for 5 min, and add Dylight 549–conjugated goat anti-mouse IgG antibody diluted with PBS at 1:1000. Incubate the plate at 37°C for 35 min.
7.Rinse the cells three times with PBS, each time for 5 min. Mount the cover glass onto a slide with 5 μl SlowFade Gold antifade reagent.
8.Observe the cells under a fluorescence microscope with an oil-immersion lens.
REAGENTS AND SOLUTIONS
Agarose-TAE gels, 1%
- 1.Weigh 1 g agarose and mix agarose powder with 100 ml 1× TAE buffer (see recipe) in a glass flask.
- 2.Microwave the flask for a few minutes until the agarose is completely dissolved, but do not over-boil the solution, as some of the buffer will evaporate and thus alter the final percentage of agarose in the gel.
Many people prefer to microwave in pulses, swirling the flask occasionally as the solution heats up.
- 3.Let agarose solution cool down to about 50°C (∼5 min), and then pour the agarose into a gel tray with a well comb in place.
- 4.Place newly poured gel at 4°C for 10 to 15 min, or let it sit at room temperature for 20 to 30 min until it is completely solid.
Store gels at room temperature in 1× TAE buffer for up to 2 weeks
Complete growth medium
- 450 ml Dulbecco's Modified Eagle Medium (DMEM; e.g., Corning, cat. no. 10-013-CV)
- 50 ml heat-inactivated fetal bovine serum (FBS; see recipe)
- 10 ml 100 mM sodium pyruvate (e.g., Thermo Fisher Scientific, cat. no. 11360070)
- 10 ml 10,000 IU/ml penicillin-streptomycin solution
- 29.2 mg/ml L-glutamine (e.g., Corning, cat. no. 30-009-CI)
- Store at 4°C for up to 2 months
Add fresh L-glutamine to a final concentration of 0.292 mg/ml every 2 weeks.
Heat-inactivated FBS
Thaw a 500-ml bottle of FBS (e.g., Thermo Fisher Scientific, cat. no. 26140-079) overnight at 4°C. If it is still frozen the next day, continue thawing at 37°C. Place the bottle in a 56°C water bath for 30 min with occasional gentle shaking. Store 50-ml aliquots at –20°C for up to 2 years.
Phosphate-buffered saline (PBS)
- 8 g NaCl (final concentration 137 mM)
- 1.44 g Na2HPO4 (final concentration 10 mM)
- 0.2 g KCl (final concentration 2.7 mM)
- 0.24 g KH2PO4 (final concentration 2 mM)
- Dissolve in 900 ml ultrapure water
- Adjust pH to 7.4 with dilute HCl if necessary
- Bring volume to 1 L with water
- Autoclave in 500-ml bottles for 20 min
- Store at room temperature or at 4°C after opening for up to 1 month
Sucrose solutions
Add 1.5 g, 2 g, 3 g, 4 g, and 5 g sucrose to five 15-ml polypropylene centrifuge tubes. Separately, prepare a buffer solution containing 50 mM Tris·Cl, pH 7.5; see Moore, 1996), 1 mM EDTA, and 0.05% (w/v) lauryl maltoside. Fill each tube containing sucrose to 10 ml with the buffer solution. Place the tubes on a tube rotator for ∼20 min until all sucrose is fully dissolved. Store solutions at 4°C for up to 2 months.
TAE buffer, 50×
- 242 g Tris base
- 57.1 ml glacial acetic acid
- 100 ml 0.5 M sodium EDTA
- Bring to 1 L with water
- Store at room temperature for up to 6 months
To make 1× TAE from 50× TAE stock, dilute 20 ml of the stock into 980 ml of nuclease-free water.
TNE buffer
- 50 mM Tris·Cl, pH 7.4 (Moore, 1996)
- 100 mM NaCl
- 0.1 mM EDTA
- Store at 4°C for up to 2 months
COMMENTARY
Background Information
The hepatitis E virus (HEV) contains a 7.2-kb single-stranded positive-sense RNA genome, which is capped and polyadenylated. In Huh7 cells, a capped 2.2-kb bicistronic subgenomic RNA is identified during HEV infection. The subgenomic RNA is utilized for translation of both open reading frame (ORF) 2 and ORF3 (Graff, Torian, Nguyen, & Emerson, 2006). The virus genome encodes three conventional ORFs: ORF1, ORF2, and ORF3. ORF1 is translated from the complete viral genome, and the polypeptide contains the putative functional domains: a methyltransferase domain (MT), a putative papain-like cysteine protease (P), a hepevirus unique (or Z-) domain (HUD), a hypervariable polyproline region (PP), a macro domain (Macro), a helicase domain (Hel), and an RNA-dependent RNA polymerase domain (RdRp) (Purdy et al., 2022). The ORF2 product is the structural protein of HEV virions, and possesses virus-neutralizing epitopes (Osterman et al., 2012). There are multiple forms of ORF2 products identified in the infected cells (Montpellier et al., 2018; Yin et al., 2018), and the protein inhibits interferon production through the N-terminal Arginine-rich domain (Lin et al., 2019). The motif also regulates the subcellular location and function of ORF2 (Hervouet et al., 2022). The ORF3 product is a small regulatory protein that is necessary for virion egress (Ding et al., 2017) and has other functions (Lin & Zhang, 2021).
There are 8 genotypes classified for Paslahepevirus balayani species HEV (Purdy et al., 2022). HEV genotypes 1 and 2 cause acute liver disease in clinical cases, while HEV genotypes 3 and 4 cause both acute and chronic infections, the latter of which are thought to occur mainly in patients with immunosuppression when infected by genotype 3 HEV (Hoofnagle, Nelson, & Purcell, 2012; Nan & Zhang, 2016). Among the four conventional genotypes of HEV, genotype 1 is mostly found in Asia (Okamoto, 2007), while genotype 2 is found to circulate in Mexico and Africa (Huang et al., 1992). Genotype 3 is mainly reported in developed countries (Okamoto, 2007), and genotype 4 is frequently reported in China and other Asian countries in recent years (Nan & Zhang, 2016; Purcell & Emerson, 2008). Genotypes 5 and 6 are identified from Japanese wild boars (Smith et al., 2014). Genotypes 7 and 8 were identified in dromedary and Bactrian camels, and genotype 7 is reported to present in an immunocompromised patient with chronic hepatitis (Lee et al., 2016; Sridhar et al., 2017). Until now, the vaccine against HEV infection has only been available in China, and off-label use of ribavirin is reported to be effective in clinical cases but not in pregnant patients (Sayed, Vercouter, Abdelwahab, Vercauteren, & Meuleman, 2015). The current strategy for the prevention of HEV relies on sanitary measures, such as providing clean water and appropriately cooking food to avoid transmission from undercooked food (Kamar et al., 2012; Nan & Zhang, 2016).
Critical Parameters and Troubleshooting
Tissue culture problems
A common issue for cell culture is contamination by bacteria, mycoplasma, and fungi. To avoid contamination, careful aseptic techniques are required, and all cell culture activities should be performed in a biosafety cabinet. Before using the cabinet, it must be exposed to UV light for at least 15 min, and the surface must be wiped with 70% ethanol. Antibiotics such as penicillin and streptomycin may be used routinely in media. Mycoplasma is a more difficult contaminant to detect and treat, as it is not visible under microscopy, and the antibiotics commonly used in cell culture do not work against it. Mycoplasma can affect both cell biology characteristics and propagation of the virus in cells, so it is important to test cultures regularly.
A protocol for mycoplasma detection is available (Uphoff & Drexler, 2002). PCR detection of a positive sample would yield a 500-bp fragment in agarose gel analysis. If the PCR test is positive, the antibiotic plasmocin (InvivoGen) can be used at a concentration of 25 μg/ml for 2 weeks. A retest of the cells is needed after antibiotic treatment. If the mycoplasma test is still positive, the cells can be treated with the antibiotic for 1 more week. For prophylactic maintenance of cell culture, plasmocin can be used at a concentration of 5 μg/ml.
Low virus yield
HEV grows poorly in cultured cells. The initial inoculation amount of HEV seems to be important for good viral propagation. Therefore, the inoculum must have enough live HEV virions and be free from inhibitors. Amphotericin B, MgCl2, and DMSO were reported to increase virus yield and can be used, as described (Chew, Situ, Wu, Yao, & Sridhar, 2022). Other cell lines or primary cells may support HEV proliferation [see review (Meister, Bruening, Todt, & Steinmann, 2019)].
The cell conditions are essential for a good viral yield. The cultured cells must be maintained in good conditions before virus inoculation. Activation of the cellular antiviral system might be a reason for low virus yield, which needs to be assessed and avoided if other possible reasons are excluded.
The virus yield is easily affected by mycoplasma contamination of the cell culture. The cultured cells should be tested routinely to make sure the culture system is free of mycoplasma. If the test is positive, the cells should be treated with plasmocin as described above.
Anticipated Results
In Basic Protocol 1, the number of HEV RNA copies in the cell culture supernatant can range from 102 to 106 copies/ml. In Basic Protocol 2, the RNA virus yield can reach up to ∼106 genomes/ml after electroporation for 5 to 6 days. The HEV virions from cell culture should band at a density of 1.15 g/cm3, about 35% sucrose, during gradient centrifugation. In Support Protocol 2, as HEV does not cause cytopathic effects, the cells should look similar to non-infected cells.
Time Considerations
For Basic Protocol 1, the cell culture preparation can be ready within 4 days. Sample centrifugation can be finished in 30 min, and viral RNA extraction can be done within 4 hr. Quantification by RT-qPCR can be finished within 3 hr. These steps can be performed within the 4 days during cell culture preparation. After inoculation, the cells need to be cultured for 5 days to allow virus proliferation before being expanded to a 75-cm2 flask. RNA virus levels in the culture supernatant can be monitored every day. The entire experiment should be completed within 2 to 3 weeks.
For Basic Protocol 2, digestion of the HEV infectious cDNA clone needs at least 4 hr and up to overnight, and electrophoresis needs 30 min, followed by DNA purification for 30 min. The in vitro transcription and RNA purification steps take 4 to 6 hr. Transfection of viral RNA and virus propagation needs ∼2 weeks. Virus concentration takes about 4 hr, and RNA virus extraction and detection take about 8 hr. In total, propagation of the virus from cDNA clones needs ∼3 weeks.
Support Protocol 1 can be finished within 1 day. Support Protocol 2 includes preparation of the cell culture lasting 4 days, virus infection and proliferation lasting 5 or 6 days, and performing the immunofluorescence assay, which takes 3 hr. Collectively, Support Protocol 2 needs about 10 days.
Acknowledgements
This work is supported by a seed grant from the University of Maryland, College Park, Maryland.
Author Contributions
Shaoli Lin : conceptualization, data curation, methodology, writing original draft, writing review and editing; Liping Yang : methodology, writing review and editing; Yanjin Zhang : conceptualization, funding acquisition, methodology, project administration, resources, supervision, validation, writing review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing not applicable—no new data generated.
Literature Cited
- Burnett, L. C., Lunn, G., & Coico, R. (2009). Biosafety: Guidelines for working with pathogenic and infectious microorganisms. Current Protocols in Microbiology , 13, 1A.1.1–1A.1.14. doi: 10.1002/9780471729259.mc01a01s13
- Chew, N., Situ, J., Wu, S., Yao, W., & Sridhar, S. (2022). Independent evaluation of cell culture systems for hepatitis E virus. Viruses , 14(6), 1254. doi: 10.3390/v14061254
- Current Protocols. (2005). Resources for international biosafety guidelines and regulations. Current Protocols in Microbiology , 00, 1B.1.1. doi: 10.1002/9780471729259.mca01bs00
- Ding, Q., Heller, B., Capuccino, J. M., Song, B., Nimgaonkar, I., Hrebikova, G., … Ploss, A. (2017). Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proceedings of the National Academy of Sciences of the United States of America , 114, 1147–1152. doi: 10.1073/pnas.1614955114
- Emerson, S. U., Nguyen, H., Graff, J., Stephany, D. A., Brockington, A., & Purcell, R. H. (2004). In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. Journal of Virology , 78, 4838–4846. doi: 10.1128/JVI.78.9.4838-4846.2004
- Emerson, S. U., Nguyen, H. T., Torian, U., Burke, D., Engle, R., & Purcell, R. H. (2010). Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. Journal of Virology , 84, 9059–9069. doi: 10.1128/JVI.00593-10
- Emerson, S. U., Zhang, M., Meng, X. J., Nguyen, H., St. Claire, M., Govindarajan, S., … Purcell, R. H. (2001). Recombinant hepatitis E virus genomes infectious for primates: Importance of capping and discovery of a cis-reactive element. Proceedings of the National Academy of Sciences of the United States of America , 98, 15270–15275. doi: 10.1073/pnas.251555098
- Graff, J., Torian, U., Nguyen, H., & Emerson, S. U. (2006). A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. Journal of Virology , 80, 5919–5926. doi: 10.1128/JVI.00046-06
- Hervouet, K., Ferrie, M., Ankavay, M., Montpellier, C., Camuzet, C., Alexandre, V., … Cocquerel, L. (2022). An Arginine-Rich Motif in the ORF2 capsid protein regulates the hepatitis E virus lifecycle and interactions with the host cell. PLoS Pathogens , 18(8), e1010798. doi: 10.1371/journal.ppat.1010798
- Hoofnagle, J. H., Nelson, K. E., & Purcell, R. H. (2012). Hepatitis E. The New England Journal of Medicine , 367, 1237–1244. doi: 10.1056/NEJMra1204512
- Huang, C. C., Nguyen, D., Fernandez, J., Yun, K. Y., Fry, K. E., Bradley, D. W., … Reyes, G. R. (1992). Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology , 191, 550–558. doi: 10.1016/0042-6822(92)90230-M
- Kamar, N., Bendall, R., Legrand-Abravanel, F., Xia, N. S., Ijaz, S., Izopet, J., & Dalton, H. R. (2012). Hepatitis E. Lancet , 379, 2477–2488. doi: 10.1016/S0140-6736(11)61849-7
- Lee, G. H., Tan, B. H., Teo, E. C., Lim, S. G., Dan, Y. Y., Wee, A., … Teo, C. G. (2016). Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology , 150(2), 355–357. e353. doi: 10.1053/j.gastro.2015.10.048
- Lin, S., Yang, Y., Nan, Y., Ma, Z., Yang, L., & Zhang, Y. J. (2019). The capsid protein of hepatitis E virus inhibits interferon induction via its N-terminal Arginine-Rich motif. Viruses , 11(11), 1050. doi: 10.3390/v11111050
- Lin, S., & Zhang, Y. J. (2021). Advances in Hepatitis E virus biology and pathogenesis. Viruses , 13(2), 267. doi: 10.3390/v13020267
- Meister, T. L., Bruening, J., Todt, D., & Steinmann, E. (2019). Cell culture systems for the study of hepatitis E virus. Antiviral Research , 163, 34–49. doi: 10.1016/j.antiviral.2019.01.007
- Montpellier, C., Wychowski, C., Sayed, I. M., Meunier, J. C., Saliou, J. M., Ankavay, M., … Cocquerel, L. (2017). Hepatitis E virus lifecycle and identification of 3 forms of the ORF2 capsid protein. Gastroenterology , 154(1), 211–223. e8. [Epub ahead of print]. doi: 10.1053/j.gastro.2017.09.020
- Montpellier, C., Wychowski, C., Sayed, I. M., Meunier, J. C., Saliou, J. M., Ankavay, M., … Cocquerel, L. (2018). Hepatitis E virus lifecycle and identification of 3 forms of the ORF2 capsid protein. Gastroenterology , 154(1), 211–223. e218. doi: 10.1053/j.gastro.2017.09.020
- Moore, D. D. (1996). Commonly used reagents and equipment. Current Protocols in Molecular Biology , 35, A.2.1–A.2.8. doi: 10.1002/0471142727.mba02s35
- Nan, Y., Ma, Z., Kannan, H., Stein, D. A., Iversen, P. I., Meng, X. J., & Zhang, Y. J. (2015). Inhibition of hepatitis E virus replication by peptide-conjugated morpholino oligomers. Antiviral Research , 120, 134–139. doi: 10.1016/j.antiviral.2015.06.006
- Nan, Y., & Zhang, Y.-J. (2016). Molecular biology and infection of hepatitis E virus. Frontiers in Microbiology , 7, 1419. doi: 10.3389/fmicb.2016.01419
- Okamoto, H. (2007). Genetic variability and evolution of hepatitis E virus. Virus Research , 127, 216–228. doi: 10.1016/j.virusres.2007.02.002
- Okamoto, H. (2011). Efficient cell culture systems for hepatitis E virus strains in feces and circulating blood. Reviews in Medical Virology , 21, 18–31. doi: 10.1002/rmv.678
- Okamoto, H. (2013). Culture systems for hepatitis E virus. Journal of Gastroenterology , 48, 147–158. doi: 10.1007/s00535-012-0682-0
- Osterman, A., Vizoso Pinto, M. G., Haase, R., Nitschko, H., Jäger, S., Sander, M., … Baiker, A. (2012). Systematic screening for novel, serologically reactive hepatitis E virus epitopes. Virology Journal , 9, 28–28. doi: 10.1186/1743-422X-9-28
- Purcell, R. H., & Emerson, S. U. (2008). Hepatitis E: An emerging awareness of an old disease. Journal of Hepatology , 48, 494–503. doi: 10.1016/j.jhep.2007.12.008
- Purdy, M. A., Drexler, J. F., Meng, X. J., Norder, H., Okamoto, H., van der Poel, W. H. M., … Smith, D. B. (2022). ICTV virus taxonomy profile: Hepeviridae 2022. Journal of General Virology , 103(9). doi: 10.1099/jgv.0.001778
- Sayed, I. M., Vercouter, A. S., Abdelwahab, S. F., Vercauteren, K., & Meuleman, P. (2015). Is hepatitis E virus an emerging problem in industrialized countries? Hepatology , 62, 1883–1892. doi: 10.1002/hep.27990
- Shukla, P., Nguyen, H. T., Faulk, K., Mather, K., Torian, U., Engle, R. E., & Emerson, S. U. (2012). Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. Journal of Virology , 86, 5697–5707. doi: 10.1128/JVI.00146-12
- Shukla, P., Nguyen, H. T., Torian, U., Engle, R. E., Faulk, K., Dalton, H. R., … Emerson, S. U. (2011). Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proceedings of the National Academy of Sciences of the United States of America , 108, 2438–2443. doi: 10.1073/pnas.1018878108
- Smith, D. B., Simmonds, P., Jameel, S., Emerson, S. U., Harrison, T. J., Meng, X. J., … Purdy, M. A. (2014). Consensus proposals for classification of the family Hepeviridae. The Journal of General Virology , 95, 2223–2232. doi: 10.1099/vir.0.068429-0
- Sridhar, S., Teng, J. L. L., Chiu, T. H., Lau, S. K. P., & Woo, P. C. Y. (2017). Hepatitis E virus genotypes and evolution: Emergence of camel hepatitis E variants. International Journal of Molecular Sciences , 18, E869. doi: 10.3390/ijms18040869
- Tanaka, T., Takahashi, M., Kusano, E., & Okamoto, H. (2007). Development and evaluation of an efficient cell-culture system for hepatitis E virus. The Journal of General Virology , 88, 903–911. doi: 10.1099/vir.0.82535-0
- Uphoff, C. C., & Drexler, H. G. (2002). Comparative PCR analysis for detection of mycoplasma infections in continuous cell lines. In Vitro Cellular & Developmental Biology—Animal, 38, 79–85. doi: 10.1290/1071-2690(2002)038<0079:CPAFDO>2.0.CO;2
- Voytas, D. (2001). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51, 2.5A.1–2.5A.9. doi: 10.1002/0471142727.mb0205as51
- World Health Organization. (2022). Hepatitis E fact sheet. Available at https://www.who.int/news-room/fact-sheets/detail/hepatitis-e
- Xu, L., Wang, W., Li, Y., Zhou, X., Yin, Y., Wang, Y., … Pan, Q. (2017). RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production. Hepatology , 65, 1823–1839. doi: 10.1002/hep.29105
- Xu, L., Zhou, X., Wang, W., Wang, Y., Yin, Y., Laan, L. J., … Pan, Q. (2016). IFN regulatory factor 1 restricts hepatitis E virus replication by activating STAT1 to induce antiviral IFN-stimulated genes. FASEB Journal , 30, 3352–3367. doi: 10.1096/fj.201600356R
- Yin, X., Ambardekar, C., Lu, Y., & Feng, Z. (2016). Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. Journal of Virology , 90, 4232–4242. doi: 10.1128/JVI.02804-15
- Yin, X., Ying, D., Lhomme, S., Tang, Z., Walker, C. M., Xia, N., … Feng, Z. (2018). Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proceedings of the National Academy of Sciences of the United States of America , 115(18), 4773–4778. doi: 10.1073/pnas.1721345115
- Zhou, X., Wang, Y., Metselaar, H. J., Janssen, H. L., Peppelenbosch, M. P., & Pan, Q. (2014). Rapamycin and everolimus facilitate hepatitis E virus replication: Revealing a basal defense mechanism of PI3K-PKB-mTOR pathway. Journal of Hepatology , 61, 746–754. doi: 10.1016/j.jhep.2014.05.026

